DNA Replication and the Genetic Code (Midterm 3 Lesson 1)
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
85 Terms
what is the cell cycle
an alternation between interphase (normal cellular activity) and mitosis (cellular division)
some terminally differentiated cells (eg mature neurons) stop dividing and enter G0 stage
when can you “see” chromosomes
right before cell division
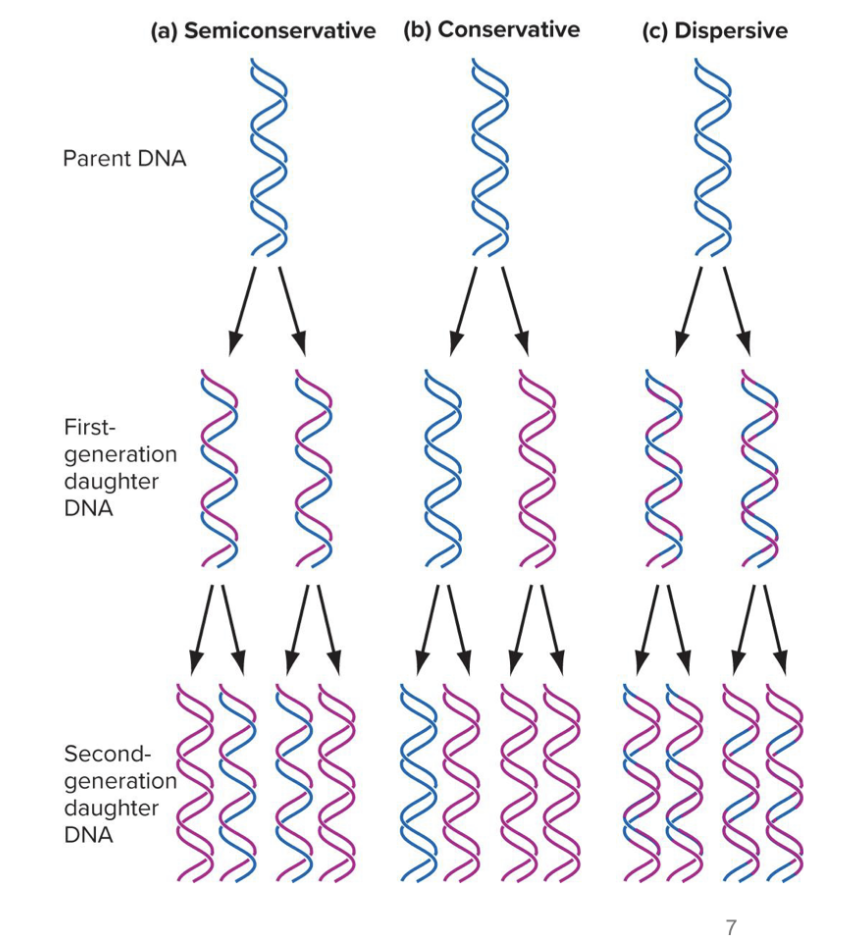
what are the 3 possible models of dna replication
Semiconservative
Conservative
Dispersive
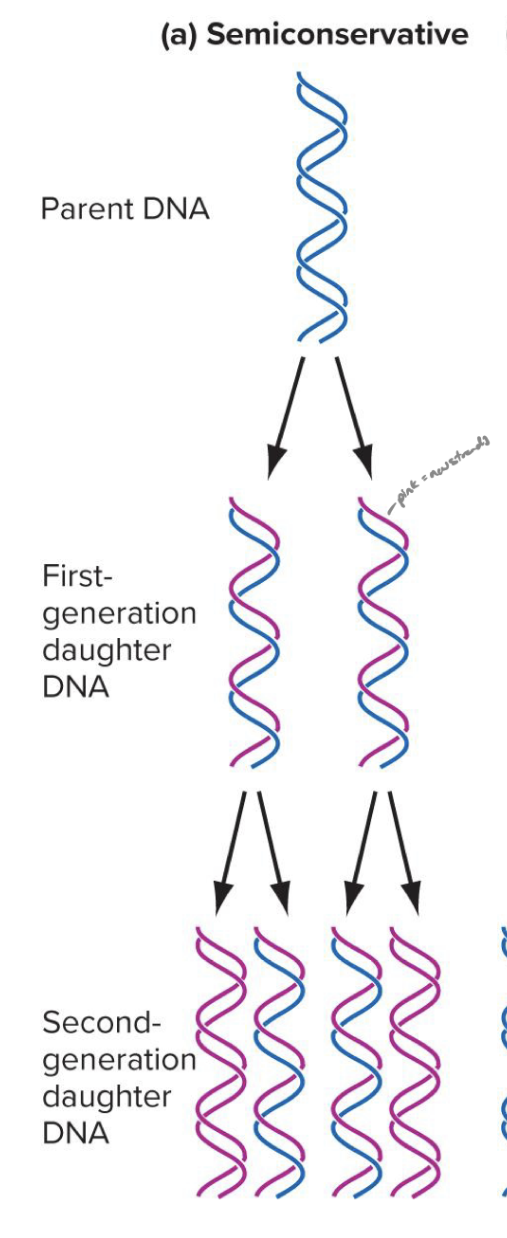
explain the semiconservative model of dna replication
it is the watson-crick model (made by the guys who first proposed the double helix model)
each new DNA molecule consists of:
One original (parental) strand
One newly synthesized strand
So after replication, each daughter DNA molecule is half old and half new — hence the term "semi" conservative.
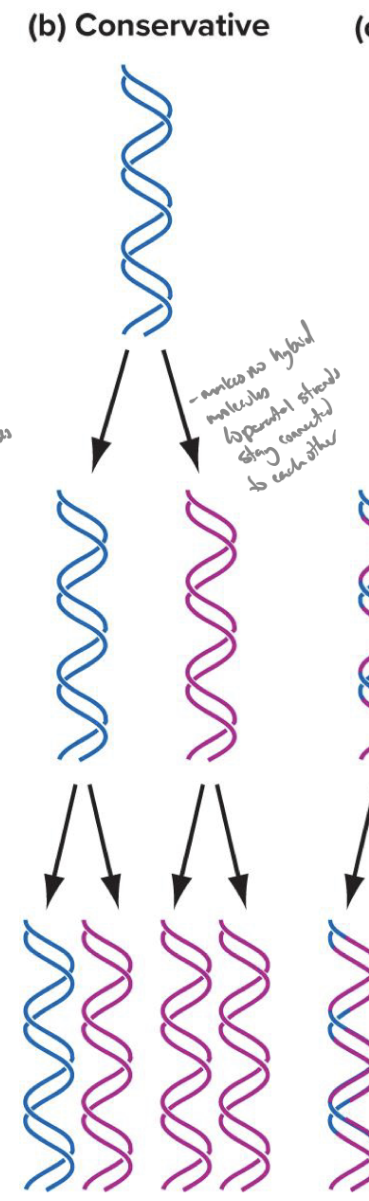
explain the conservative model of dna replication
parental double helix remains intact, both strands of daughter helicies are newly synthesized
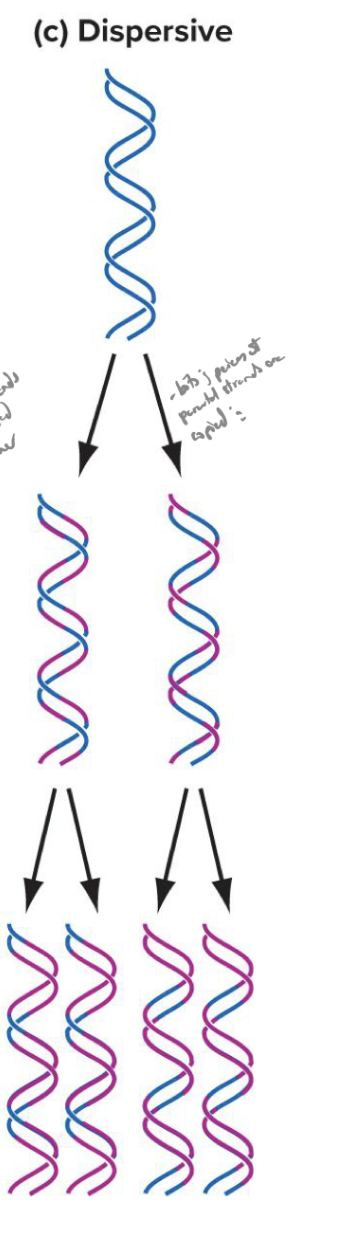
explain the dispersive model of dna replication
both strands of daughter helicies contain og and newly synthesized dna
explain the Meselson-Stahl experiment
grew e.coli with diff isotopes of nitrogen (N14 and N15)
bacteria incorperate that N into dna (into nitrogenous bases)
isolated dna after diff numbers of cell divisions
detected isotopes based on density (centrifugation)
when N14 is put through centrifuge, the band created is up high (light), N15’s band is low (heavy)
put the N15 dna strands in N14 medium (so any daughter dna would be N14 unlike parent N15 strand)
after first generation, new double helices were all in middle of N14 and N15 bands → showed double helicies were a mixture
after second gen, saw there were some strands that had ONLY N14 in them (some strands located up higher than the mixed ones) → shows they’re not dispersive or all dna would be a mixture
confirmed dna replication is semi-conservative
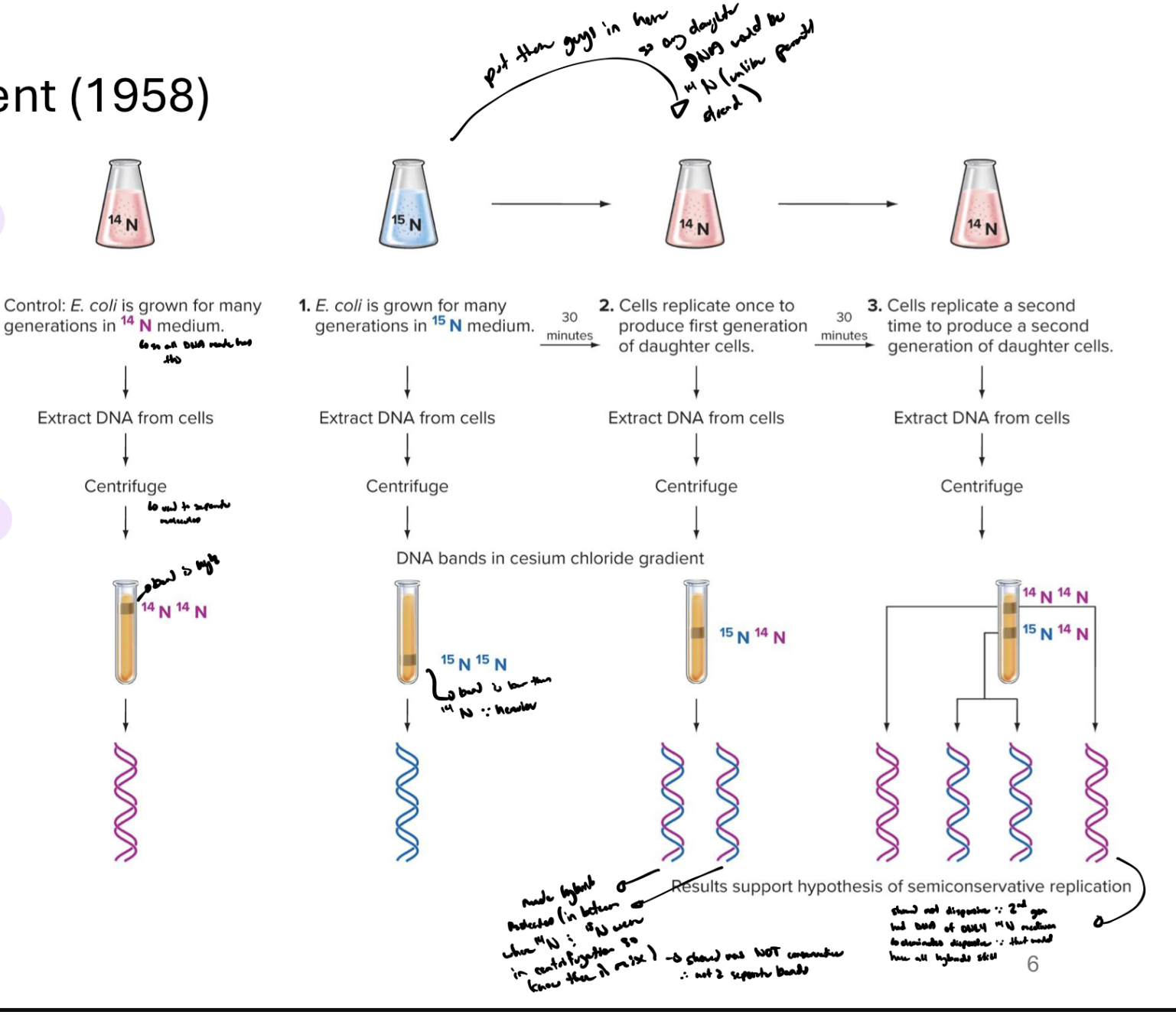
how did the meselson-stahl experiment show that the new dna produced was semiconservative and not dispersive
bc 2nd gen had dna of ONLY 14N medium (one parental medium)
which bands would you expect to see from the meselson-stahl experiment if replication was conservative or dispersive
conservative = 2 diff bands in tube (N14 and N15) all throughout replications
dispersive = only one band in middle of 14N and 15N (everything would be mixed)
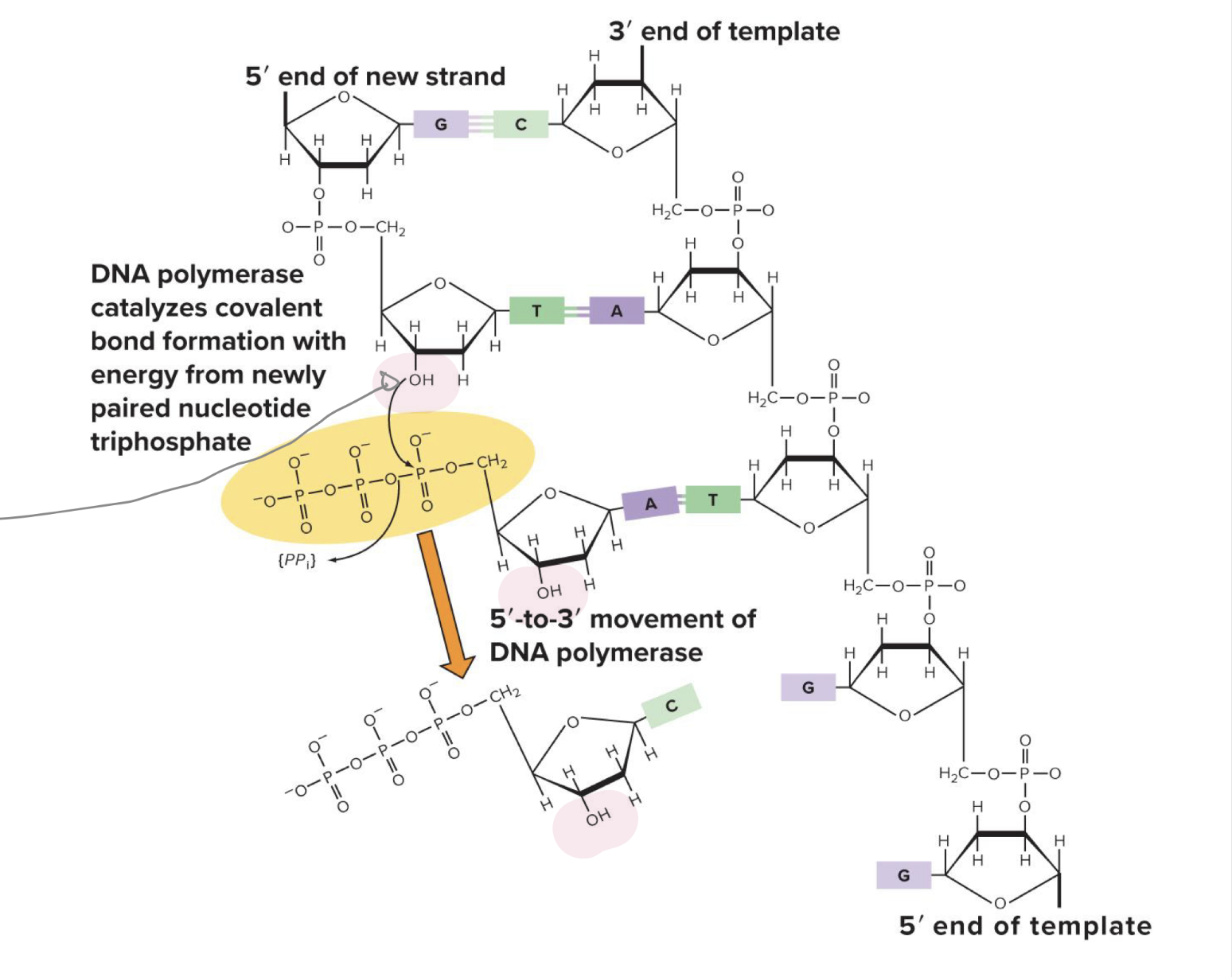
which direction does DNA synthesis go in
5’ to 3’
what direction is the template strand of dna read in
3’ to 5’
where are new nucleotides added on the growing strand
to the 3’-OH of the growing strand
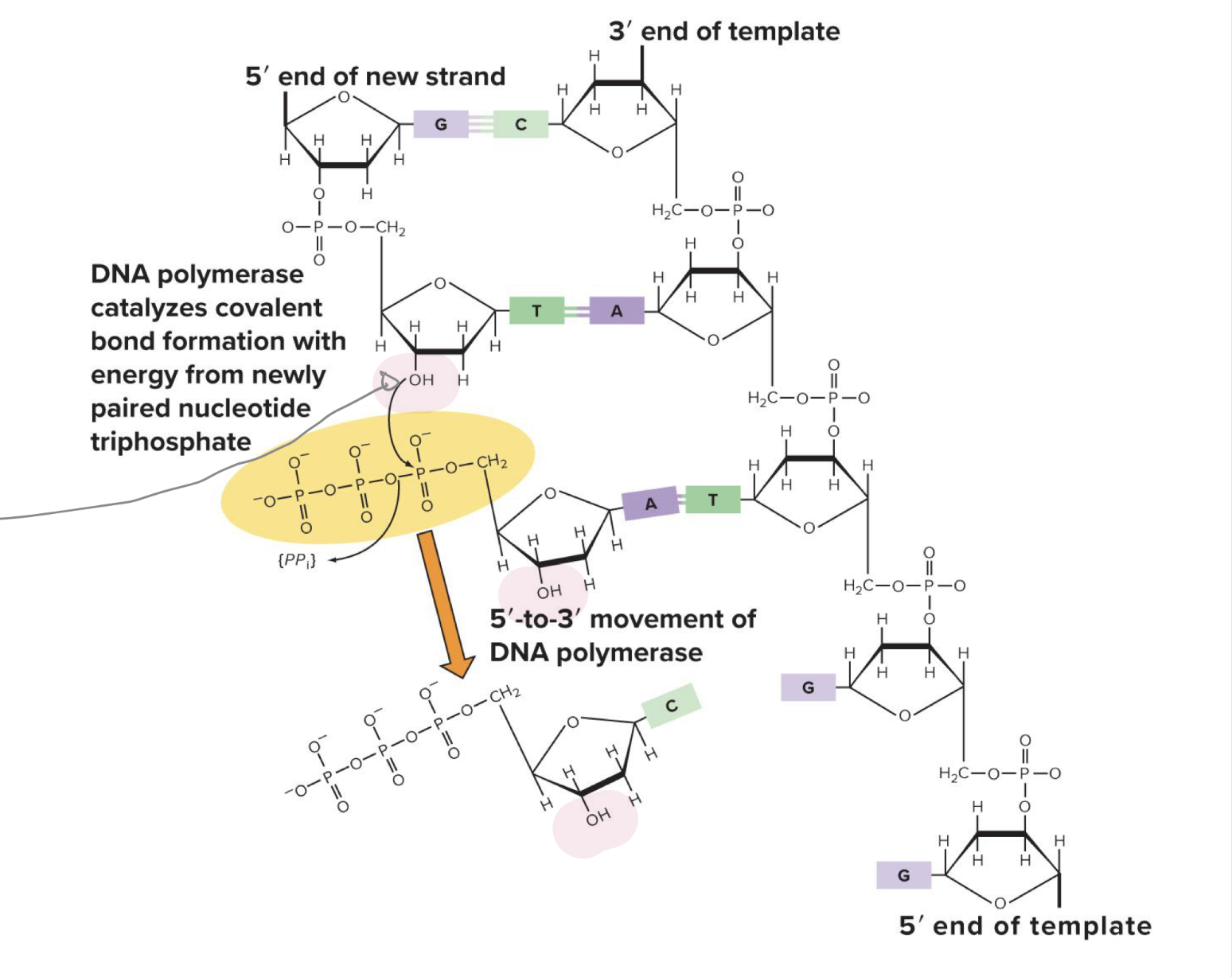
which direction does DNA polymerase build new dna molecules in
5’ to 3’ direction
what are the 3 requirements for DNA pol activity? explain each
Four dNTPs (di nucleotide triphosphates)
get incorporated into growing chain
need the 4 (A,C,T,G) to properly build
cleaving of phosphate bonds provides E for dna pol activity
Single-stranded template (needs something to copy from)
other enzymes unwind a dsDNA (double stranded dna) molecule to exposed ssDNA (single stranded dna) segments
Primar with exposed 3’-OH end
dna pol cannot start a new strand → can only add nucleotides to existing strand
what are the 2 stages of dna replication
initiation
elongation
provide an overview of initiation in dna replication
enzymes open the double helix
provide an overview of elongation in dna replication
enzymes connect correct sequence of nucleotides on newly formed dna strands
E for dna synthesis comes from high-E phosphate bonds in dNTPs
dna pol is the enzyme that catalyzes new phosphodiester bonds
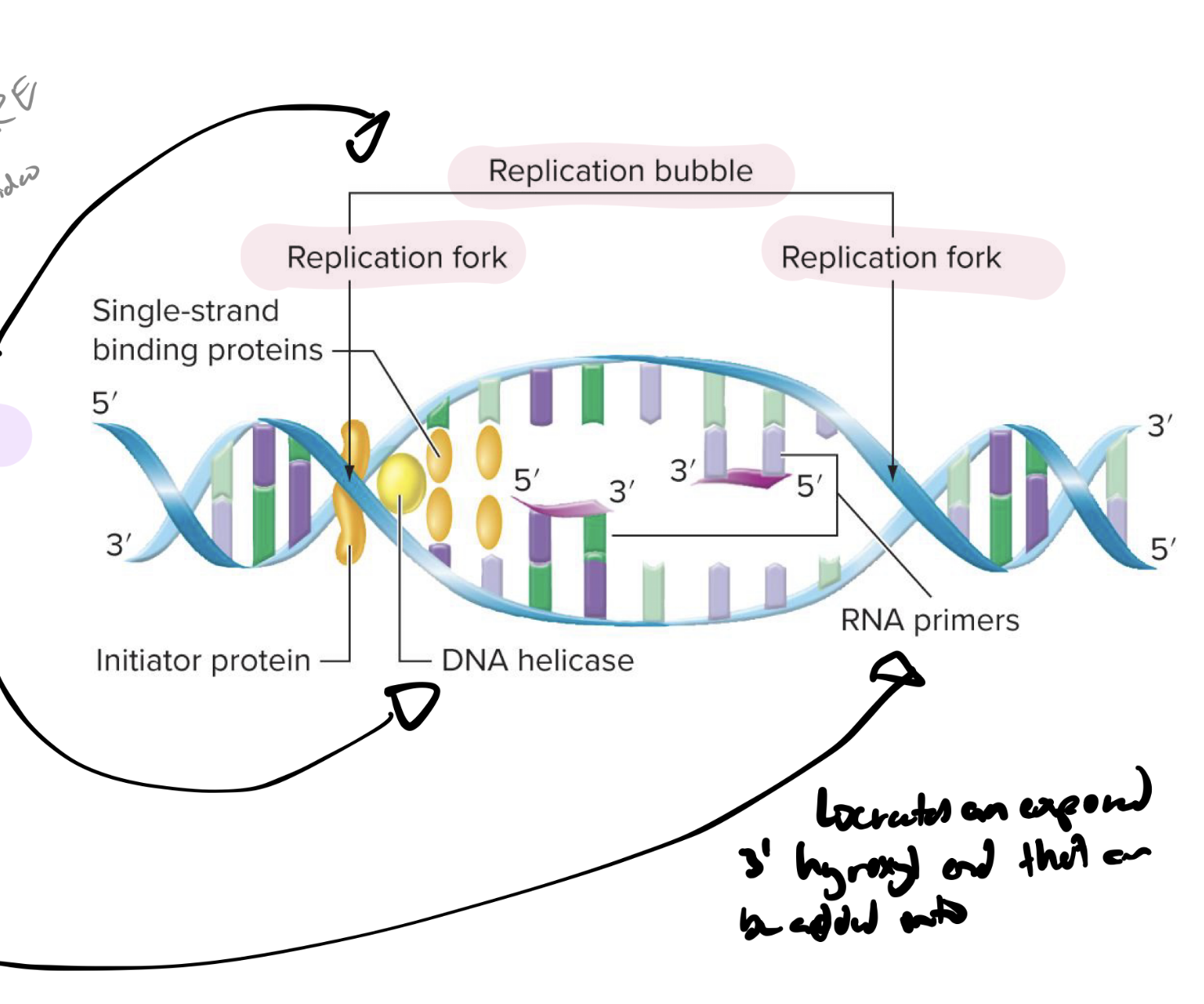
explain the steps for initiation of dna replication in prokaryotes
initiator protein binds to origin of replication
origin of replication = short sequence of specific nucleotides (where initiator protein binds)
DNA helicase unwinds helix
replication bubble forms w a replication fork at each end (replicaton proceeds along the fork)
single-stranded binding (SSB) proteins keep dna helix open
and help stabilize the single-stranded dna
primase synthesizes rna primers
what do rna primers do
create an exposed 3-OH (hydroxyl) end that can be added onto by dna pol in dna replication
what is rna? how does it compare to dna?
ribonucleic acid
less stable than dna
sugar in rna is ribose instead of deoxyribose
uracil is used instead of thymine
can form a double-strand with DNA OR RNA through hydrogen bonding
what does dna polymerase 3 do
adds nucleotides to rna primer to make dna
which dna sequence is the nucleotide sequence in the offspring dna copied from
the template strand
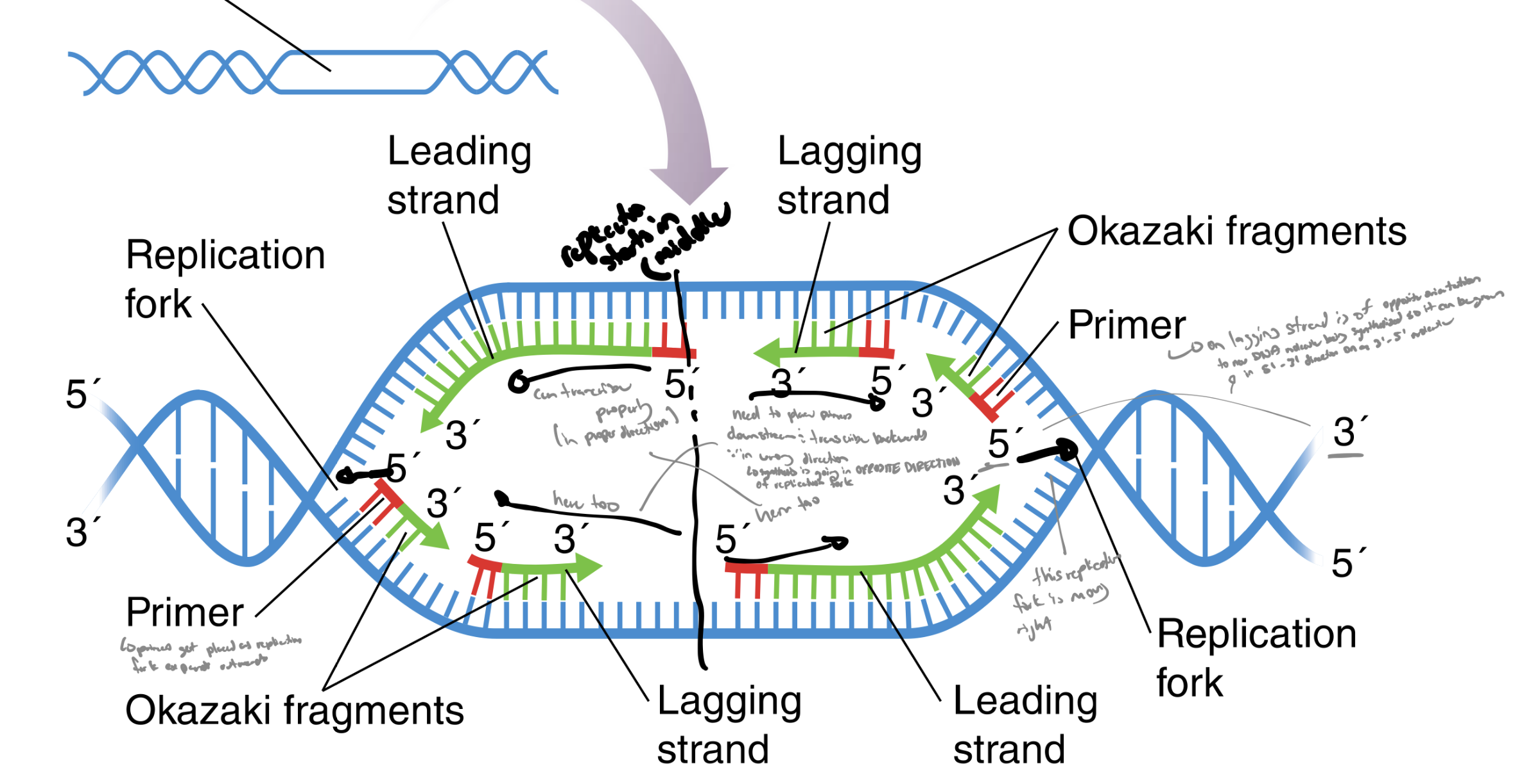
which type of synthesis does the leading strand have in dna replication
continuous synthesis
which type of synthesis does the lagging strand have in dna replication
discontinuous synthesis
what are okazaki fragments
short dna fragments of the lagging strand
what does dna pol 1 do
replaces rna primer with dna nucleotides
explain the steps in elongation in dna replication in prokaryotes
reference…
dna ligase
leading strand
template strand
lagging strand
dna pol 3
dna pol 1
okazaki fragments
rna primers
nucleotides are added to rna primer to synthesize a new strand of dna (adding)
catalyzed by dna pol 3
adds nucs to rna primer to make dna
nucleotide sequence is copied from the template strand
leading strand has continuous synthesis in the direction of the replication fork (produces a 5’ to 3’ strand)
lagging strand has discontinuous synthesis to produce a 3’ to 5’ strand by synthesizing short 5’ to 3’ fragments
called okazaki fragments
dna pol 1 replaces rna primer w dna nucleotides (so there’s no rna in the dna)
dna ligase covalently joins adjacent okazaki fragments to complete the lagging strand (so they’re not just chunks)
how does the replication fork move (reference leading and lagging strands)
in opposite directions
the leading strand moves continuously from one rna primer
the lagging strand is synthesized using repeated primers placed as the replication fork moves (has to wait for 5’ primers to be placed as the replication fork moves downstream and then replicates dna back towards the center of the replication bubble) towards middle of replication bubble
note: all dna is still synthesized in the 5’ to 3’ direction
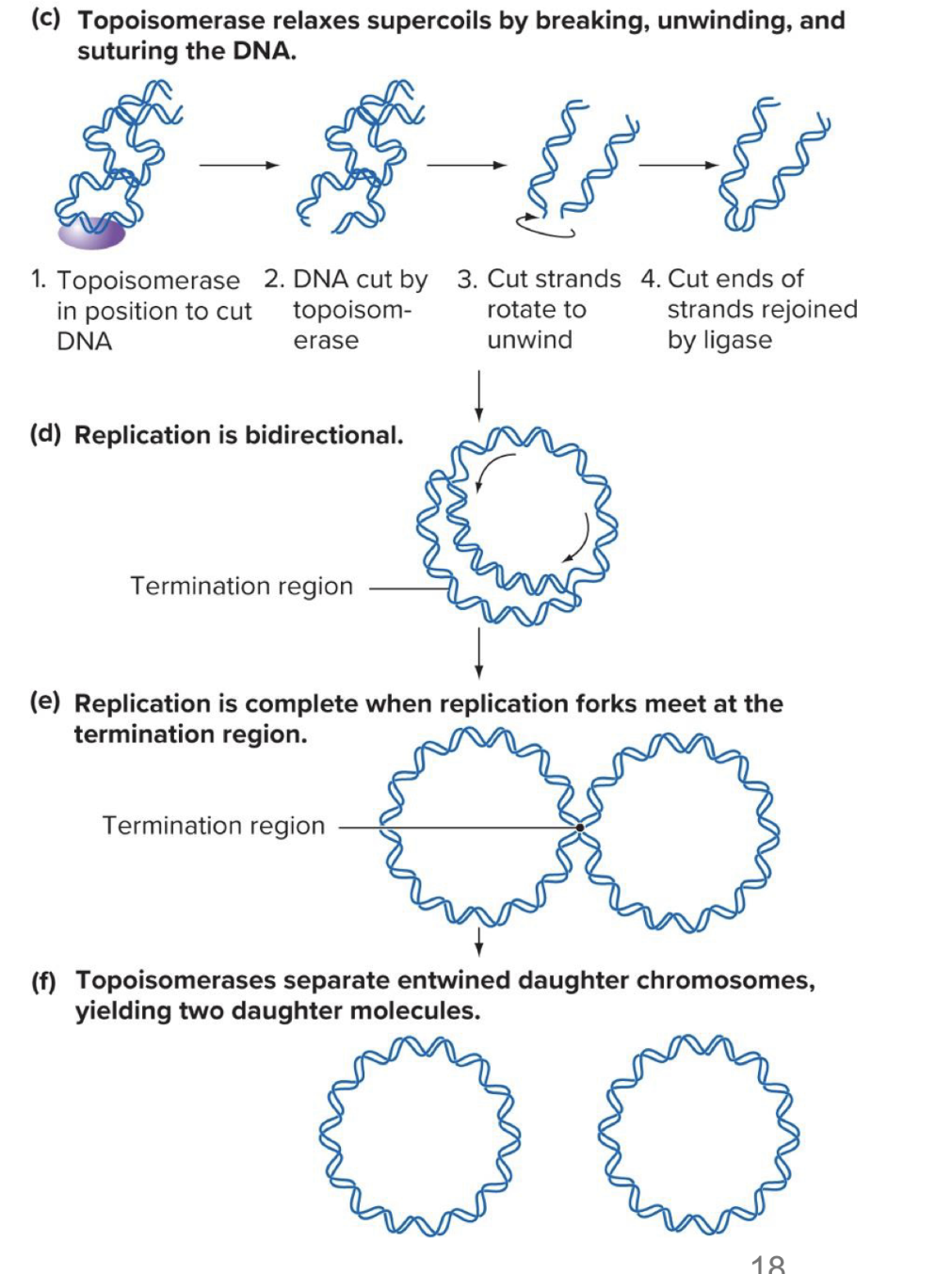
what does dna topoisomerase do
relaxes supercoils in prokaryotes
cuts back sugar phosphate backbone
what does dna ligase do
seals unwound broken dna strands in prokaryotes
which processes allow for dna replication occur in circular bacterial chromosomes
replication proceeds in 2 directions from a single origin of replication (dont forget, circular chromosomes)
synthesis occurs in both directions until replication forks meet
unwinding of dna created supercoiled dna ahead of the replication fork (basically super tangled)
would stop pol from being able to transcribe
dna topoisomerase relaxes supercoils
does so by cutting super phosphate backbone
causes dna strands to unwind
unwound broken strands are sealed tg again by dna ligase
dna replication occurs bidirectionally
completes when replication forks meet at the termination region (last point of contact btwn the 2 dna molecules)
topoisomerases separate the entwined daughter chromosomes, making 2 daughter molecules
what do exonucleases do
removes nucleotides
double checks to make sure proper nucleotides are being added to new dna molecules, if not then exonucleases them (takes them off)
what do polymerases do
make polymers (eg dna)
what does proofreading mean in dna synthesis
means it can detect if the wrong nuc was added to the strand → minimizes mutations from occuring
what does dna polymerase 3 do in prokaryotes in regards to polymerase and exonuclease activity, along w their primary fx
major enzyme responsible for synthesis of new strands
5’ to 3’ polymerase actitivy
3’ to 5’ exonuclease activity, proofreading ability
what does dna polymerase 1 do in prokaryotes in regards to polymerase and exonuclease activity, along w their primary fx
responsible for removal of rna primers and gap filling
5’ to 3’ polymerase activity
3’ to 5’ exonuclease activity for proofreading
5’ to 3’ exonuclease activity for primer degradation
removes rna primers in 5’ to 3’ direction
what is the problem with humans having large chromosomes? what is our solution for this?
you have to replicate all your dna in S phase
would take too long if each chrom only had one origin of replication (and one replication bubble)
to make it go faster, eukaryotes initiate dna replication at several points along the chromosome
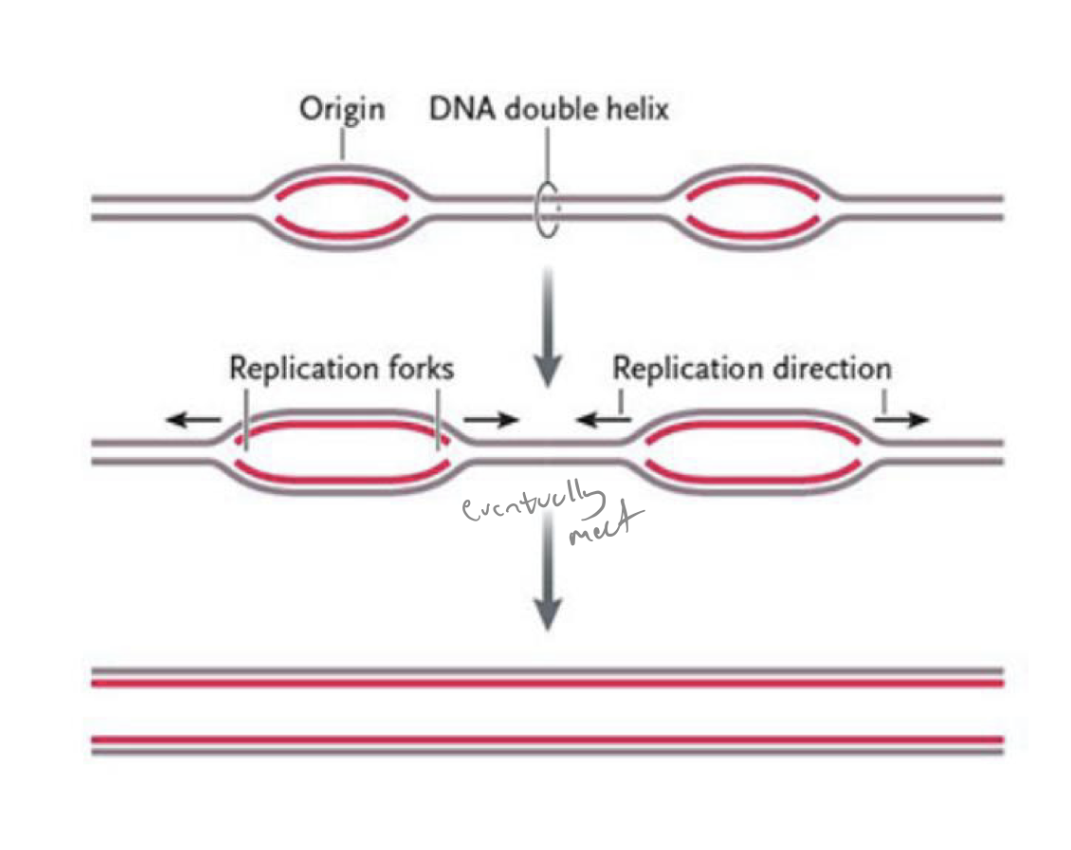
t/f: prokaryotes have only one origin of replication
true (but eukaryotes typically have multiple)
what is the problem w having linear chromosomes
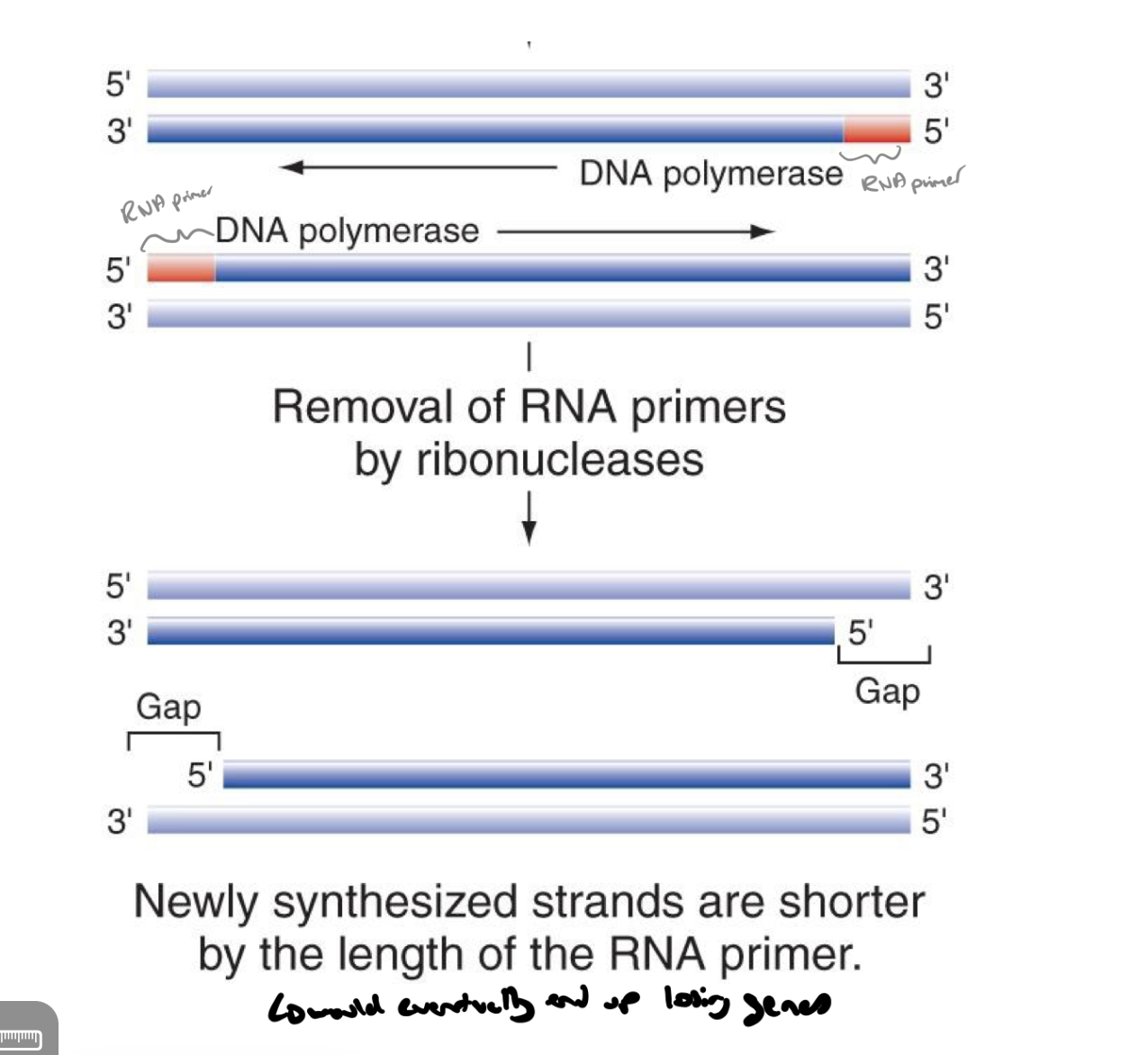
the removal of rna primers by ribonucleases results in newly synthesized dna strands being shorter (by the length of the rna primer → would eventually result in losing genes)
called the end-replication problem
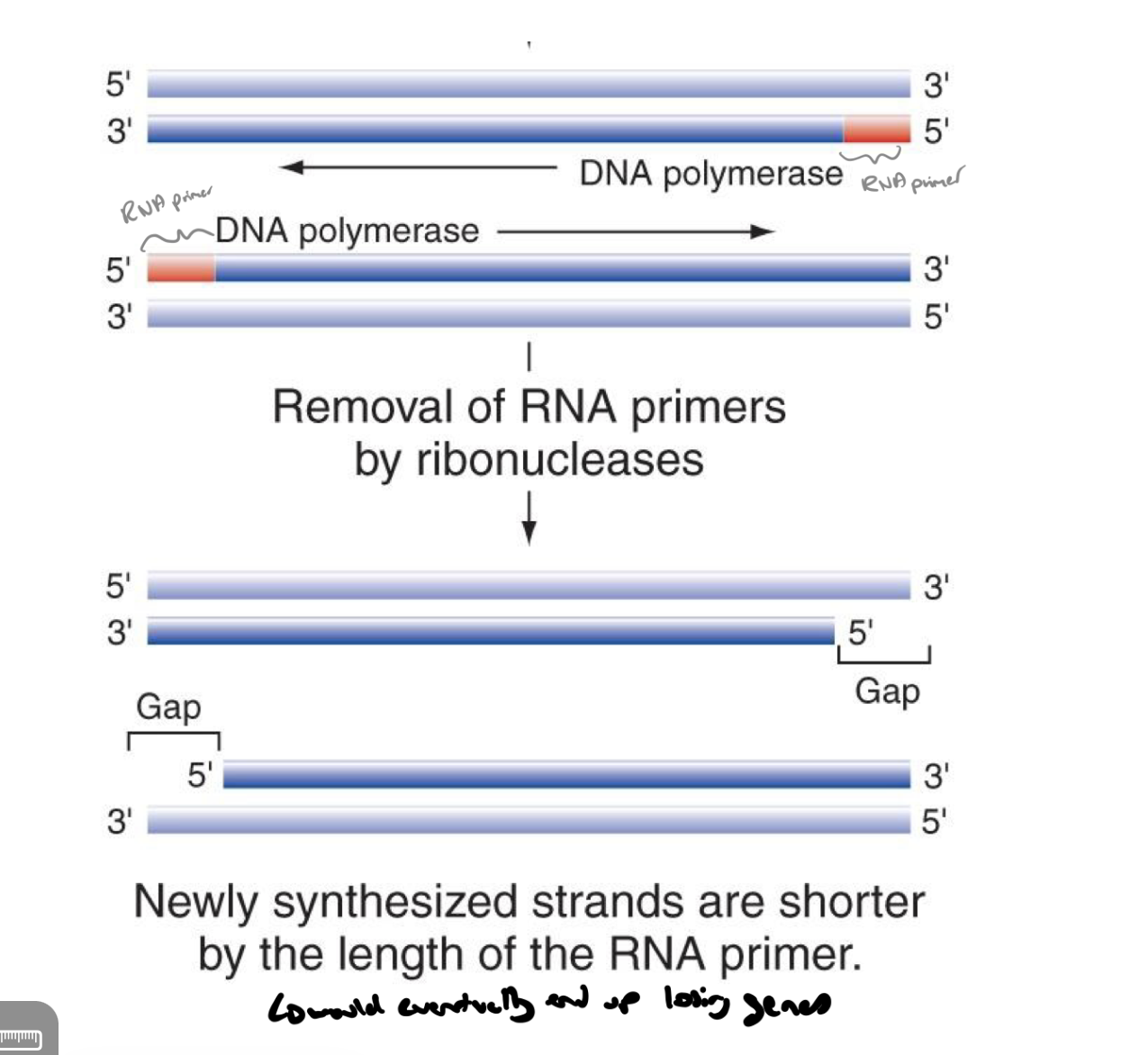
how do eukaryotes combate linear chromosome depletion
telomeres
what are telomeres
regions of repeated non-coding dna at end of linear chromosomes
contains repeated units of TTAGGG * 250-1500 (in humans, they’ll vary across species)
non-coding regions of dna that can be depleted by the removal of rna primers so when the chroms shorten that shortening doesn’t eat into the acc dna
shorten in each cell division
“absorbs” the loss so that critical info (genes) are not immediately affected
what is telomerase and how does it work
has an rna sequence that compliments the telomere sequence
EXTENDS/MAINTAINS TELOMERES
parent dna strand is elongated using complementary telomerase sequence
primase and dna pol synthesize remainder of complimentary strand
in other words…
Telomerase is a special enzyme that extends the ends of linear chromosomes — specifically, the telomeres, which are repetitive DNA sequences at the tips of chromosomes
Each time a cell divides, DNA polymerase cannot fully replicate the very end of the lagging strand (this is known as the end-replication problem). Without telomerase, telomeres would get shorter with every cell division — eventually leading to cell aging or death
note: telemerase is not in all cells (mostly just germ and stem cells ; it is not located in somatic cells → they j die when their telomeres are gone)
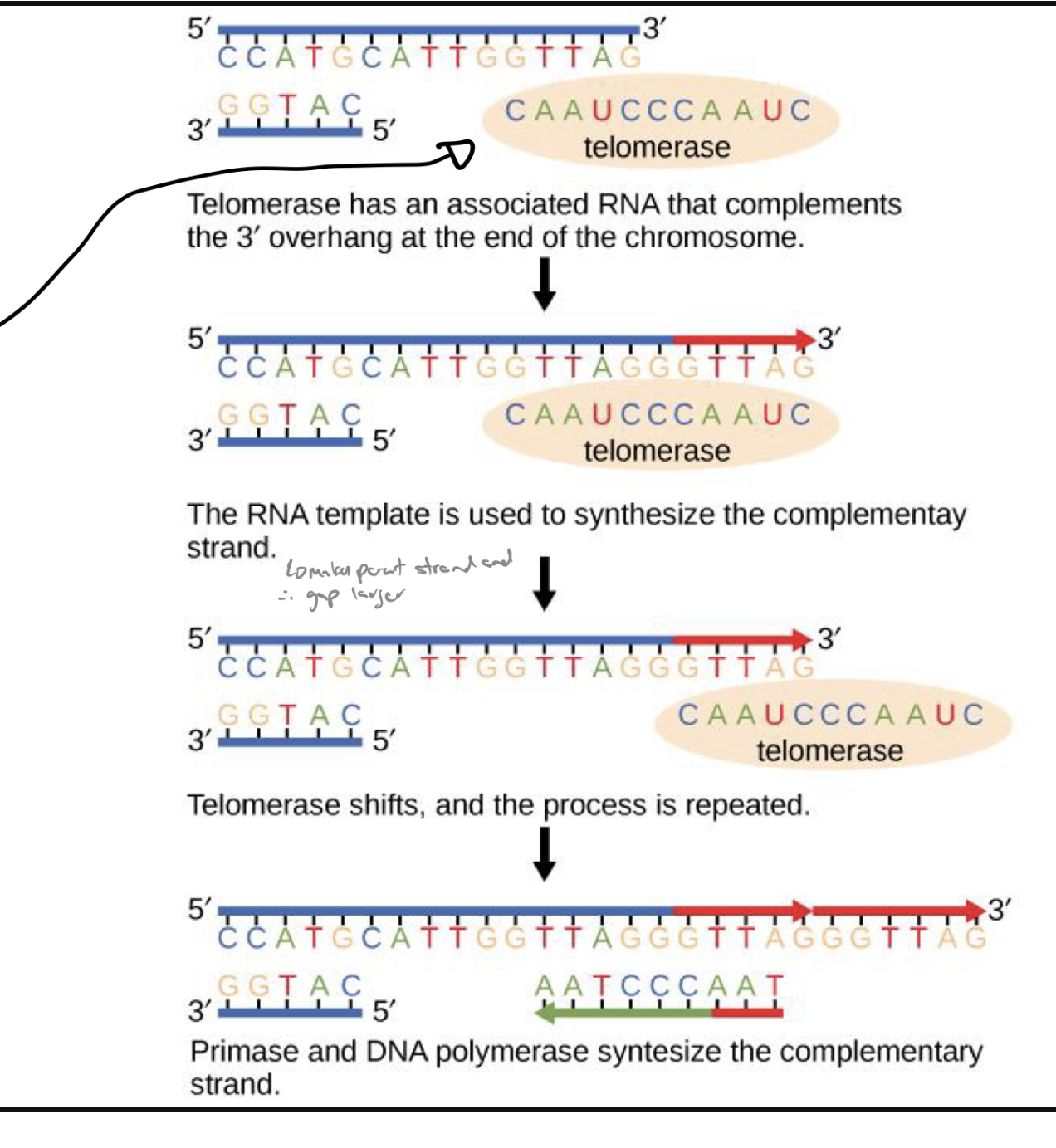
which dna strand is synthesized by primase and dna pol
the remainder of the complementary strand (the newly created strand)
what does telomerase consist of
a protein component and rna component
explain how telomerase works (reference telomerase elongation and translocation
translocation: protein in telomerase brings rna component to telomere where it binds to telomere sequence on the old strand of dna (leaving the new strand of dna w an exposed overhang)
elongation: exposed dna sequence has nucleotides bind to it and extend the complimentary dna strand
makes it so primers will bind to telomere and THOSE regions of dna wont be transcribed but the coding regions will be
process repeats
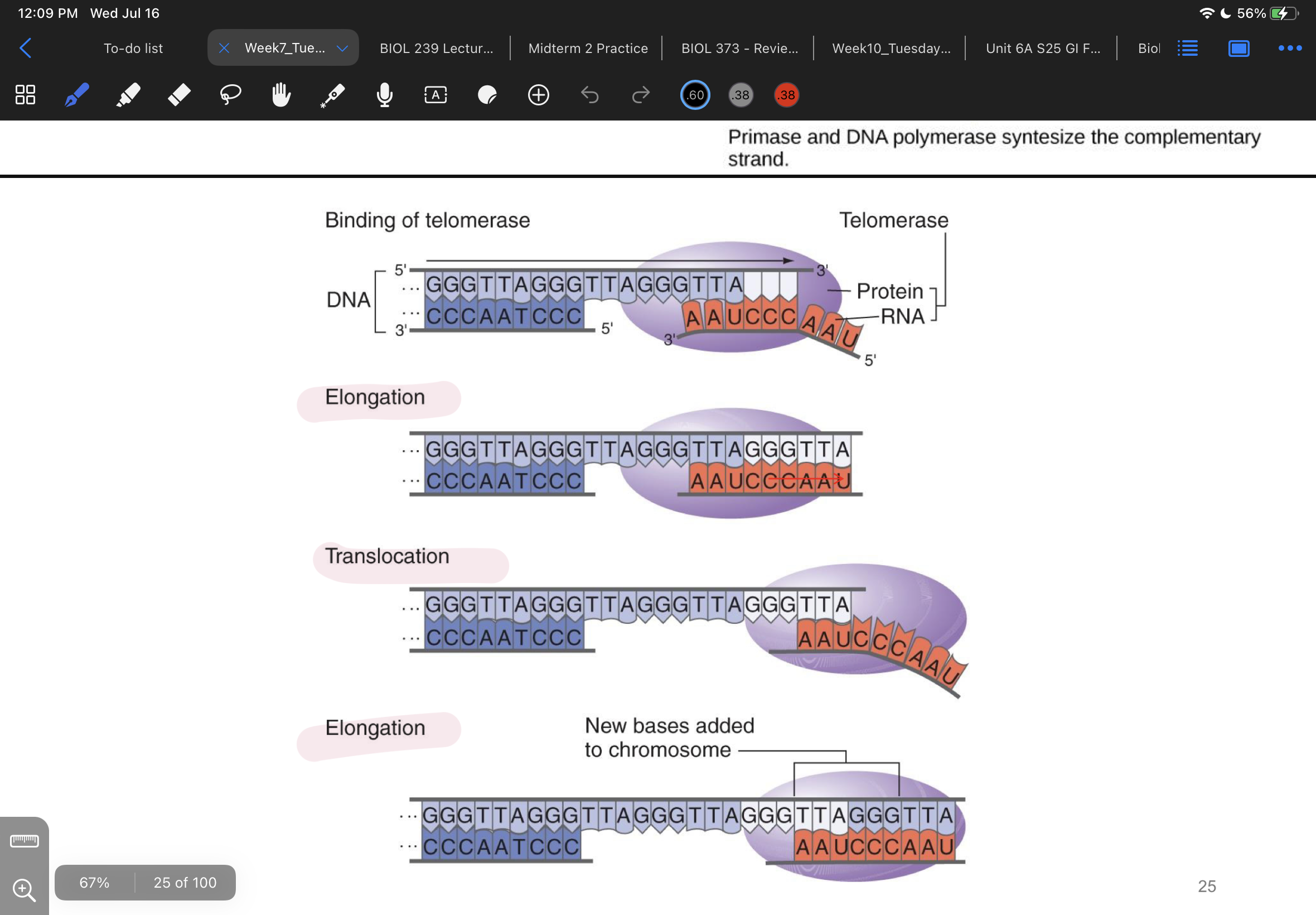
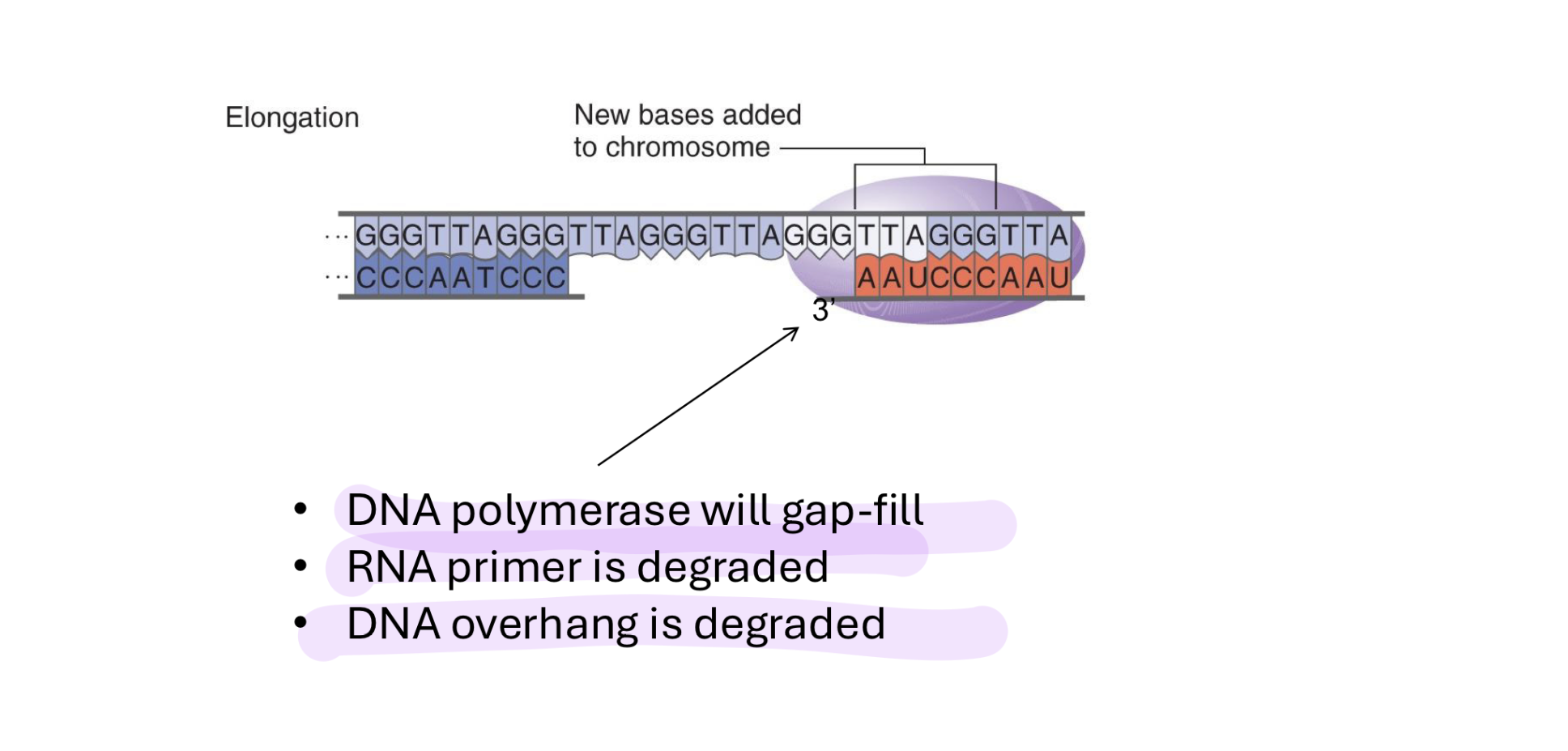
what happens to telomerase after several rounds of elongation
dna pol gap-fills (completes the other (new) strand so it doesnt just have nothing there)
from 5’ to 3’ end using primer on far side of the telomerase
rna primer is degraded
dna overhang is degraded
which cells in humans express and dont express telomerase
reproductive cells do and a few kinds of stem cells do
adult somatic cells dont
what is senescence
the eventual information (gene) loss in chroms with continuous cell division (degradation with age) → happens at less than 50 generations in a culture
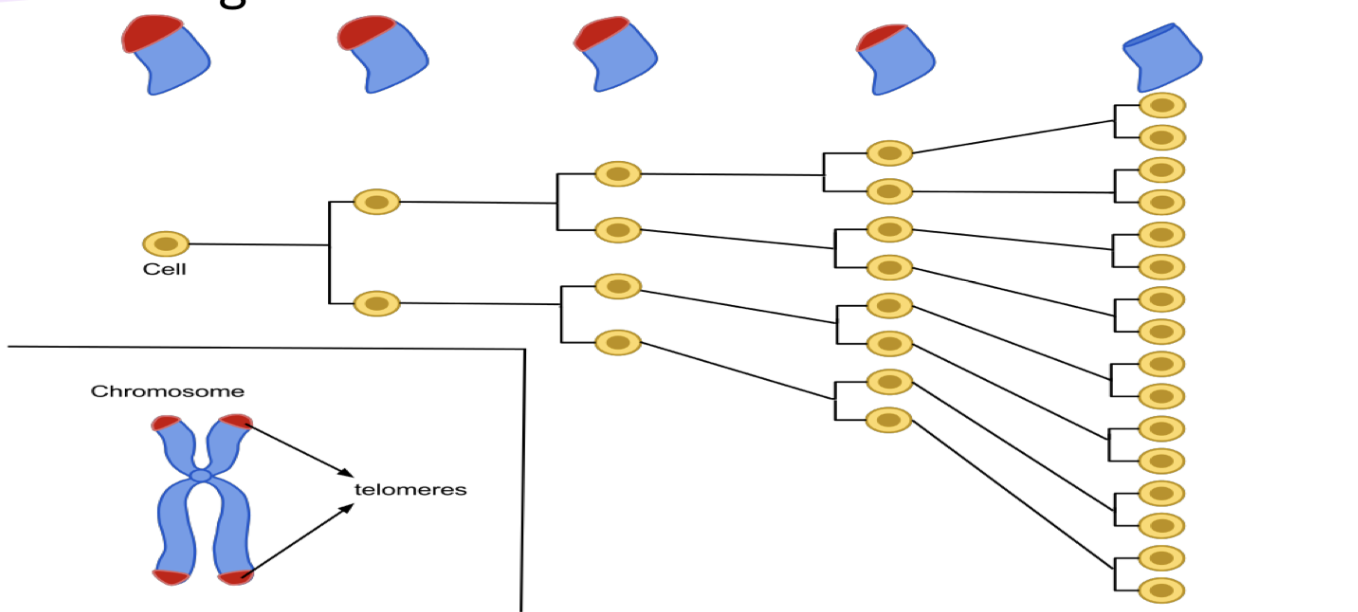
what happens to animals w out any telomeres left on their chroms (even if they are the same age)? what does reintroducing telomerase do
look much older
have shorter life span
reintroducing telomerase spurs almost a complete recovery
BUT re-activating it in somatic cells greatly inc risk of cancer
telomeres shortening limits amount of times cells can divide
w out that the cell can divide a lot more than usual
inc chances mutations will occur
how does dna lead to gene products
transcription and translation
what does transcription do
creates rna from dna
what is an rna transcript
output of transcription
serves directly as mRNA in prokaryotes; processed to become mRNA in eukaryotes
what does translation do
creates protein from rna
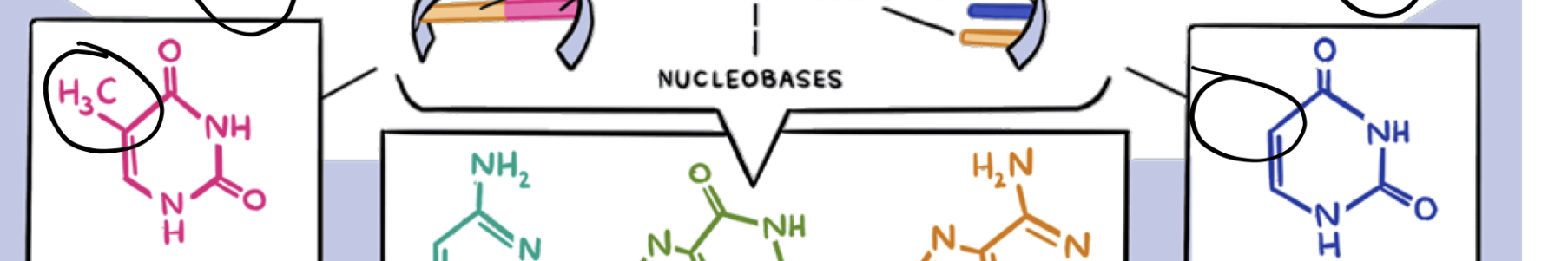
what is the diff between dna and rna
dna:
deoxyribonucleic acid
usually double stranded sugar phosphate
contains nucleotide/nucleobase thymine
rna:
ribonucleic acid
usually single-stranded sugar phosphate
contains nucleotide/nucleobase uracil
which molecules are these? say which is which specifically
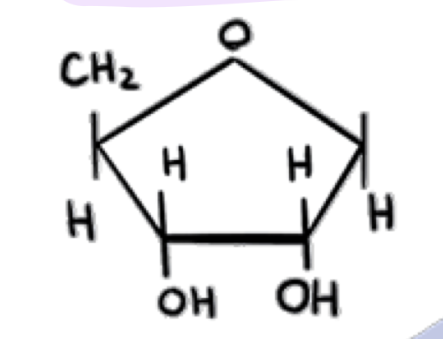
what is this
deoxyribose (on rna)
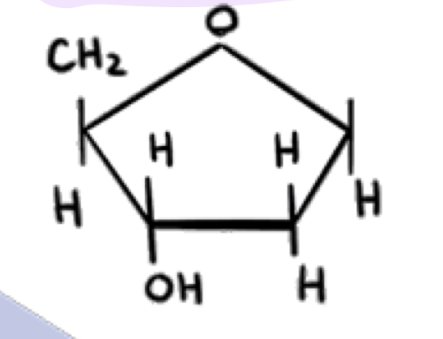
what is this
deoxyribose (found on dna)
does deoxyribose or ribose have the shorter lifespan? what about between uracil and thymine
ribose and uracil have a shorter half-life than deoxyribose and thymine of dna
what is the primary structure of proteins
the AA sequence
what is the secondary structure of proteins
local folding
what is the teritary structure of proteins
overall shape of chain (including beta pleated sheets and alpha helices and stuff)
what is the quaternary structure of proteins
multiple chains tg
what determines the AA sequence in proteins
the rna sequence
how rna bases read (not direction, method)
in groups of 3 AAs (called codons)
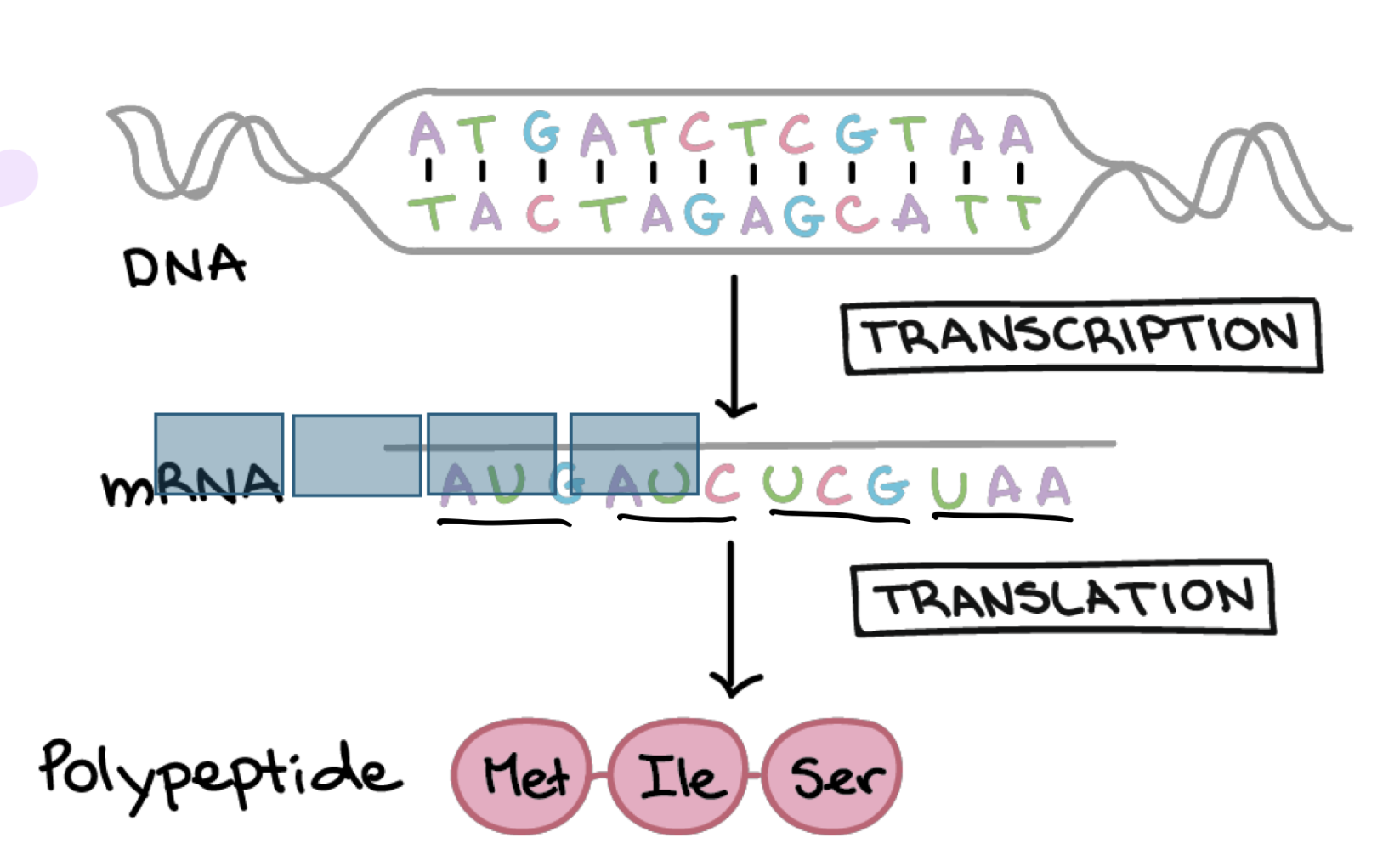
how many possible values are there for each of the 3 bases individually in any individual codon
4 (one for each base in rna)
how many possible codons are there
4×4×4 (4 possibilities for each base, 3 bases in codon) = 64
what is AUG
the start codon → first codon translated into an AA
what are the stop codons
UAA, UAG, UGA (can check data sheet on test)
what is the template strand of dna
the dna that will actually bind / create base pairs with AAs from newly synthesized rna molecule
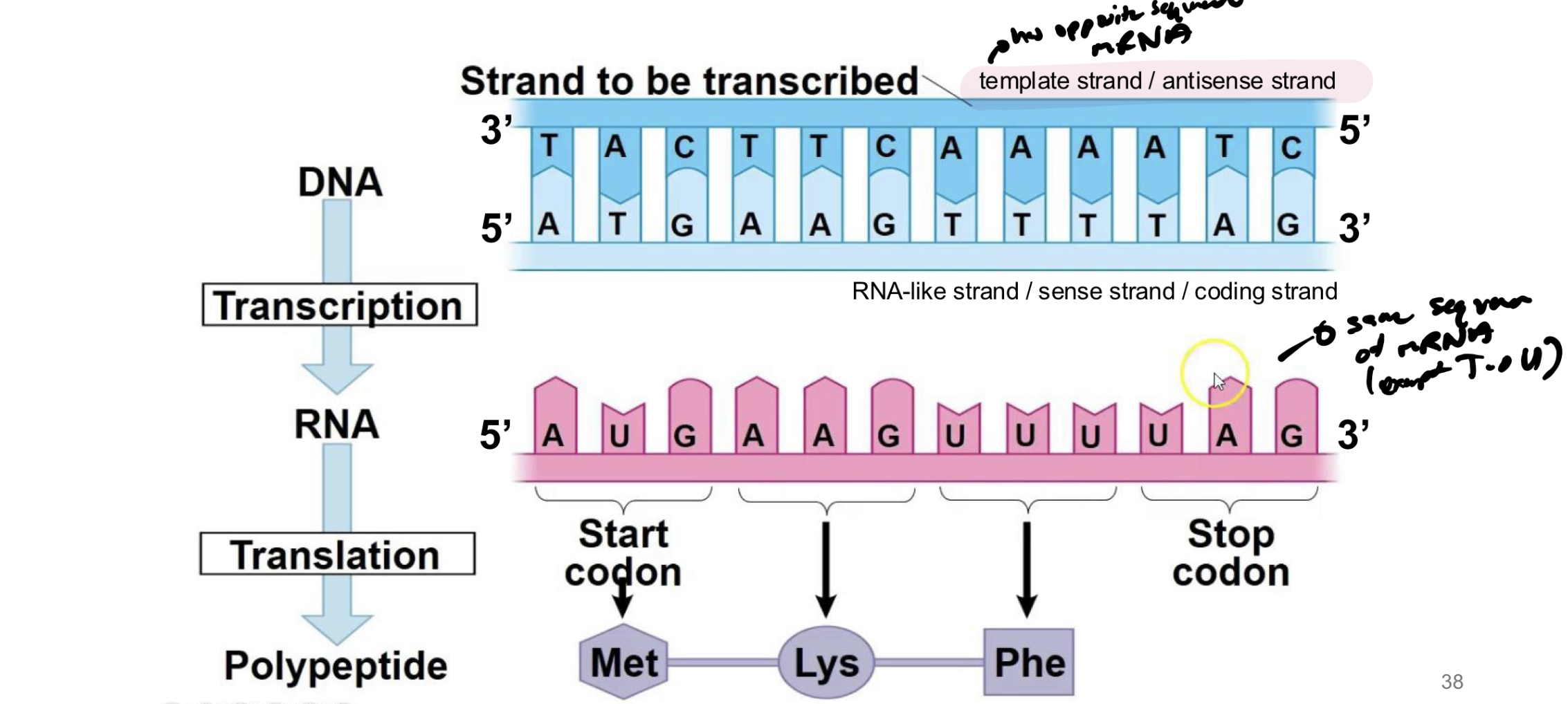
what is the antisense strand
the template strand of dna
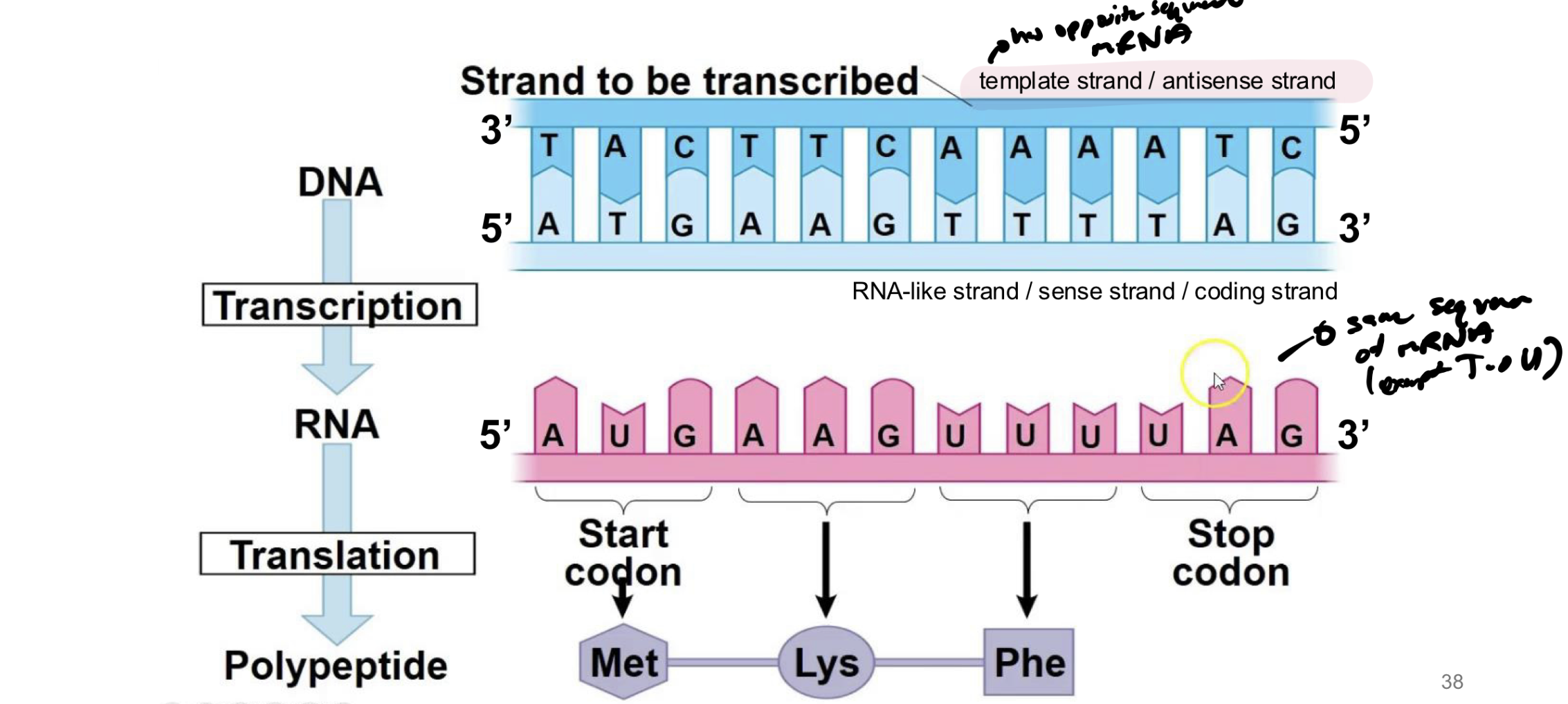
what is the sense strand/coding strand of dna
the strand of dna that was previously bound to the template → will be identical to the new RNA strand produced (except T→U)
which direction is DNA read in translation and is mRNA read in translation
DNA is read in 3’ to 5’ to synthesize new mRNA strand from 5’ to 3’
mRNA is read from 5’ to 3’ direction and synthesizes new protein in that direction too (even though no 5’ or 3’ on new protein)
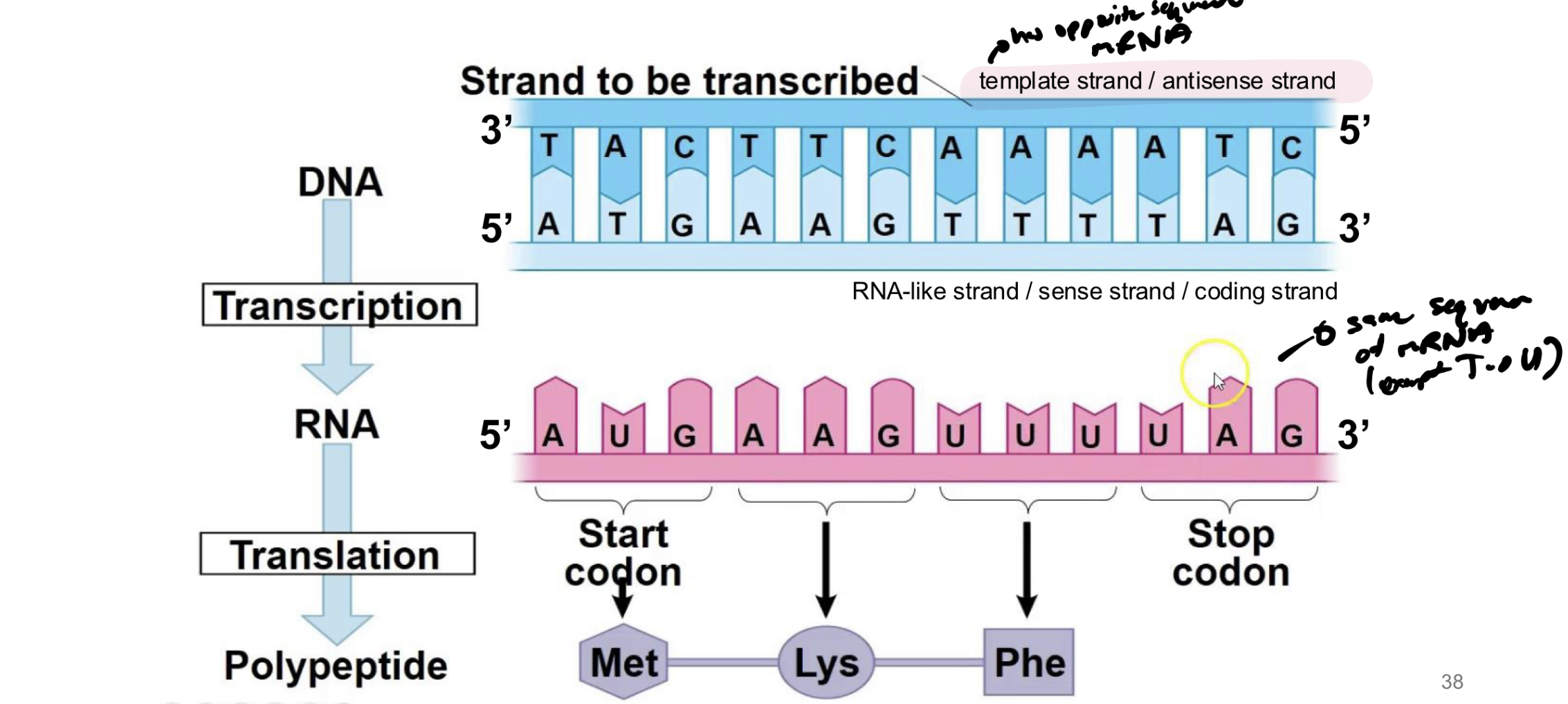
t/f: all rna molecules start with AUG
false → that is where the reading frame starts
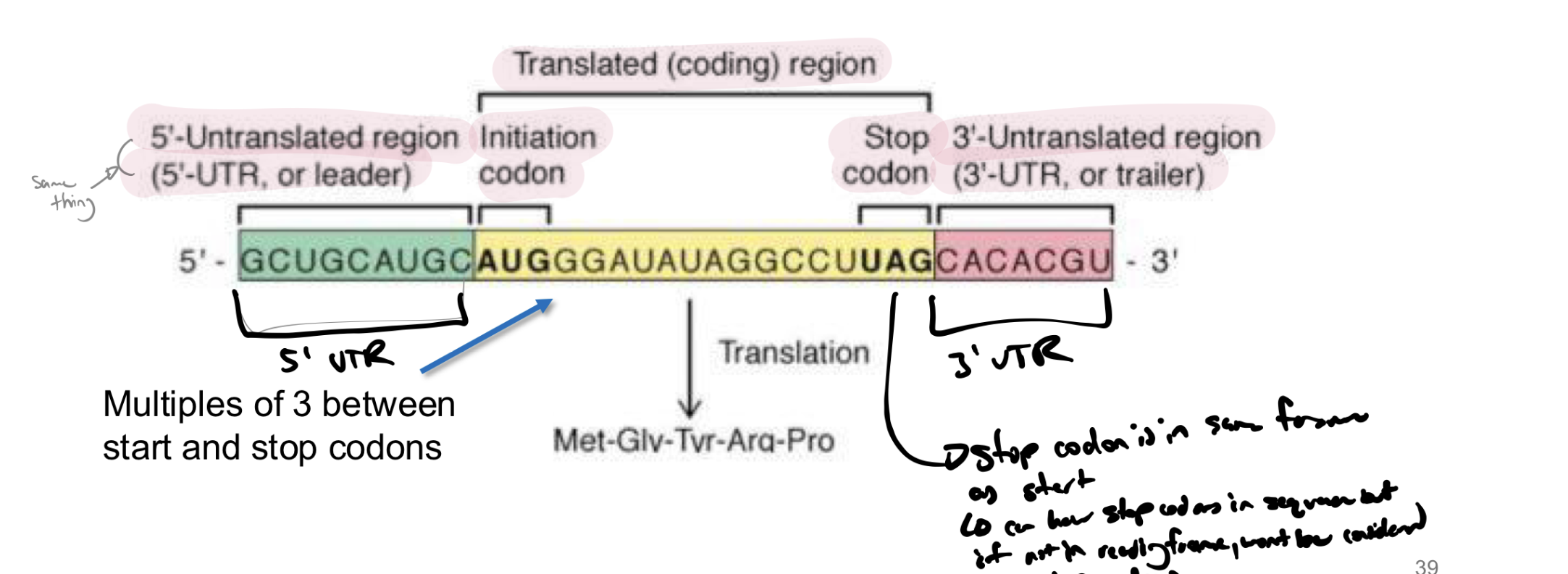
what is the reading frame in translation
rna molecule does not start with AUG → the reading frame is from AUG to a stop codon
note: stop codon must be IN FRAME with the start codon
how do we know each nuc is part of only one codon (explain the observation and conclusion)
observation: each point mutation (mutation where one nuc is swapped for another) affects only one AA
conclusion: each nuc is part of only one codon
if it was part of multiple then would have multiple protein mutations per point mutation
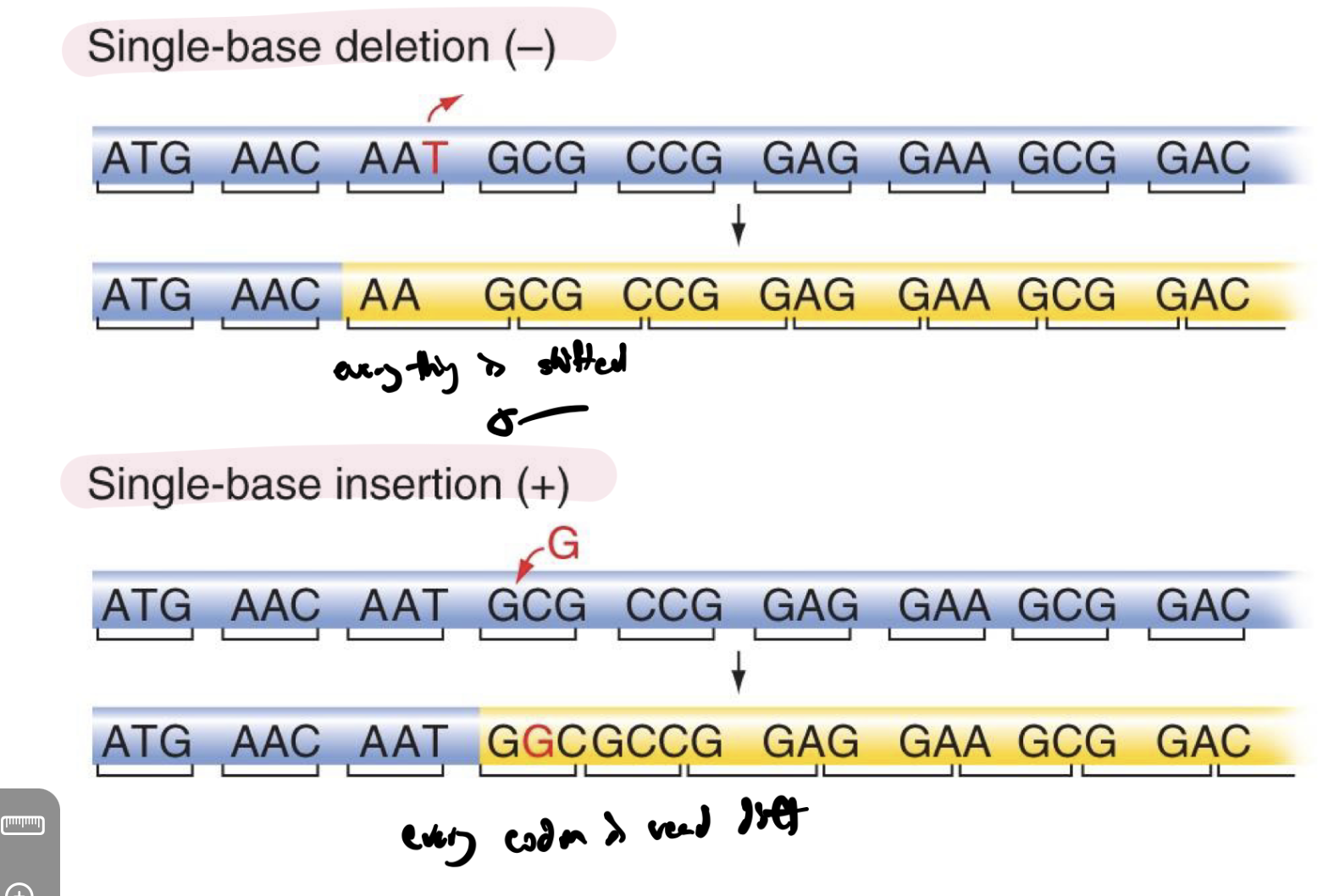
what are frameshift mutations and what are some examples of frameshift mutations?
mutations that change the reading frame of rna from then-on
changes AA sequence
eg deletion and insertion
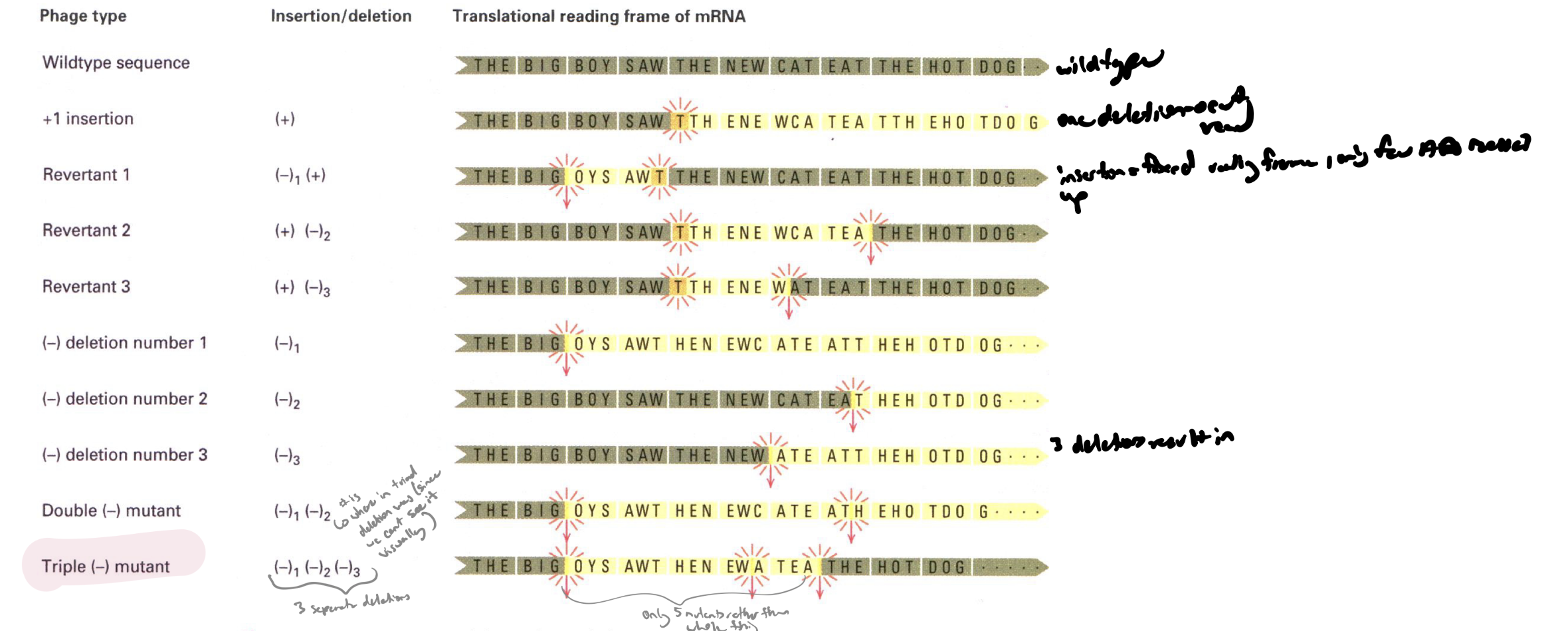
what is intragenic suppression
restoration of gene fx by one mutation cancelling the other in the same gene (eg equal # of insertions and deletions or 3 insertions/3 deletions) how did peole know which codon codes for w
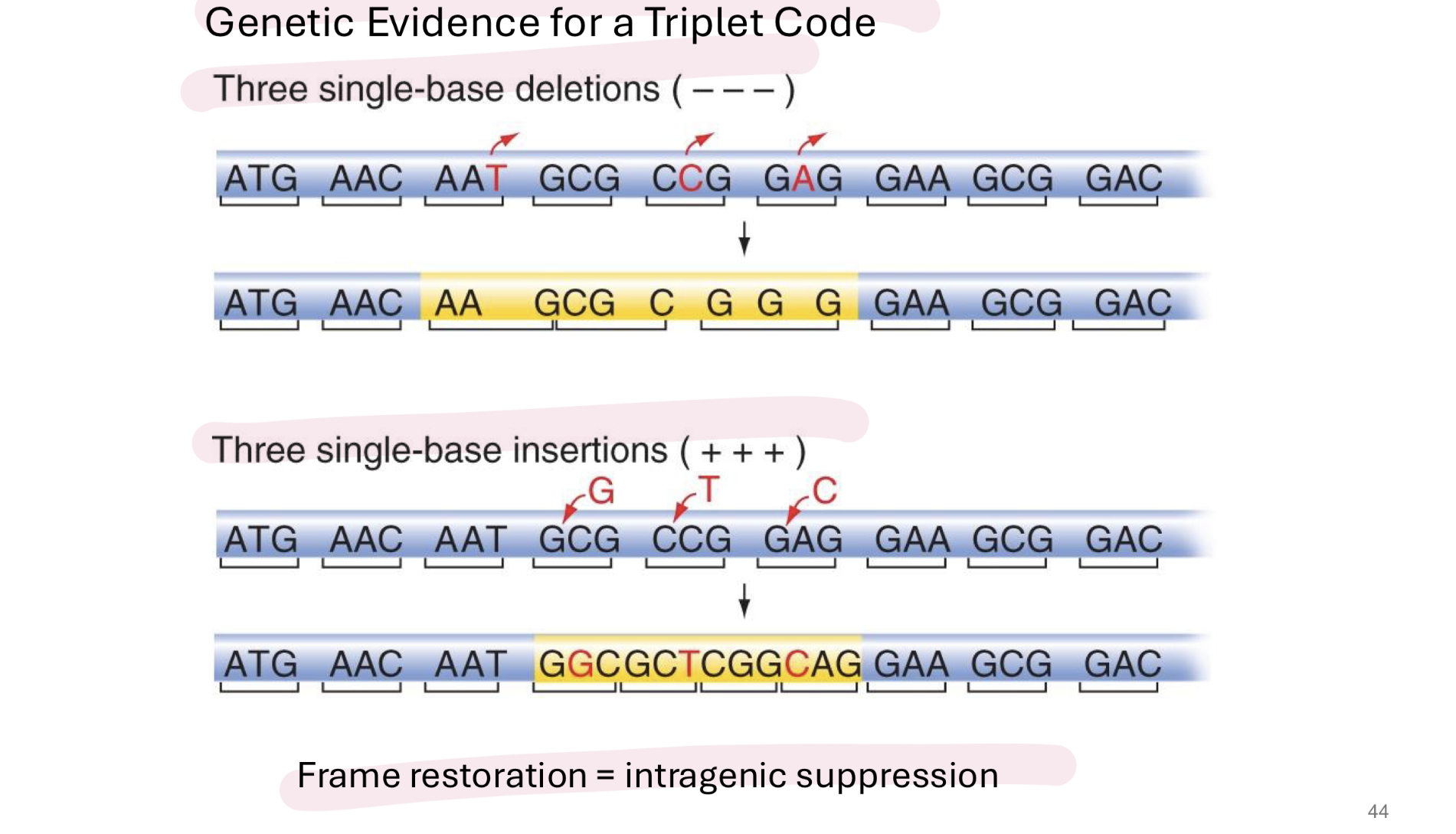
how do we know that codons are 3 nucs long
3 single based deletions or 3 single base insertions result in frame restoration → example of intragenic suppression
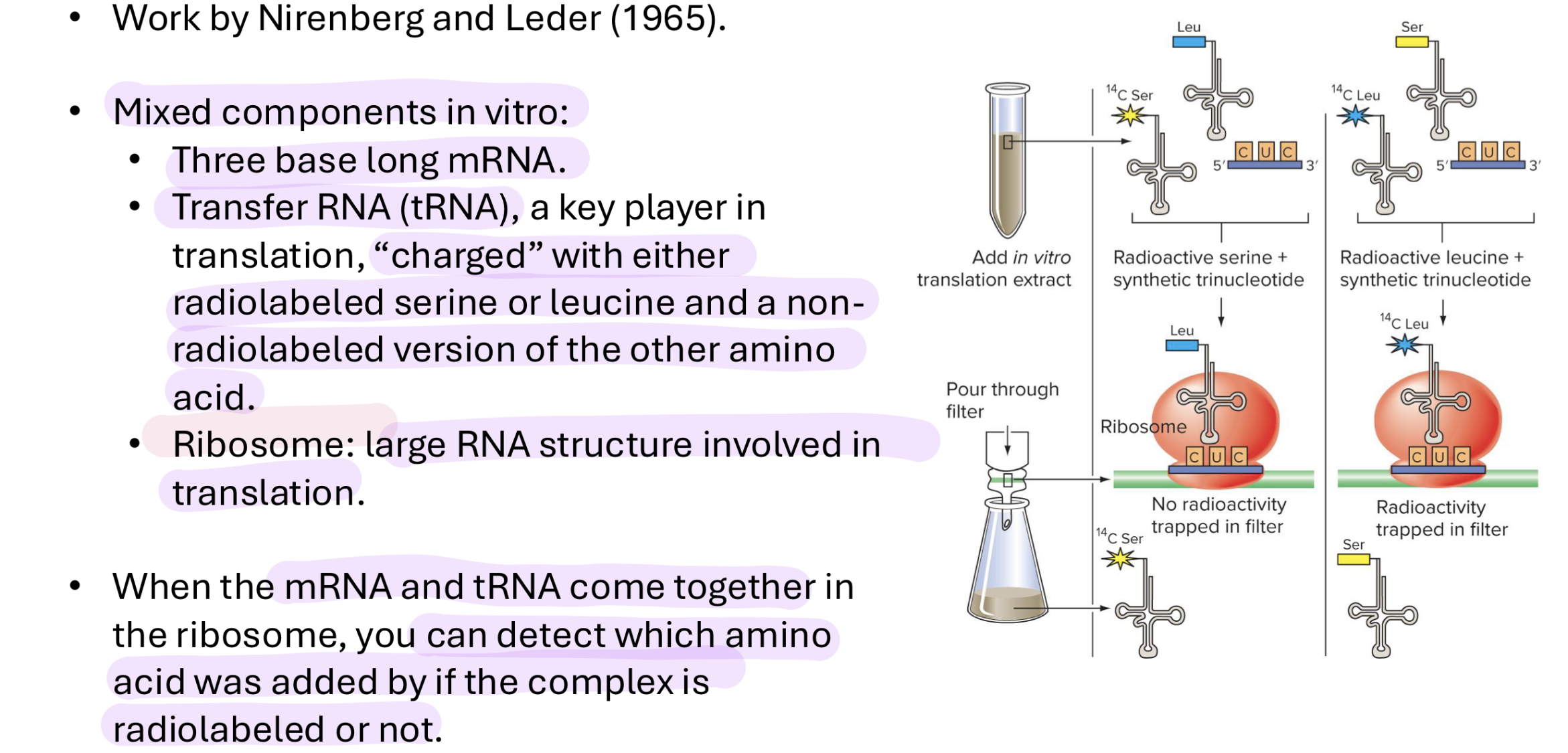
how did people figure out which codon codes for which AA
added artificial poly-U-mRNAs to cell-free translation systems and tracked AA in polypeptide product
added make mRNA with a bunch of radioactive Us into an environment where they could be translated and tracked the AA product
did this for all types of rna bases (all 4)
then introduced another base into the culture → could now have multiple products depending on the order of the 2 bases (eg Ser or Leu for UC) → made them alternate (UCUCUC)
“charged” tRNA w either radiolabeled serine or leucine and non-radiolabeled of the other AA
could then see which AA was added by if the complex was radiolabeled or not
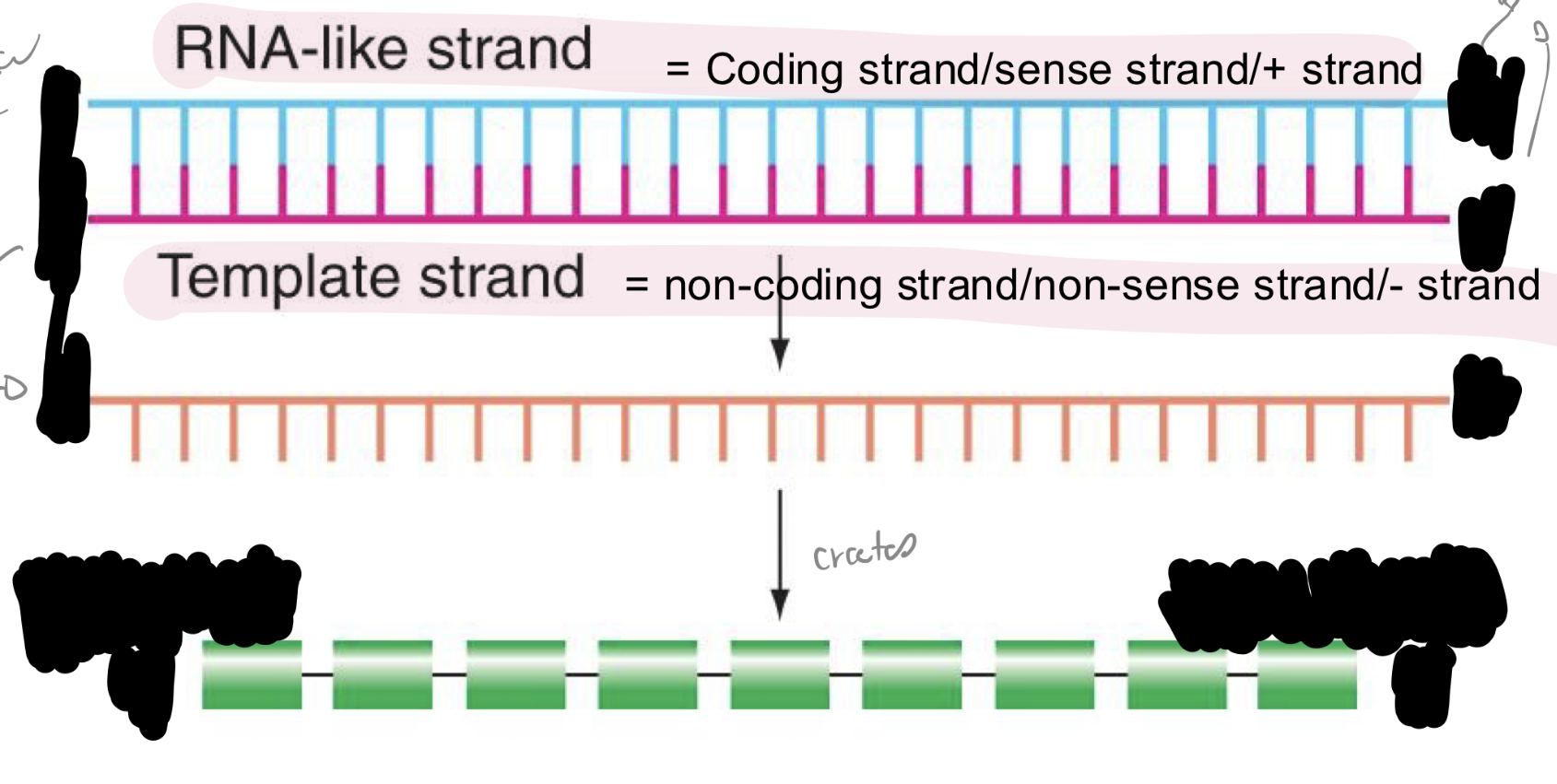
explain the relationships of polarities in DNA, mRNA, and polypeptides
note: moving from 5’ to 3’ of each mRNA, each successive codon is interpreted into AA starting w N-terminus and ending w C-terminus
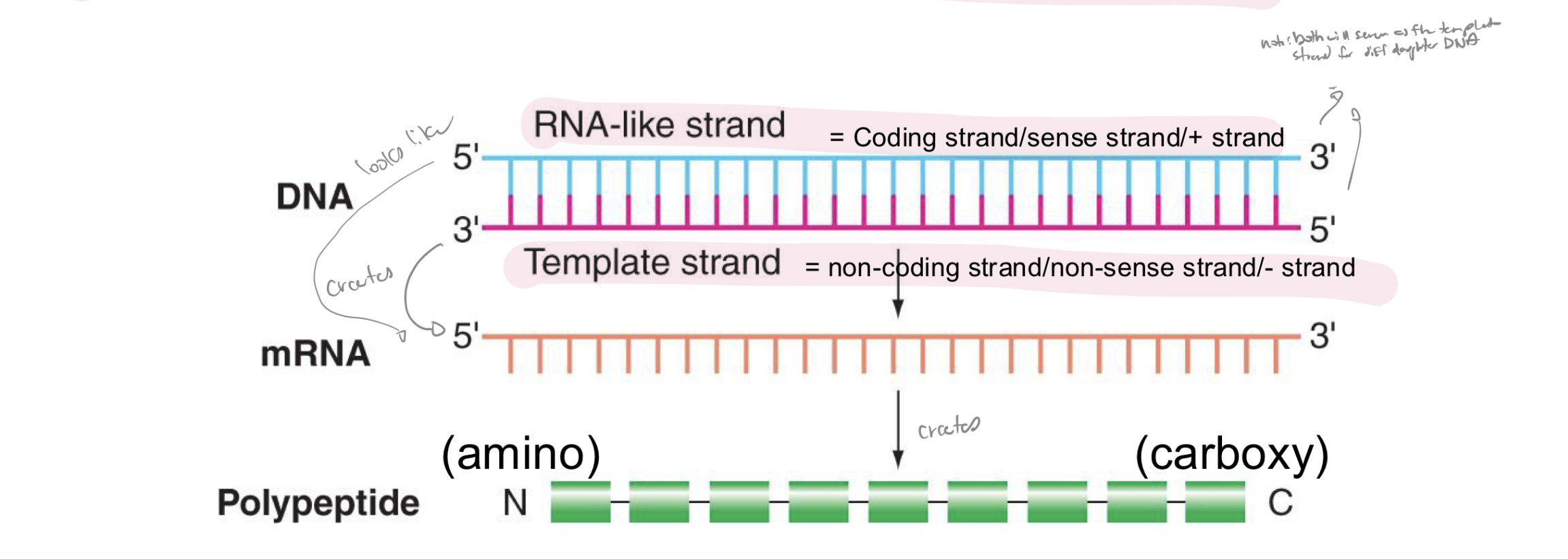
t/f: the genetic code is universal
false → it is ALMOST universal across species