19. Antibody Effector Functions
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
34 Terms
What are antibody isotypes, and why do they differ?
Five major isotypes: IgG, IgM, IgD, IgA, IgE.
Called isotypes/classes; each has distinct structure and function.
Structural differences include number/type of glycans and constant region features.
These structural variations determine their effector functions in immunity.
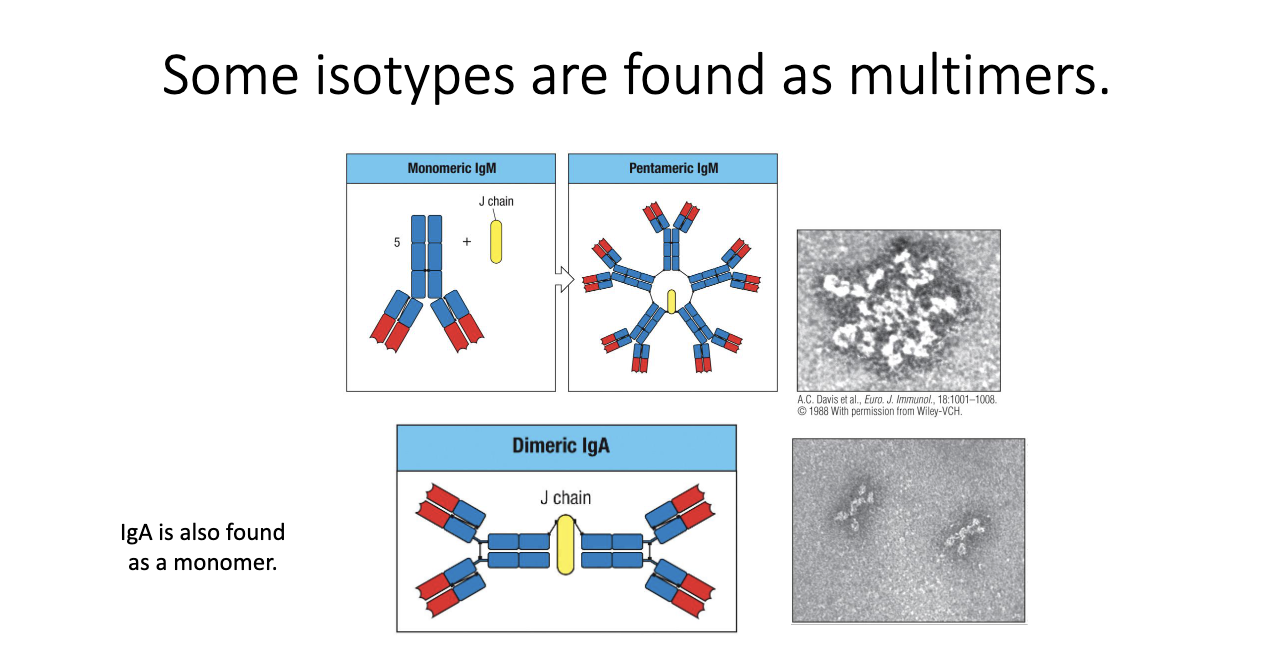
Why do IgM and IgA form multimers, and what does the J chain do?
IgM forms pentamers via the J chain → 5 antibodies linked.
IgA forms dimers (also J chain–dependent); found especially at mucosal surfaces (ex: intestines).
Purpose: increase avidity (total binding strength) to compensate for lower affinity monomers.
IgM: almost always pentameric.
IgA: can be monomeric or dimeric depending on tissue context.
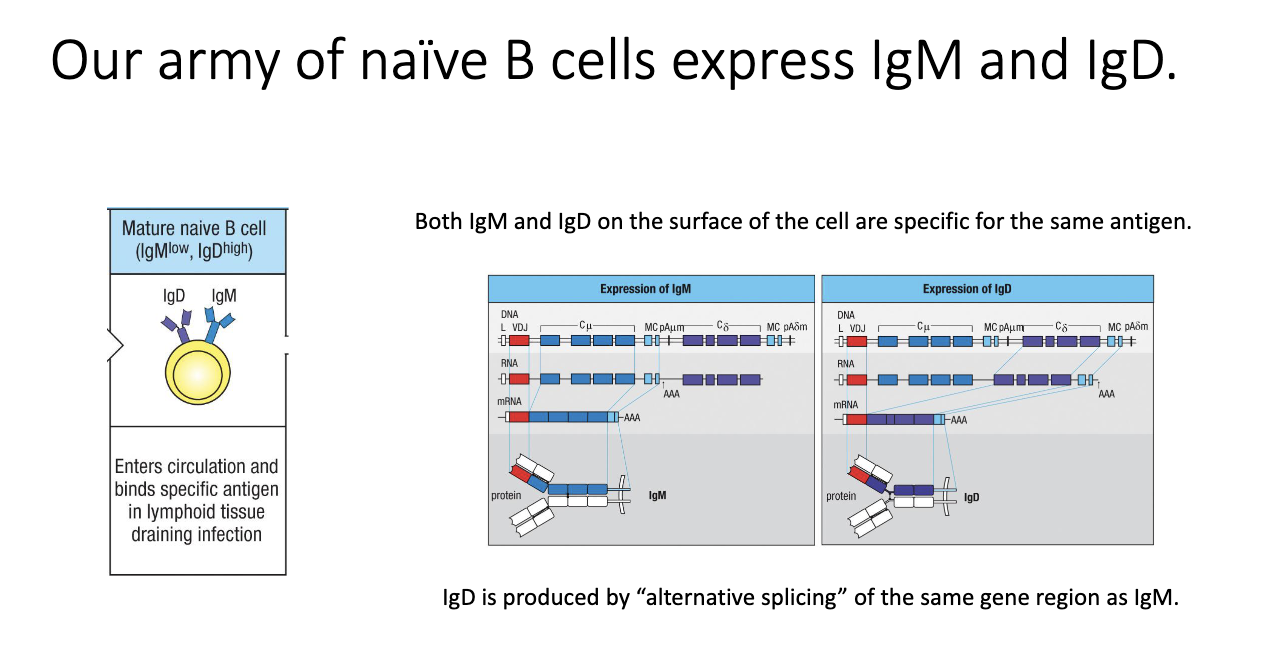
Why do naïve B cells express both IgM and IgD, and how is this achieved?
Naive B cells uniquely co-express IgM and IgD.
Both forms recognize the same antigen.
Achieved via alternative splicing of a shared transcript containing both constant regions.
Purpose of IgD is still unclear, but it is biologically required.
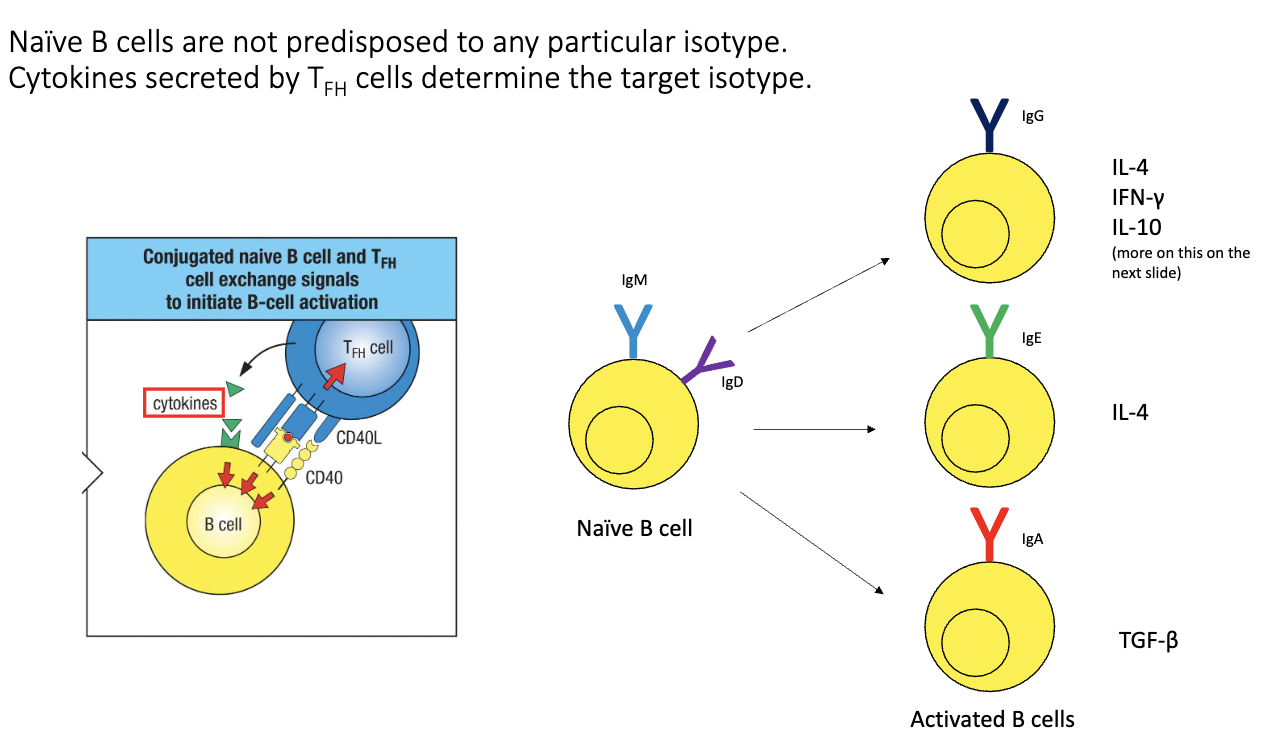
How do B cells switch from IgM/IgD to other isotypes?
Driven by signals from TFH cells:
Antigen-specific TCR–MHC II recognition.
CD40–CD40L co-stimulation.
Cytokines → determine isotype outcome.
Examples:
IL-4 → IgE
TGF-β → IgA
Dendritic cell → T cell → B cell signaling ensures isotype matches infection type.
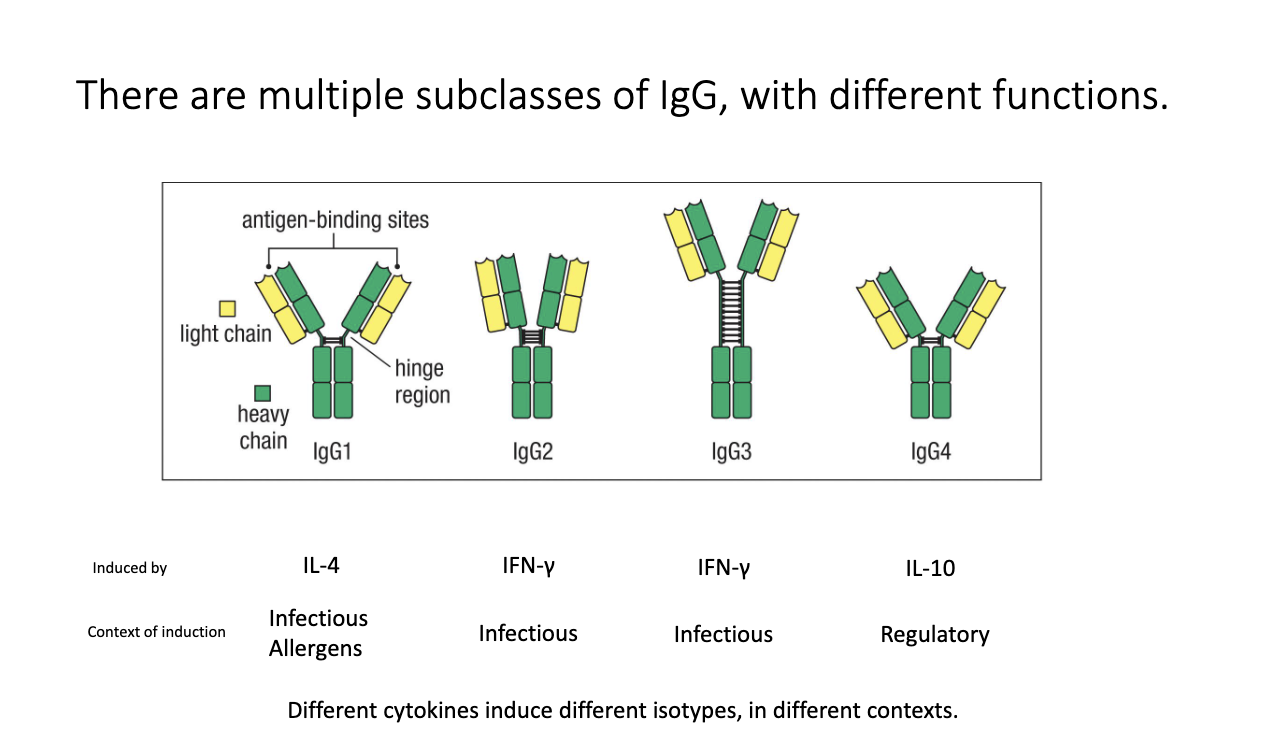
How do IgG subclasses differ, and what induces them?
Humans have four IgG subclasses: IgG1, IgG2, IgG3, IgG4.
Each has distinct hinge structures + effector functions.
Cytokine induction:
IL-4 → IgG1 (infectious and allergens)
IFN-γ → IgG2, IgG3 (infectious/anti-viral)
IL-10 → IgG4 (regulatory/anti-inflammatory)
Subclasses participate in different immune contexts.
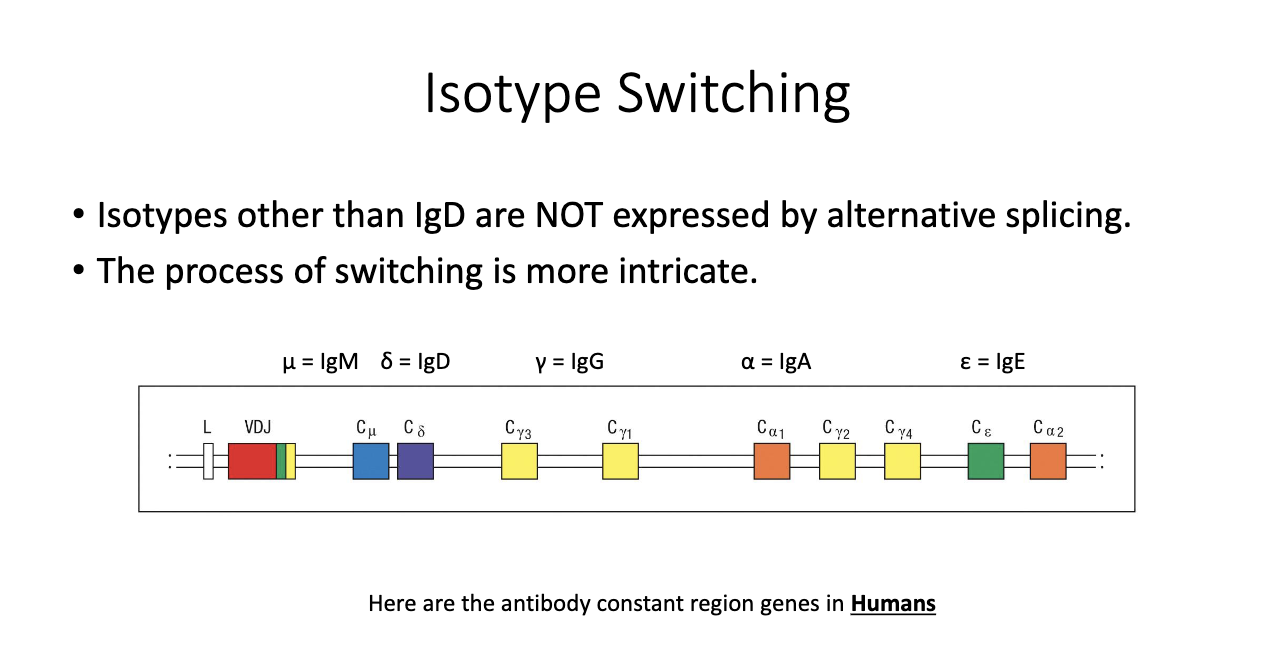
How are constant region genes arranged in the human genome, and why does it matter?
Genomic order (simplified): M → D → G3 → G1 → A1 → G2 → G4 → E → A2.
Result of gene duplication events during evolution.
Different isotypes evolved distinct roles despite shared ancestry.
Only IgM/IgD are close enough for alternative splicing; others require DNA recombination.
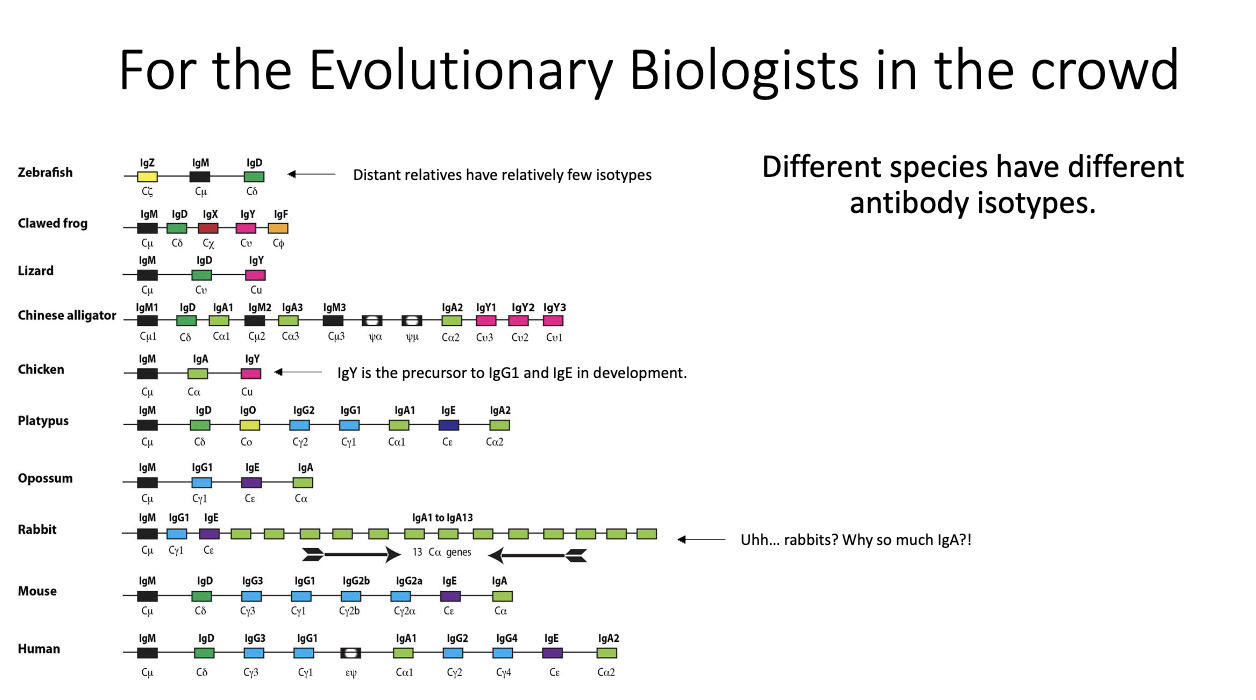
What does cross-species variation in antibody isotypes reveal?
Isotype repertoires vary widely between species (e.g., rabbits have 13 IgA genes).
IgY in birds is precursor to mammalian IgG1 + IgE.
All mammals have exactly one IgE, suggesting strong selection pressure.
Indicates rapid evolution driven by host–pathogen interactions.
Why is IgD produced by alternative splicing but other isotypes require switching?
IgM and IgD lie adjacent in the genome → allow alternative splicing into two transcripts.
Other constant regions are separated by thousands of base pairs, preventing this mechanism.
All switching beyond IgM/IgD is after VDJ recombination and B-cell activation.
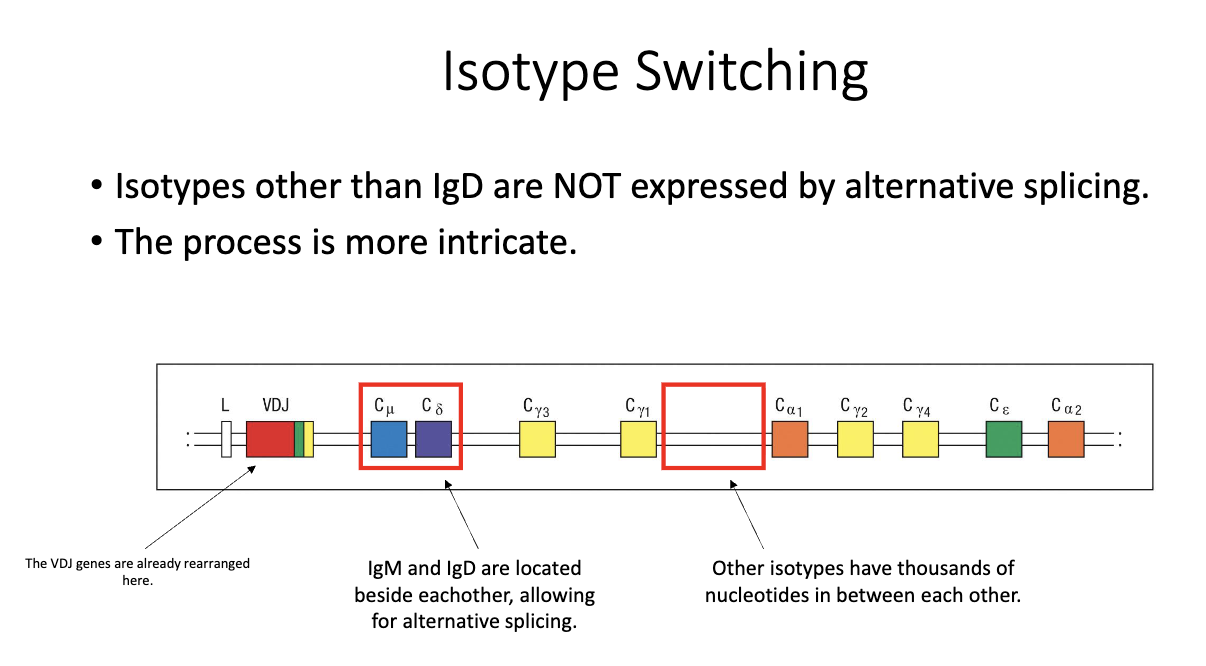
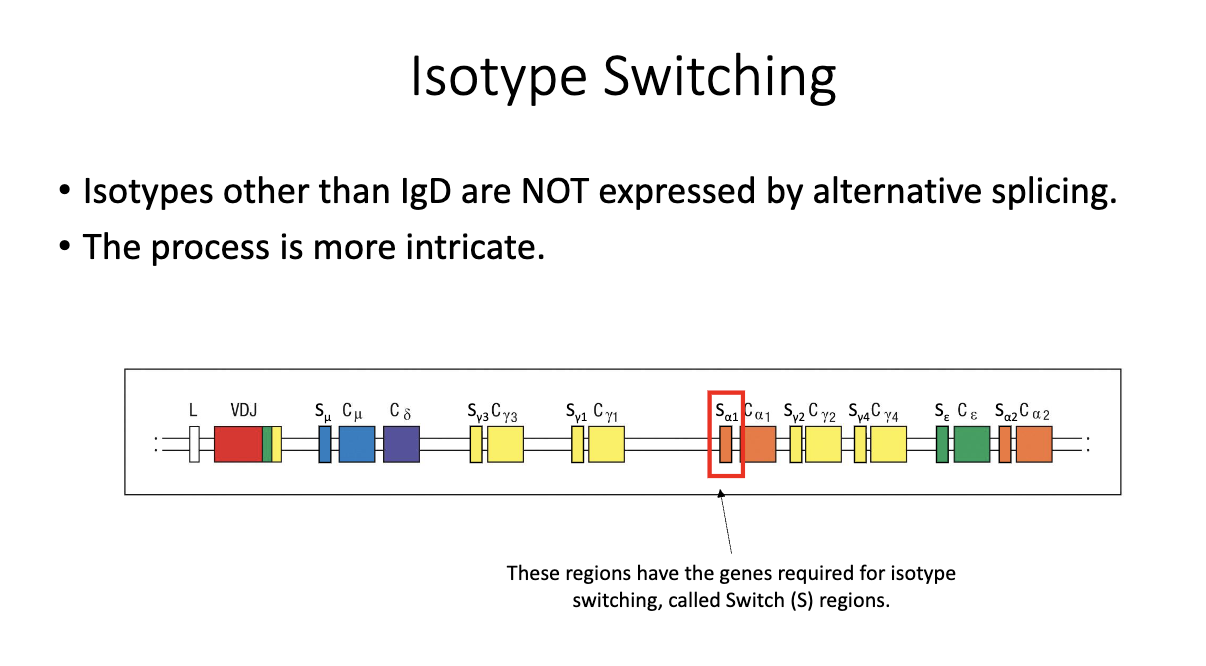
What are switch (S) regions, and why are they important for isotype switching?
Located upstream of each constant region (except δ/IgD).
Not translated; function is purely regulatory.
Required for the recombination process that enables class-switch recombination (CSR).
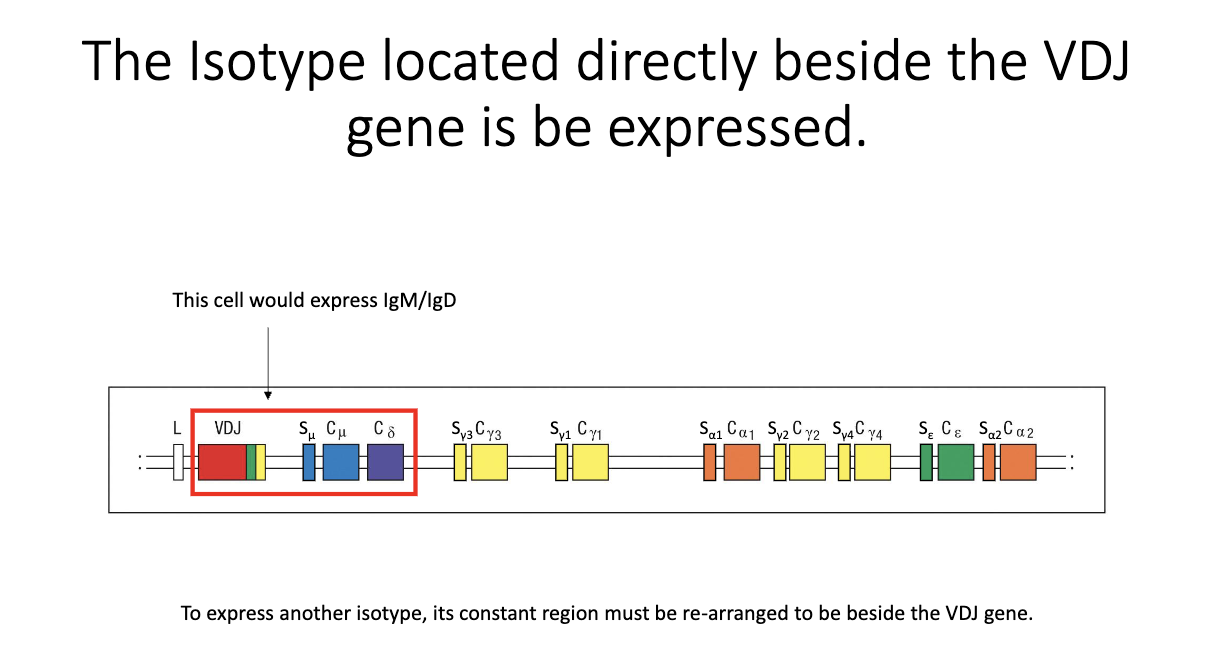
How does a B cell begin producing a new isotype at the DNA level?
Only the constant region directly adjacent to VDJ can be transcribed.
Naive state: IgM and IgD flank the VDJ → both expressed.
To express a new isotype, the relevant constant region must be moved next to VDJ.
Achieved by cutting out intervening DNA between the switch regions → irreversible CSR event.
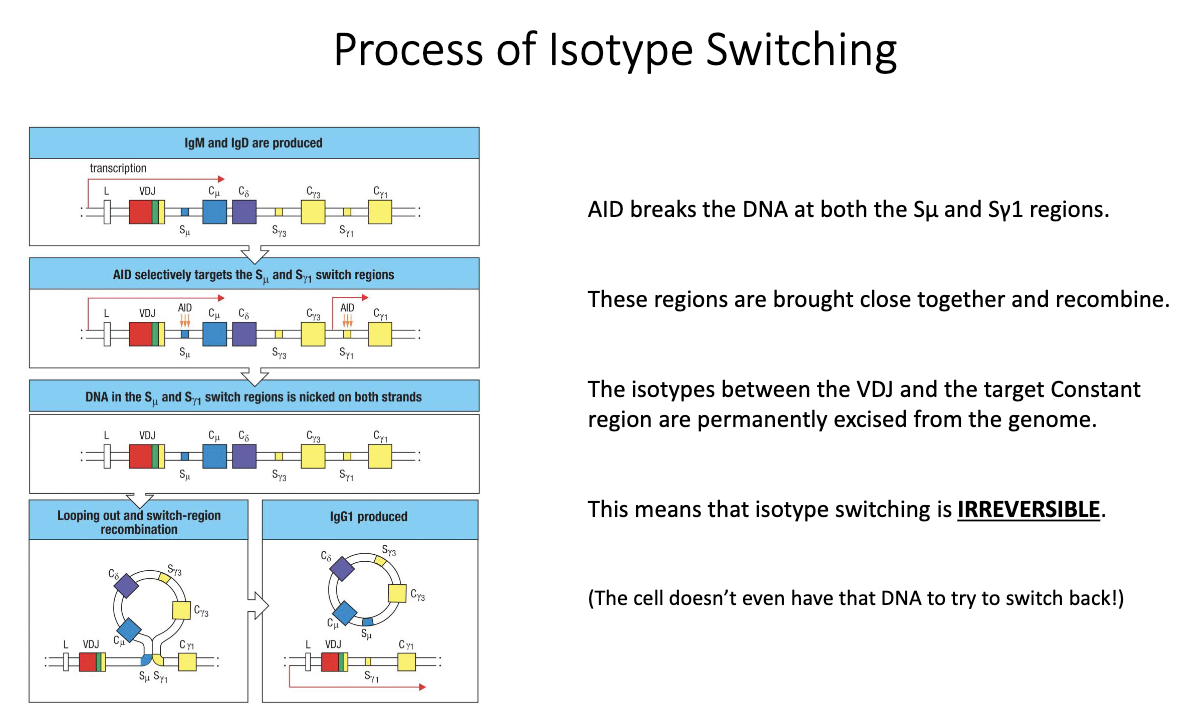
How does AID mediate class-switch recombination (CSR) to generate new isotypes?
Transcription begins upstream of VDJ, passing through IgM/IgD.
AID (upregulated after TFH help) introduces mutations/breaks in switch (S) regions.
Cytokines determine which S region AID targets (e.g., IL-4 → Sγ1, IFN-γ → Sγ3).
AID creates double-stranded breaks → DNA ends are joined; intervening DNA loops out as a switch circle.
New constant region is placed beside VDJ → irreversible isotype switch.
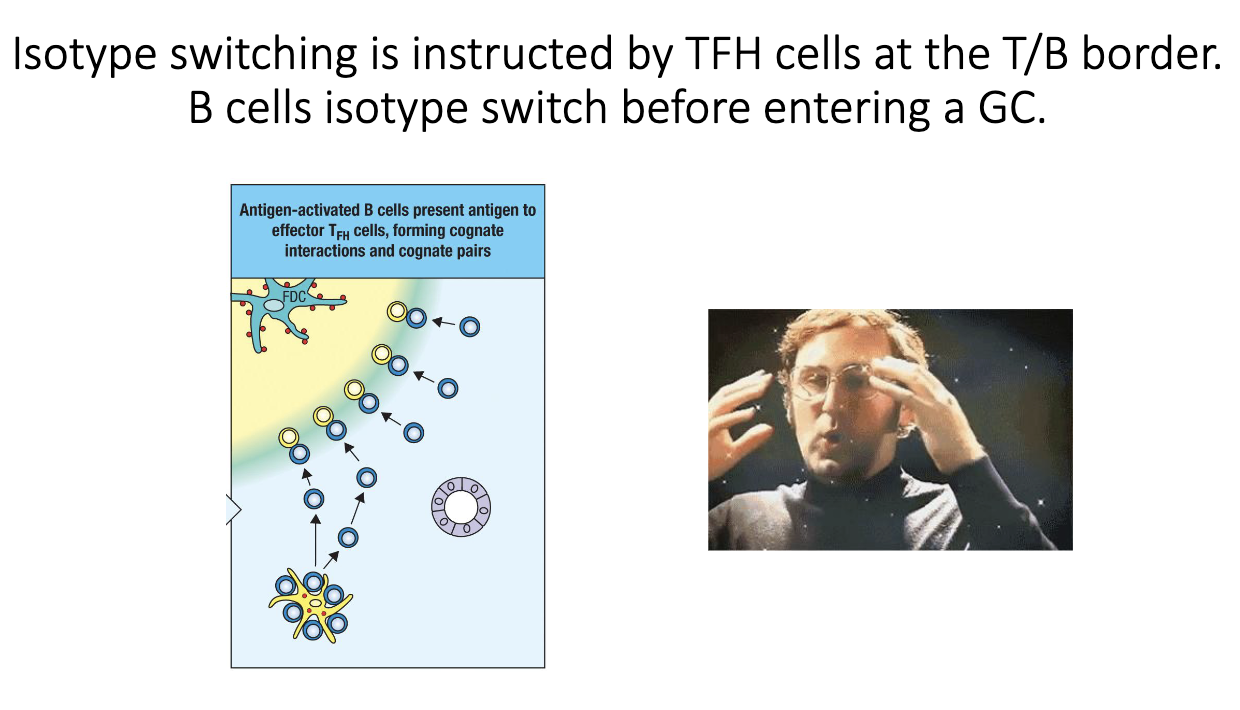
Does isotype switching occur inside germinal centers?
Textbooks say yes, but this is outdated.
Coróla Vinuesa (2019) showed CSR occurs before B cells enter germinal centers.
Switching happens at the T–B border, immediately after TFH–B cell interaction.

When do B cells isotype switch relative to germinal center entry?
CSR is triggered during the T cell–B cell “hug” at the T–B border.
By the time a B cell migrates into the germinal center, it has already switched.
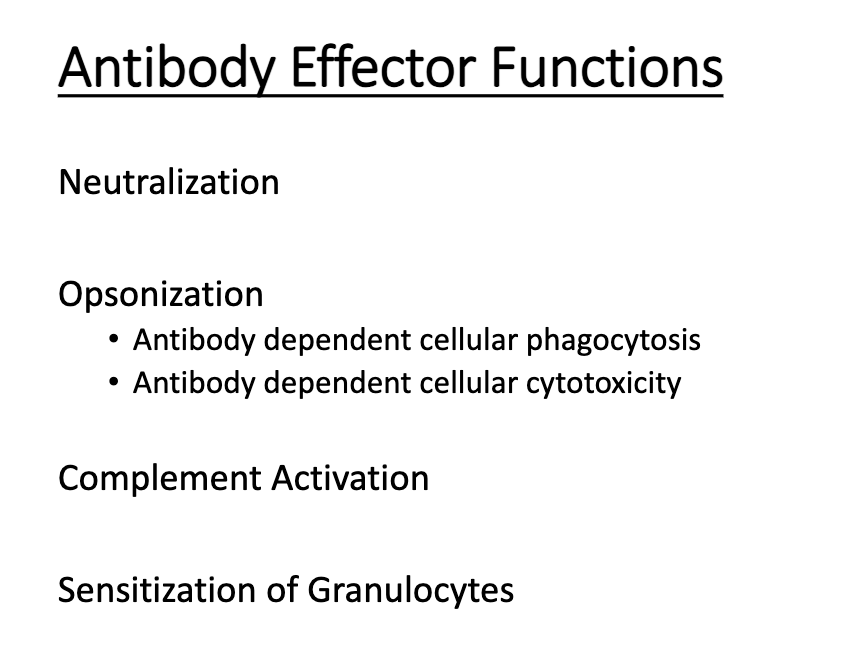
What are the four major pathogen-eliminating functions of antibodies?
Neutralization
Opsonization
Complement activation
Granulocyte sensitization
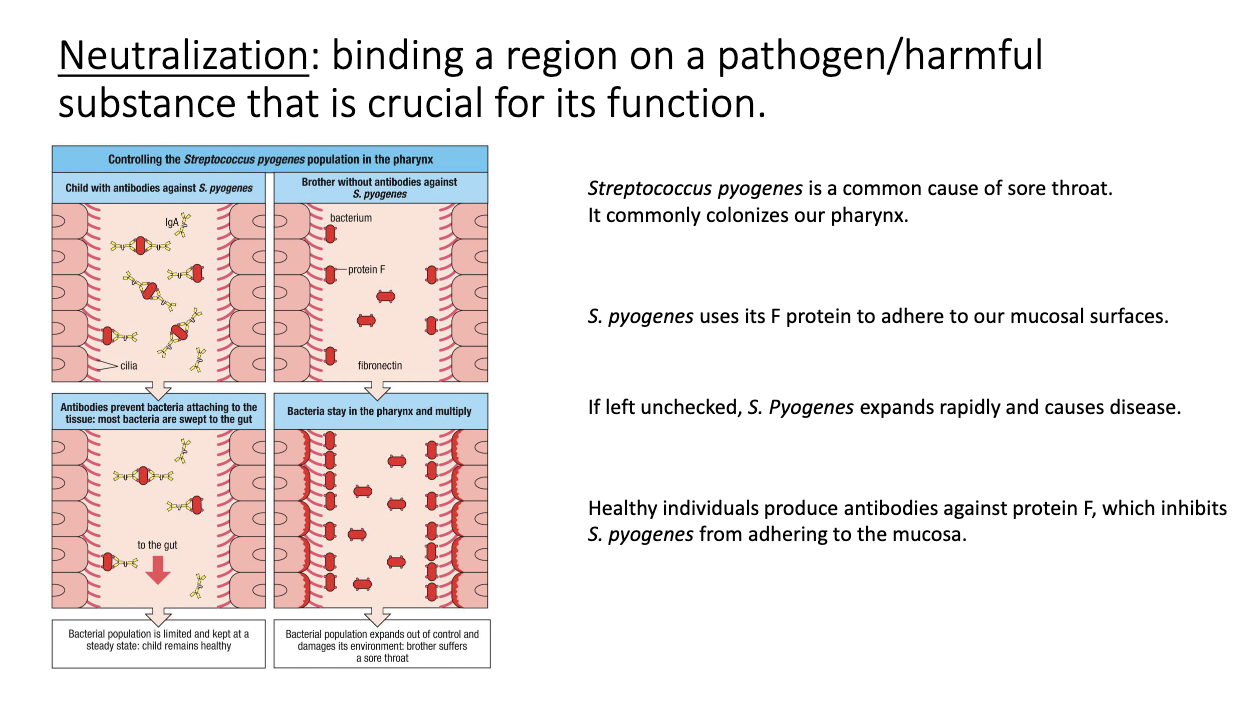
How does antibody-mediated neutralization work?
Antibody binds a pathogen’s entry/attachment site → blocks access to host cells.
No additional immune machinery needed.
High-affinity antibodies can permanently neutralize targets.
Classic example: mucosal IgA blocking influenza or bacterial adhesins.

How do antibodies neutralize bacterial pathogens like Strep pyogen?
Bacteria use adhesins (e.g., F protein) to bind mucosal surfaces.
IgA binds these F-protein → prevents attachment/colonization.
Pathogen is cleared before causing symptomatic infection.
Why do IgM pentamers rely on avidity for neutralization?
Early IgM is low affinity (pre–germinal center).
Pentamer structure gives 10 binding sites → strong avidity even with weak individual interactions.
Compensates for poor affinity to still block pathogens effectively.

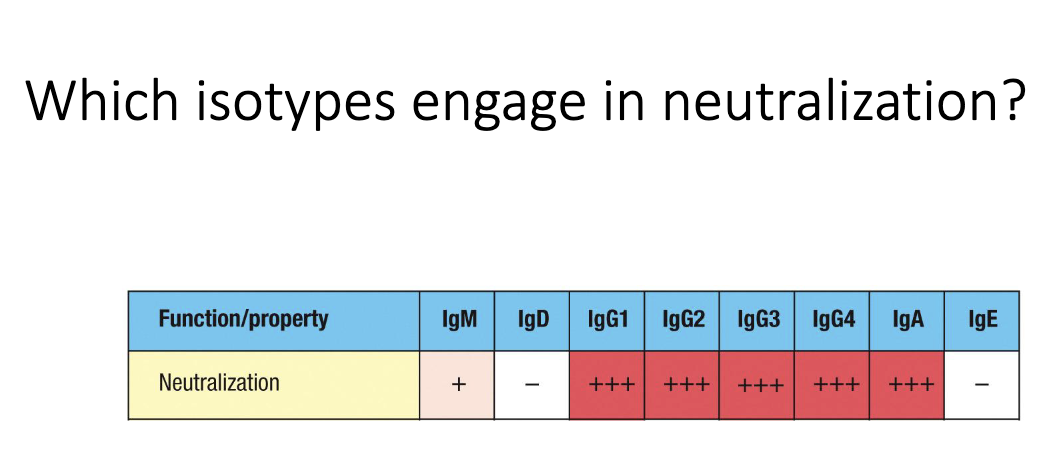
Which antibody isotypes are most effective at neutralization?
IgG (systemic) and IgA (mucosal) are strongest neutralizers.
IgM provides weaker but still meaningful neutralization via high avidity.
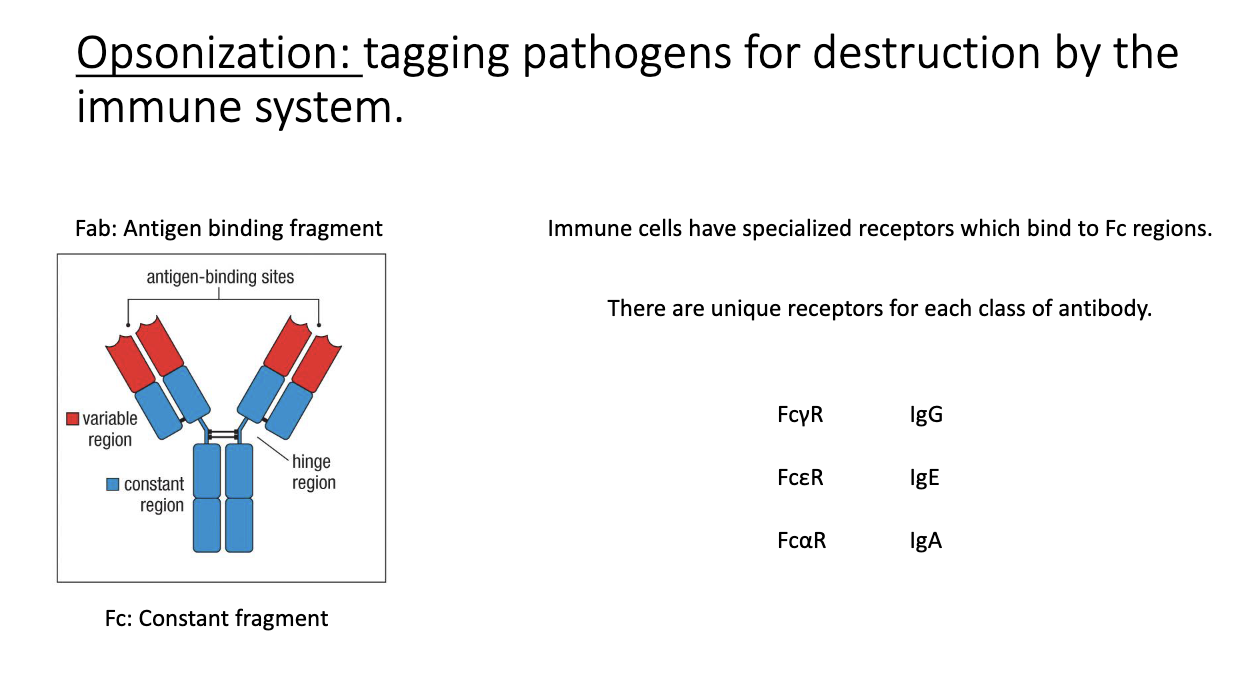
What is opsonization, and how do FC receptors participate?
Antibody binds a pathogen, and immune cells detect the Fc region via Fc receptors.
Fc receptor naming: Fc + isotype Greek letter + R (e.g., FcγR for IgG).
Leads to phagocytosis and destruction of the antibody-coated microbe.
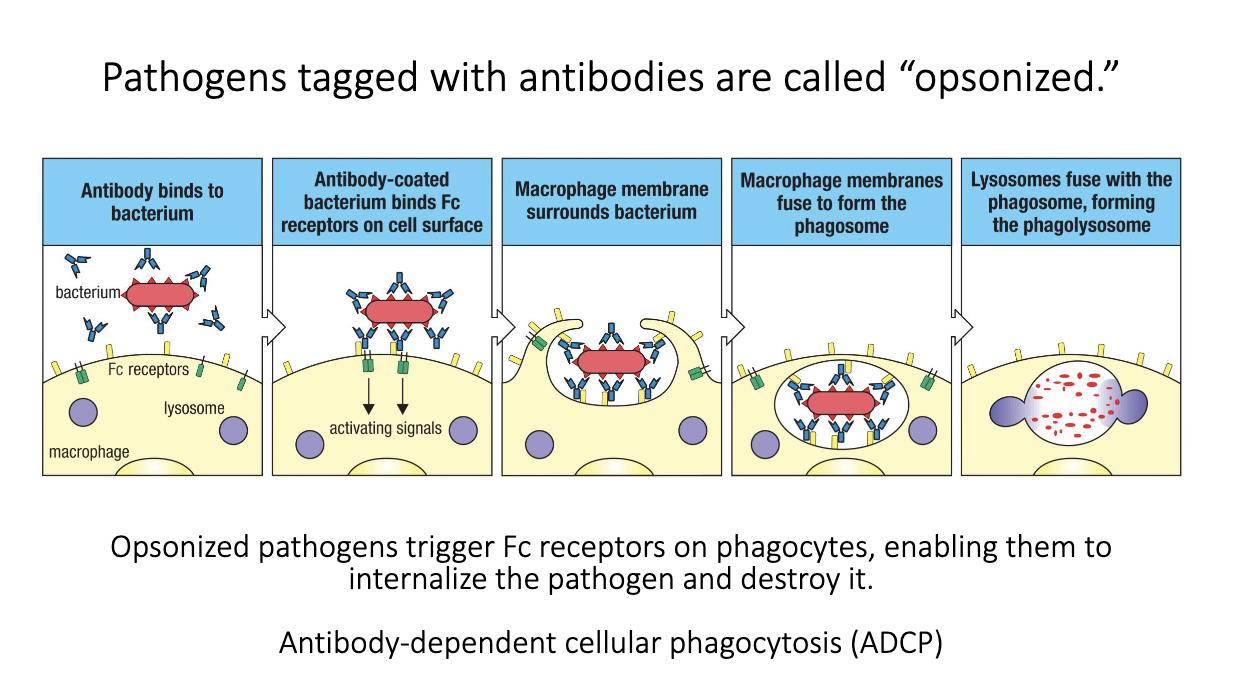
What is antibody-dependent cellular phagocytosis (ADCP)?
Pathogen coated with antibodies = opsonized.
Fc receptors on phagocytes bind Fc regions of those antibodies.
FcγR signaling tells the cell to engulf and destroy the target.
Leads to phagolysosome fusion and killing of the pathogen.
Antibodies augment innate immunity by making microbes easier to phagocytose.
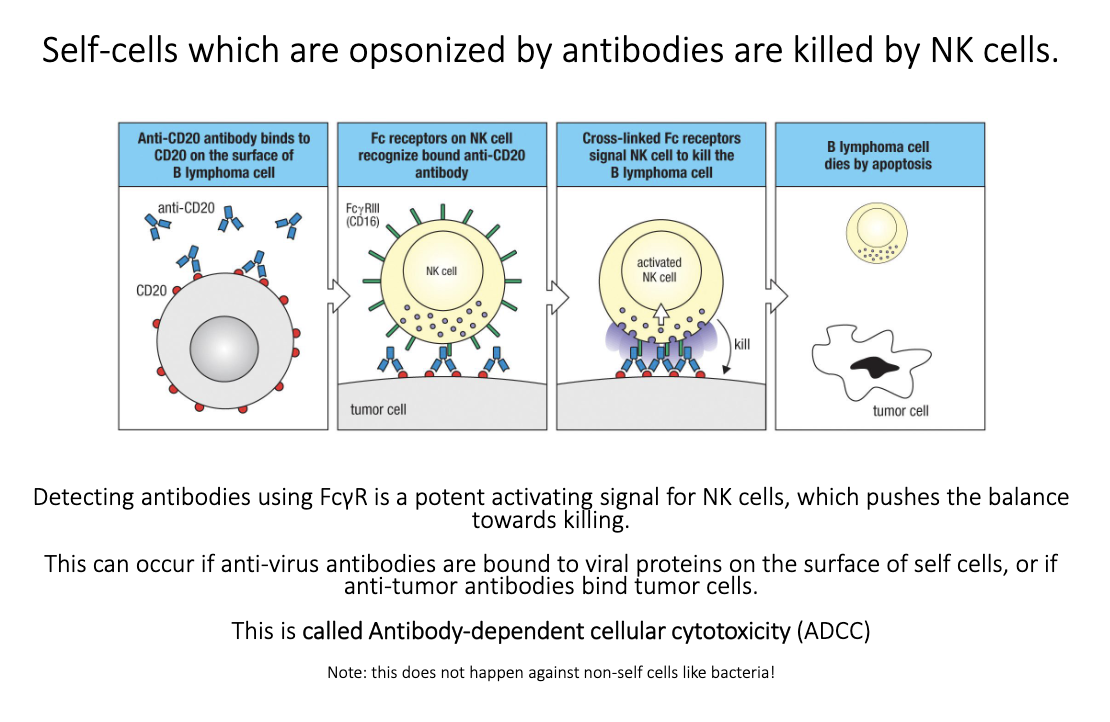
How does antibody-dependent cellular cytotoxicity (ADCC) work?
Antibodies bind antigens on infected or cancerous host cells.
NK cells use FcγRIII to recognize bound IgG.
Fc engagement → strong activating signal → target cell killing.
Used therapeutically (e.g., anti-CD20 antibodies killing B-cell cancers).
Does not work on bacteria because NK killing uses perforin/granzymes → apoptosis, which bacteria can’t undergo.
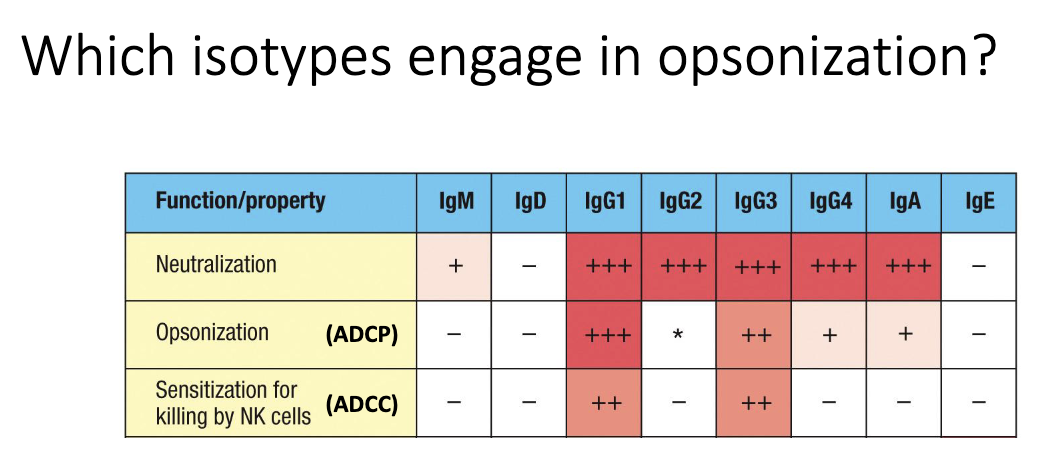
Which isotypes are effective at opsonization?
Primarily IgG1 and IgG3.
These bind Fcγ receptors strongly, enabling ADCP and ADCC.
IgG2 and IgG4 bind FcγRs poorly → weak opsonizing ability.
Which antibodies activate complement, and how?
IgM is the strongest complement activator (ideal structure for C1 binding).
IgG1 and IgG3 also activate complement effectively.
Complement binding to antibody-coated surfaces → classical pathway → pathogen lysis.
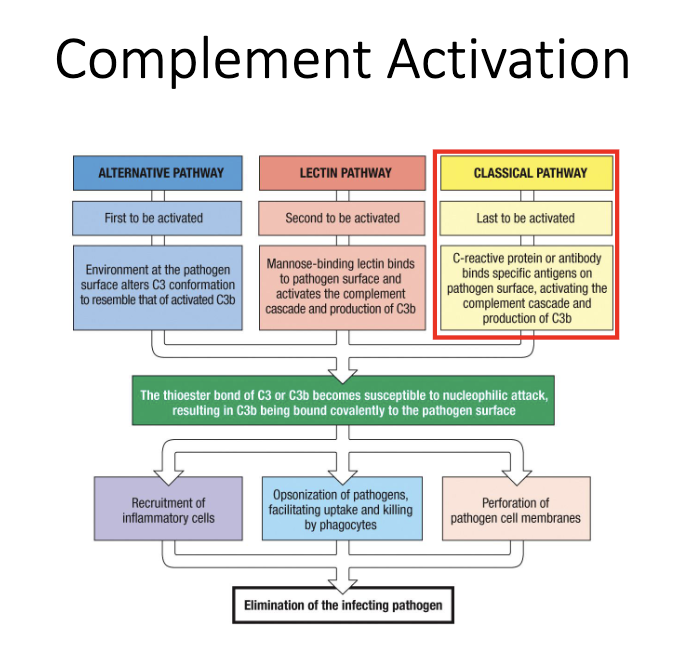
Why is IgM exceptionally good at fixing complement?
Pentameric “planar” structure matches C1q binding sites perfectly.
C1q engagement rapidly triggers the classical complement cascade.
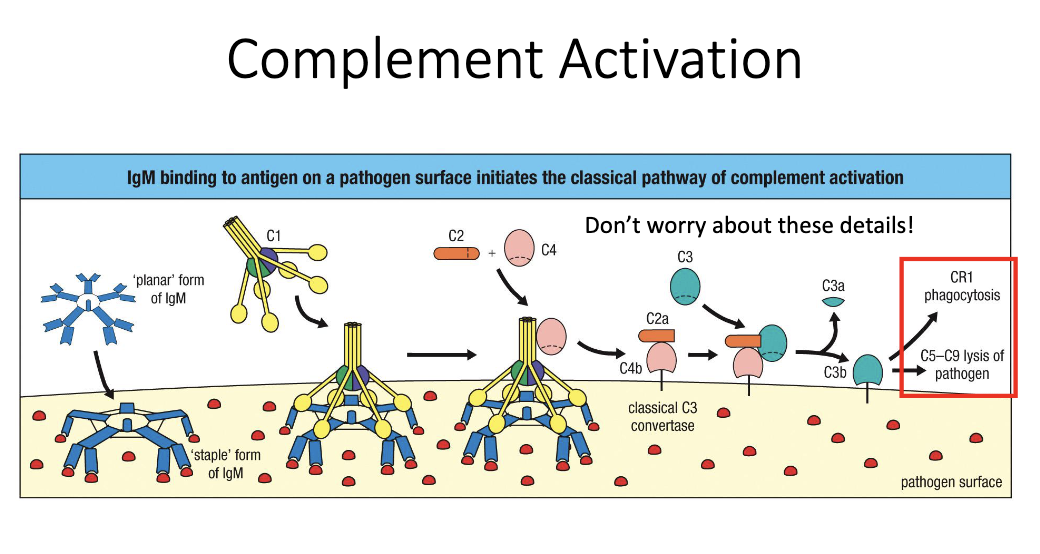
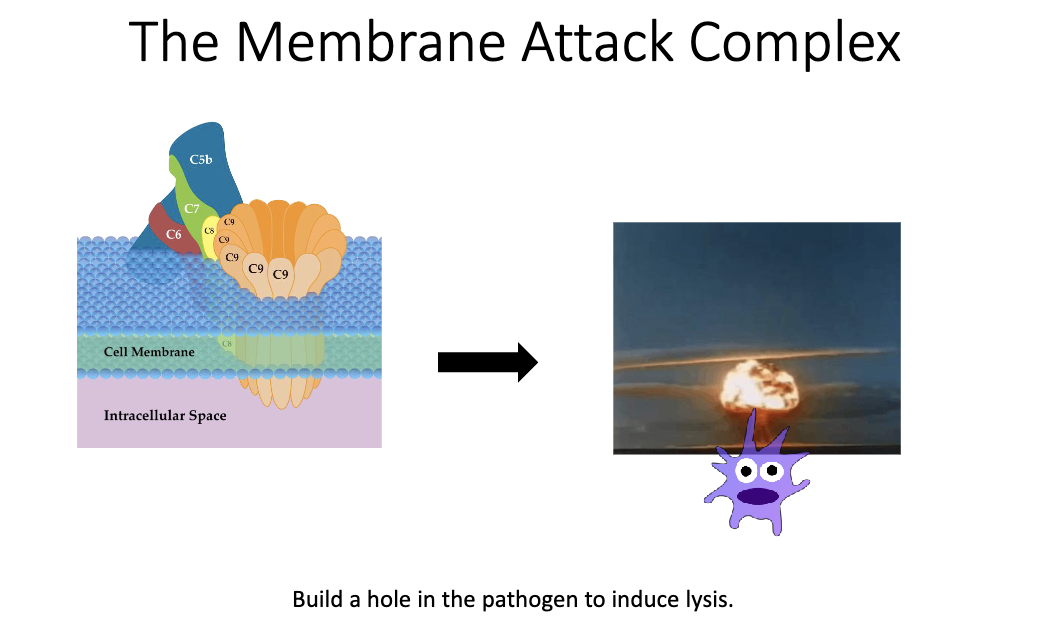
What is the end result of antibody-driven complement activation?
Complement cascade forms the MAC pore in microbial membranes.
Leads to osmotic lysis and death of the pathogen.
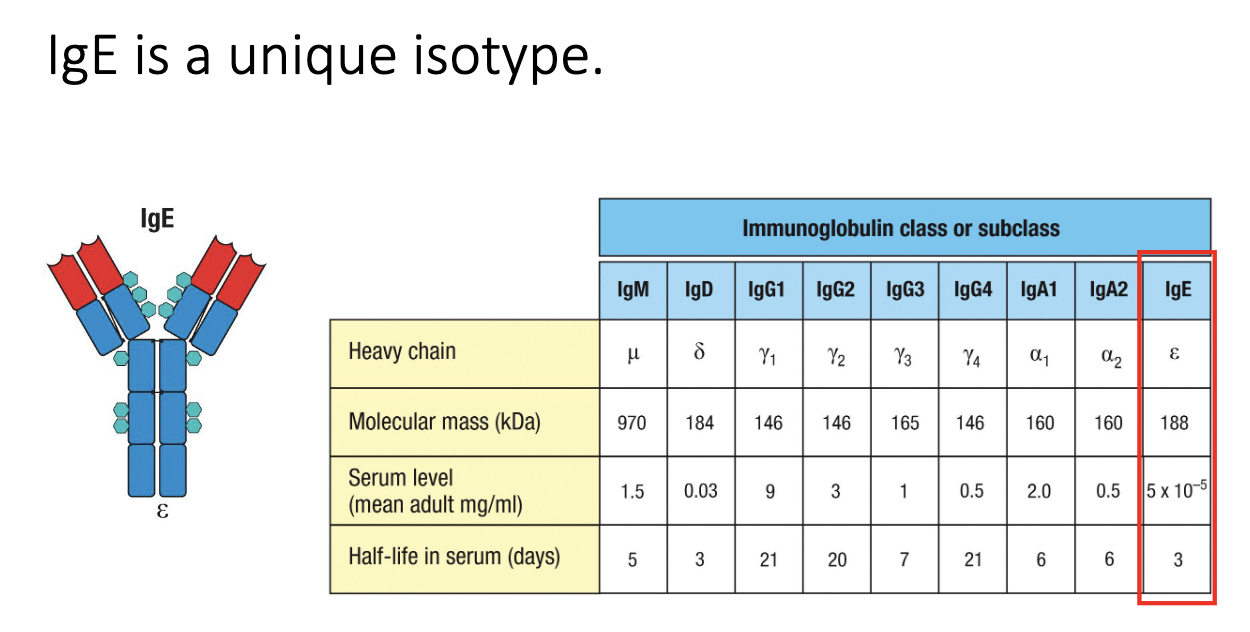
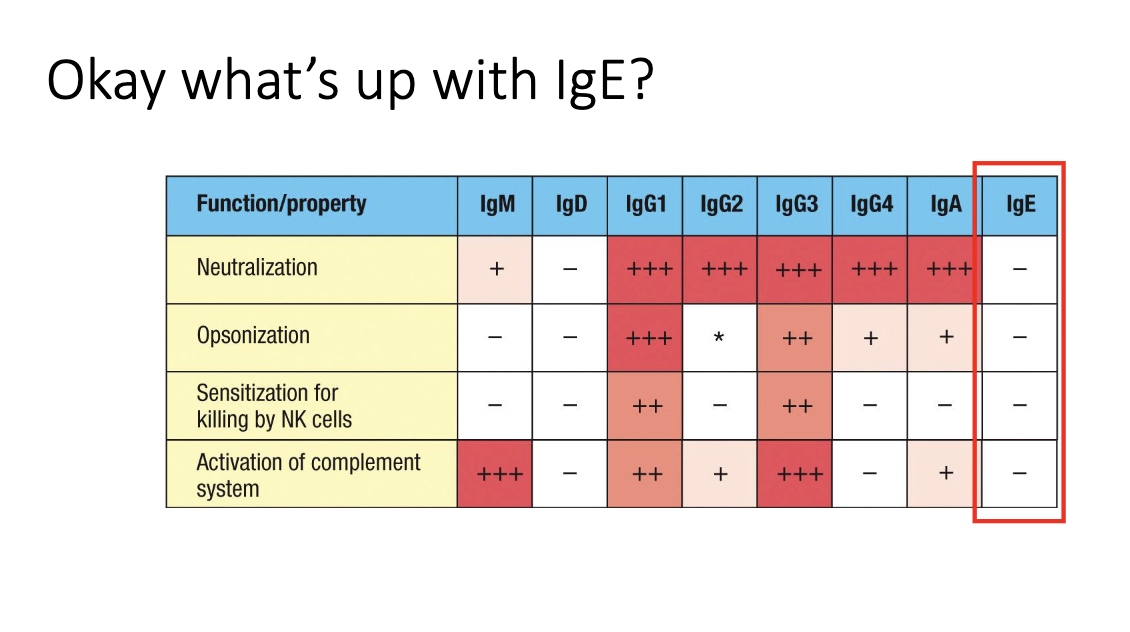
Why doesn’t IgE carry out neutralization or opsonization in circulation?
Serum IgE levels are extremely low (ng/mL range).
Too scarce to meaningfully neutralize or opsonize microbes.
Its role is instead mediated through FcεRI-bound IgE on granulocytes.
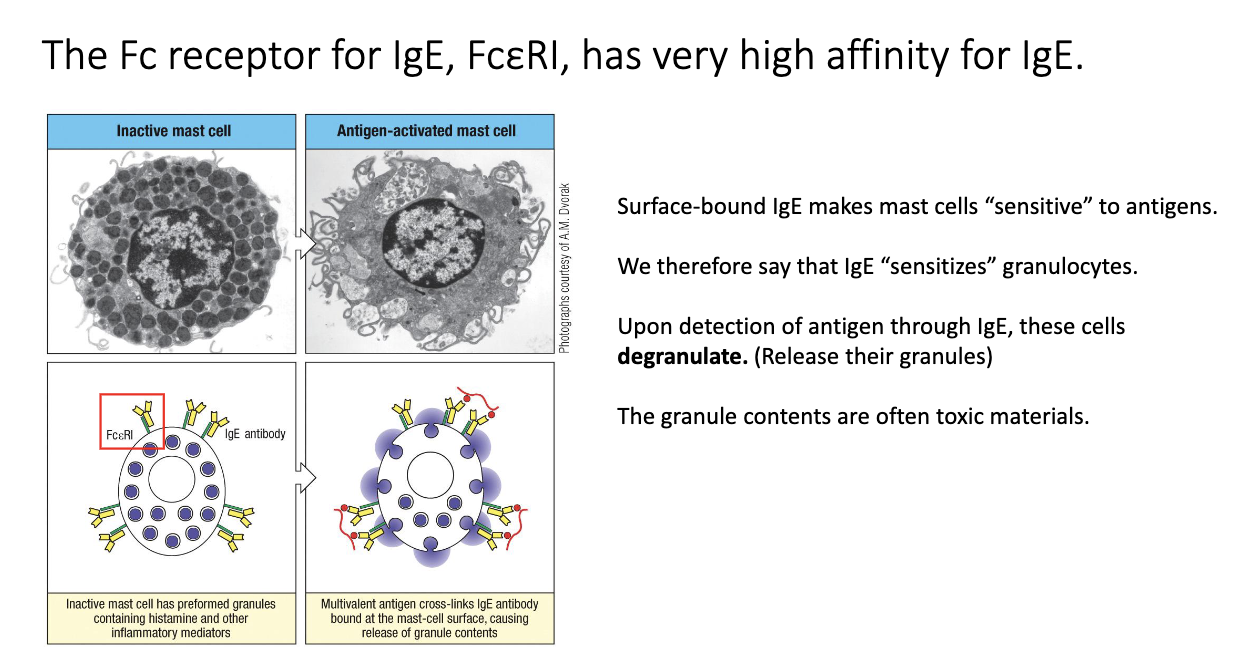
How does IgE sensitize mast cells and trigger degranulation?
IgE binds FcεRI on mast cells with very high affinity → remains for ~100+ days, compared to in serum IgE lasts for 7 days.
Antigen crosslinks IgE–FcεRI complexes → rapid degranulation (<1 hour).
Releases histamine, proteases, cytokines → drives allergy and anaphylaxis.
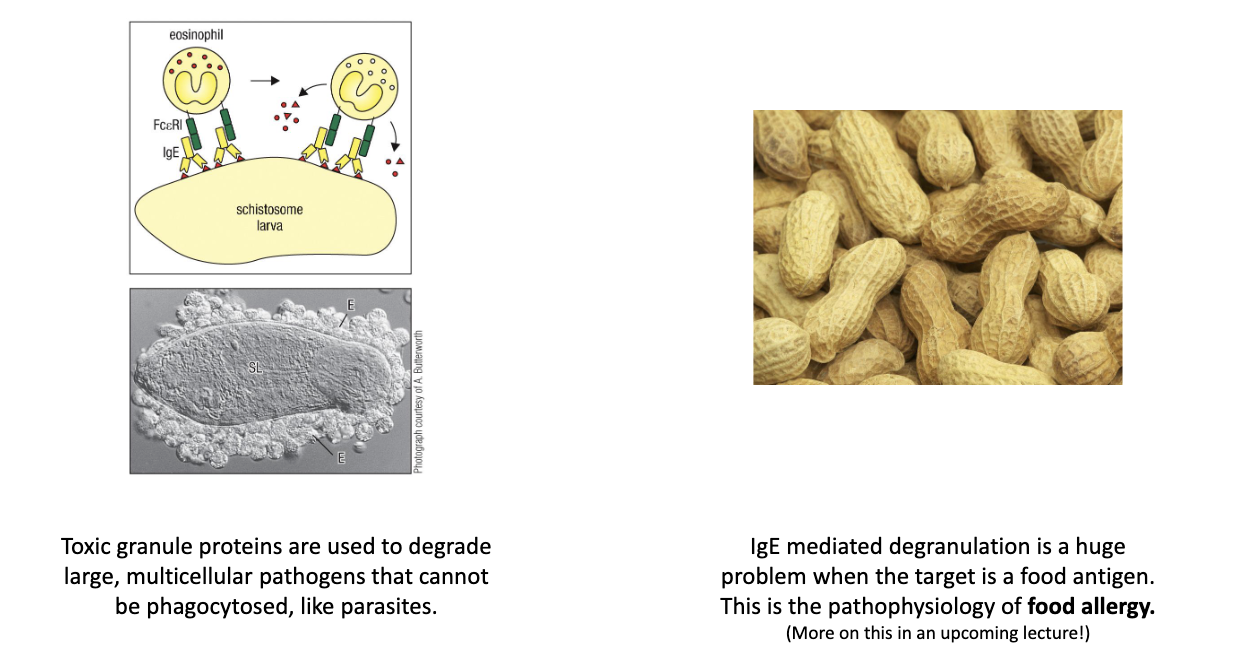
Same mechanism used to attack parasites via eosinophils/mast cells.
How does IgE cause allergic reactions and anaphylaxis?
Pre-bound IgE on mast cells crosslinks upon re-exposure to allergen.
Triggers massive degranulation → vasodilation, itching, shock.
Same biology evolved for anti-parasite defense.
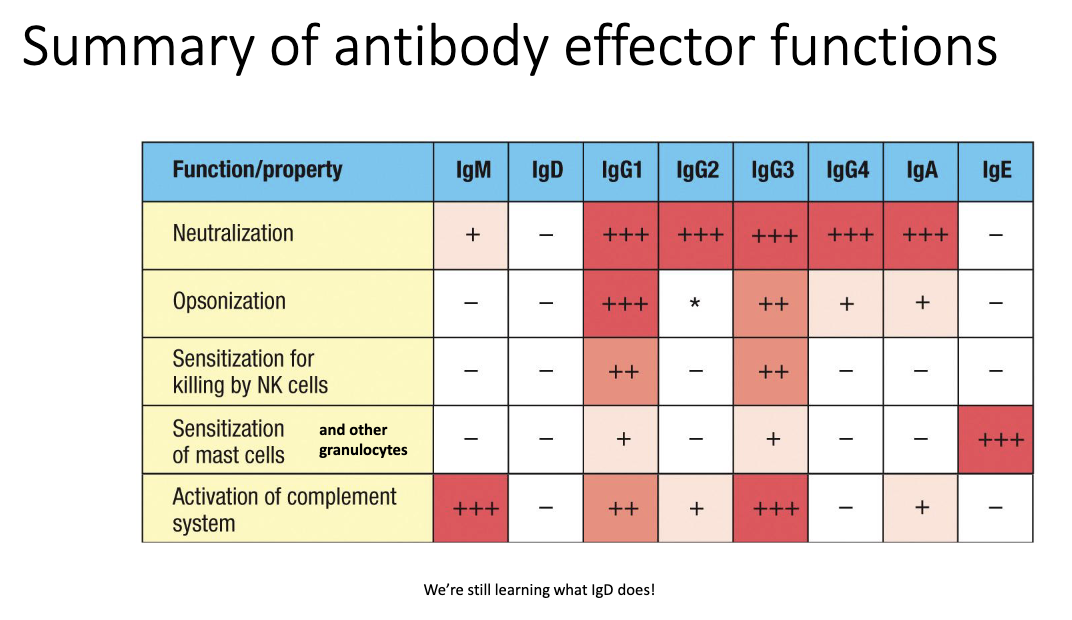
Which antibody isotype sensitizes mast cells and granulocytes?
IgE (via FcεRI).
IgD remains functionally unclear.
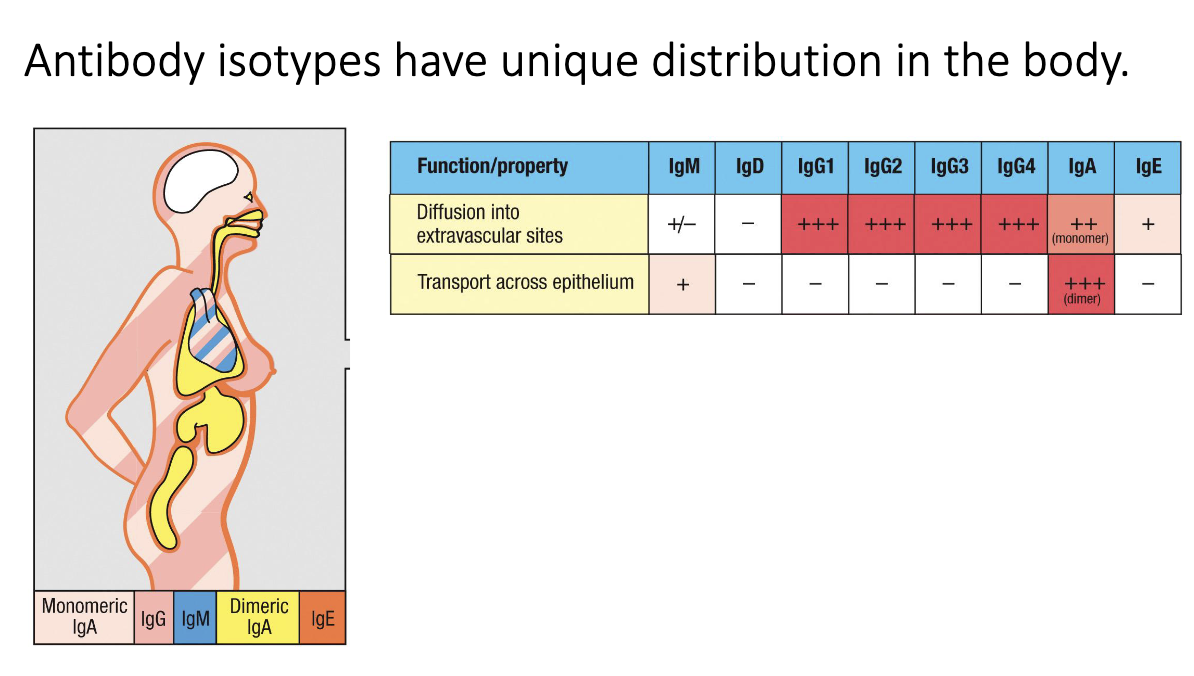
How are antibody isotypes distributed throughout the body?
Dimeric IgA → mucosal surfaces (gut, lung, urogenital tract).
IgE → bound to mast cells/skin.
IgG & monomeric IgA → systemic circulation.
Brain/CNS normally antibody-free → antibodies here indicate pathology.
Ex: MS results from antibodies in brain/CNS.
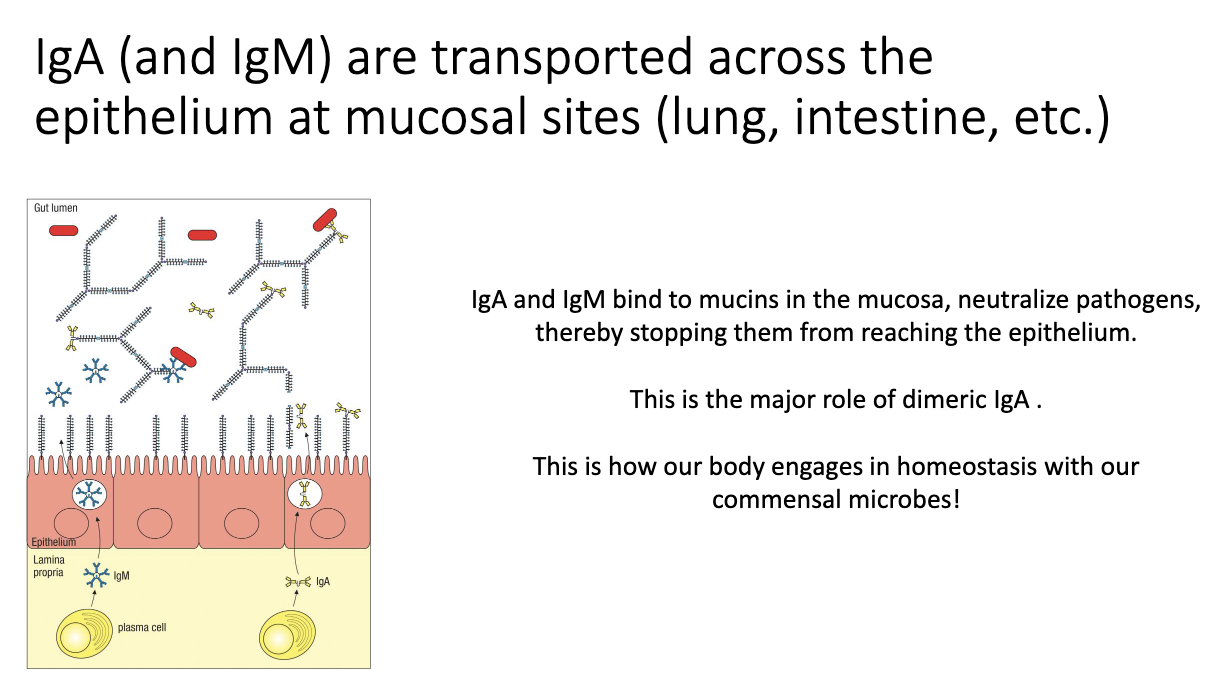
How are IgA and IgM transported into mucosal lumen?
Plasma cells secrete IgA/IgM.
Epithelial cells use the poly-Ig receptor to capture them.
Antibodies are transported and released into the lumen to bind microbes.
Maintains gut homeostasis by keeping bacteria out of host tissues.
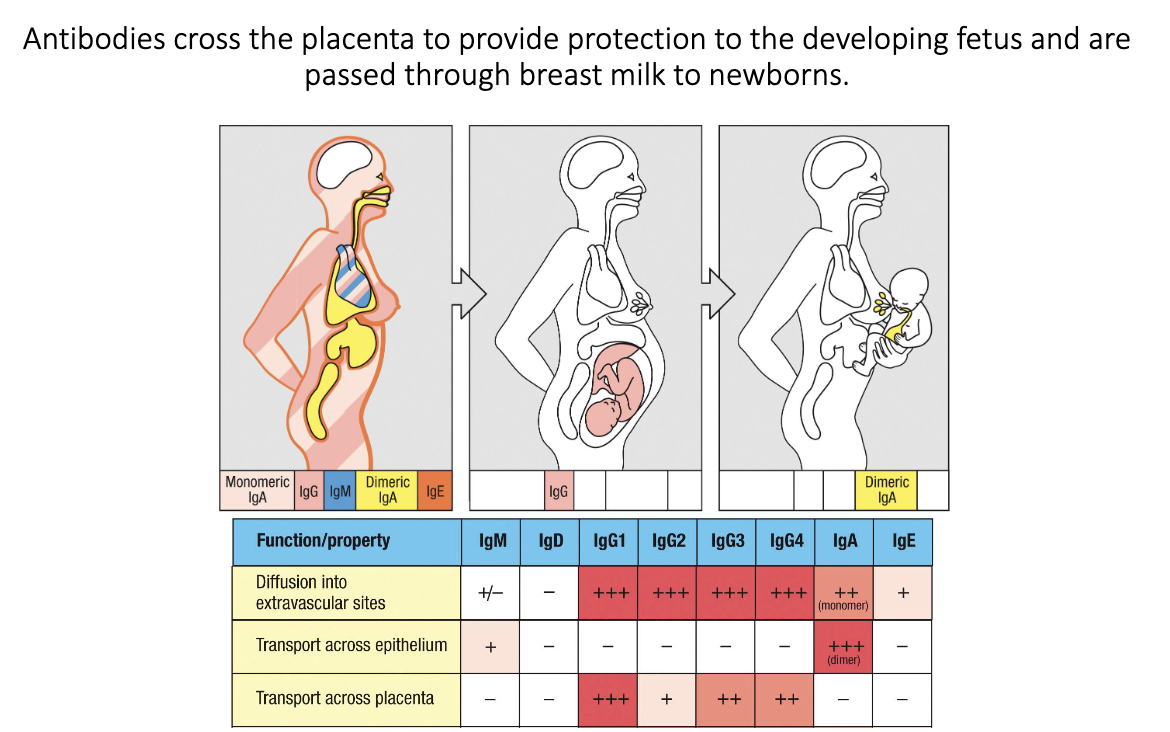
How do mothers pass antibodies to their babies?
IgG crosses the placenta → fetal protection in utero.
IgA from breast milk coats infant gut after birth.
“Passive transfer” of antibodies from parent to fetus/child allows for protection before fetus/infant antibody production matures.
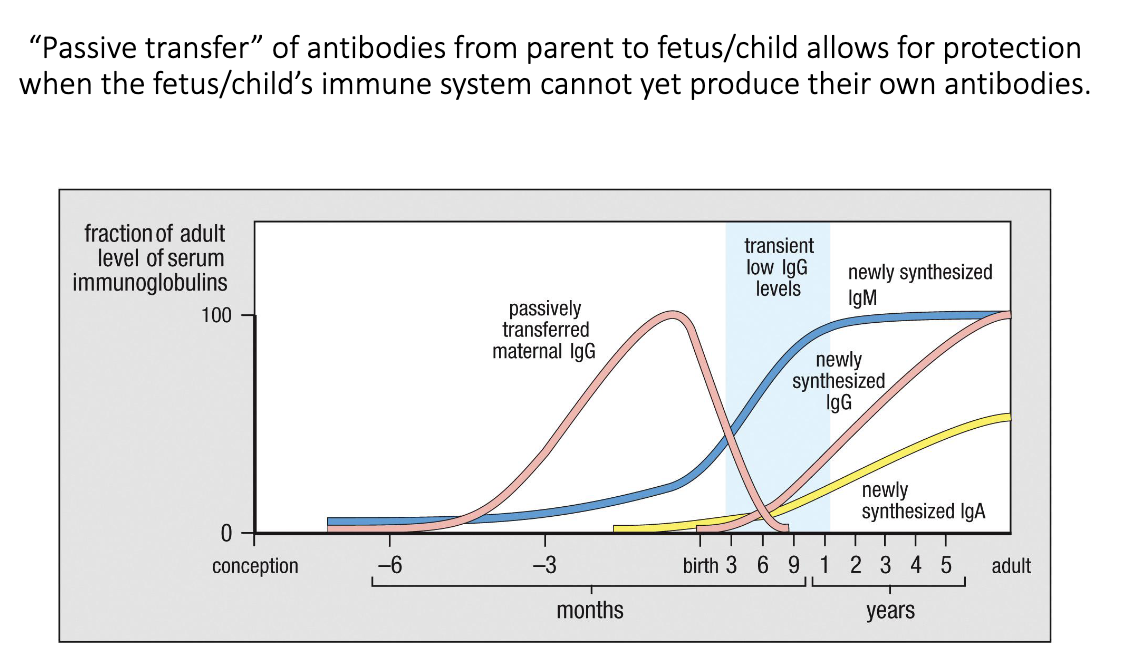
What is passive transfer of maternal antibodies, and how does it protect newborns?
During pregnancy, maternal IgG crosses the placenta to the fetus.
After birth, the newborn briefly has low antibody levels until its own IgG/IgA production ramps up.
During this window, maternal IgA from breast milk protects the infant’s gut.
Combined, these mechanisms provide early-life immunity before the baby’s immune system matures.
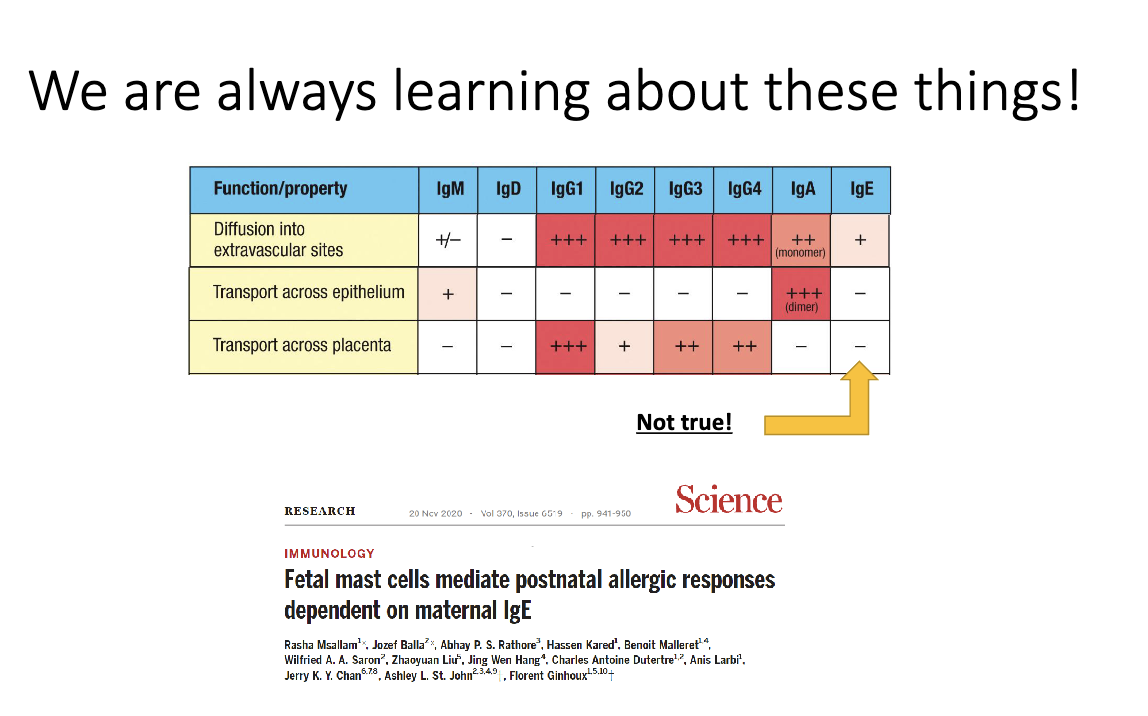
Can maternal IgE cross the placenta?
Historically thought no, but 2020 data show yes.
Fetal mast cells can acquire maternal IgE.
Provides a potential mechanism for inheritance of allergy risk.