Lecture #13.1 | Oncogenes as Transcription Factors: Fos/Jun
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
Nucleosomes
made of histone octamers (2x H2A, H2B, H3, H4) with Lys-rich tails at the N-terminal domain (NTD)
DNA wraps itself around and the tails protrude (n-terminal domains)
diameter of 10 nm
H1 histone is an open segment then there is a linker segment until another histone
What does it mean the tails stick out?
tails are modified by acetylation, methylation, and phosphorylation on Lys/Arg residues that changes the charge
Lys/Arg: Positively charged
Histone code hypothesis
different modifications determine if the chromatin is open (active) or closed (inactive).
Acetylation of lys
leads to open chromatin because it decreases the positive charge on the Lys in the histone tails.
Methylation
generally leads to repressed transcription, with the exception of H3K4me3, which is a mark for active transcription.
Promoters
contain specific DNA sequences (response elements, RE) to which TFs bind.
Promoters are immediately upstream of the transcription start site (+1, ~5 kb).
RE = TF binding sites (TFBS), typically 6-15 bp.
Enhancers
(~500-1000 bp) contain multiple TFBS.
They can be farther from +1 (up to 100 kb) and can be upstream (e.g., -10 kb) or downstream of +1 (e.g., +10 kb).
Pioneering TF
bind the histone-bound DNA, opening up the chromatin and making it easier for other TFs to bind.
first thing that binds
Co-activators
recruit histone acetyltransferases (HATs).
HAT activity leads to acetylation of histone tails, resulting in open chromatin.
Acetylation decreases the positive charge on the Lys in the histone tails.
Co-repressors
recruit histone deacetylases (HDACs), leading to deacetylation of histone tails.
Non-acetylated histone tails result in closed chromatin.
Mediator Complex
After TFs bind enhancers and recruit co-activators, the Mediator complex is recruited to coordinate with the General Transcription Factors (GTFs).
GTFs orient RNA polymerase in the correct direction for transcription.
DNA bending brings enhancers close to promoters.
RNA polymerase straddles +1, the start site of transcription.
GTFs (TFII proteins, including TATA-binding protein) bind the core promoter (-60 bp to +60 bp) containing the TATA box.
RNA Pol II transcribes protein-coding genes.
The Mediator complex coordinates with the GTFs.
Summary of transcription initiation
Activators bind DNA in a sequence-specific fashion in promoters/enhancers.
Activators recruit co-activators, which modify the nucleosomes (nucleosome remodeling complexes move nucleosomes out of the way, requiring ATP).
Activators and co-activators recruit the General Transcription Factors (GTFs) – Mediator, TFIID, RNA Pol II.
Transcription is initiated once the C-TD of RNA Pol II is phosphorylated by TFHII (one of the 9 subunits is a kinase).
Promoters often stay “marked” for transcription even after RNA Pol II leaves the promoter.
Primer promoter for reactivation
Oncogenes function as transcription factors (TFs).
They possess a modular structure with a DNA-binding domain (DBD), a dimerization domain, and an activation domain.
They bind specific DNA response elements (RE) in the regulatory regions of genes, typically as dimers.
REs often consist of half-sites.
TFs are often members of families of related proteins that form homodimers and heterodimers, increasing the complexity of gene regulation.
Binding of dimers increases the stability of the TF bound to DNA.
The activation domain interacts with co-activators, the Mediator complex, and GTFs (including RNA Pol II), and sometimes co-repressors.
What is meant by the modular nature of TFs?
Well defined activation domain and DNA binding domain
forms a homodimer with a combination of 2 factors
complexity is created by dimerization of half sites
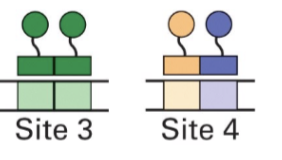
small changes in the motif or spacing lead to different transcriptional outcomes

AGGTCA
core recognition motif/half-site for a class of transcription factors, especially nuclear receptors
How does this relate to the ST Pathway
TFs are phosphorylated in response to the signal transduction (ST) pathway (e.g., MAPK, JNK, PKC, etc.).
Phosphorylation may activate or inhibit the activity of a TF.
Phosphorylation may affect:
Nuclear localization
Protein dimerization
DNA binding
Co-activator/co-repressor recruitment
Protein stability
C-fos activation
MAPK phosphorylates TFs TCF and SRF, which bind the SRE in the c-Fos promoter
initiates transcription
What is c-fos?
proto-oncogene and a transcription factor.
Originally identified as a viral oncogene, v-Fos.
It is an immediate early response gene activated by growth factors in serum.
Does not need to wait to increase its activity, its an immediate response
c-Fos is phosphorylated in response to growth factors (immediate), and its RNA/protein levels are increased (early).
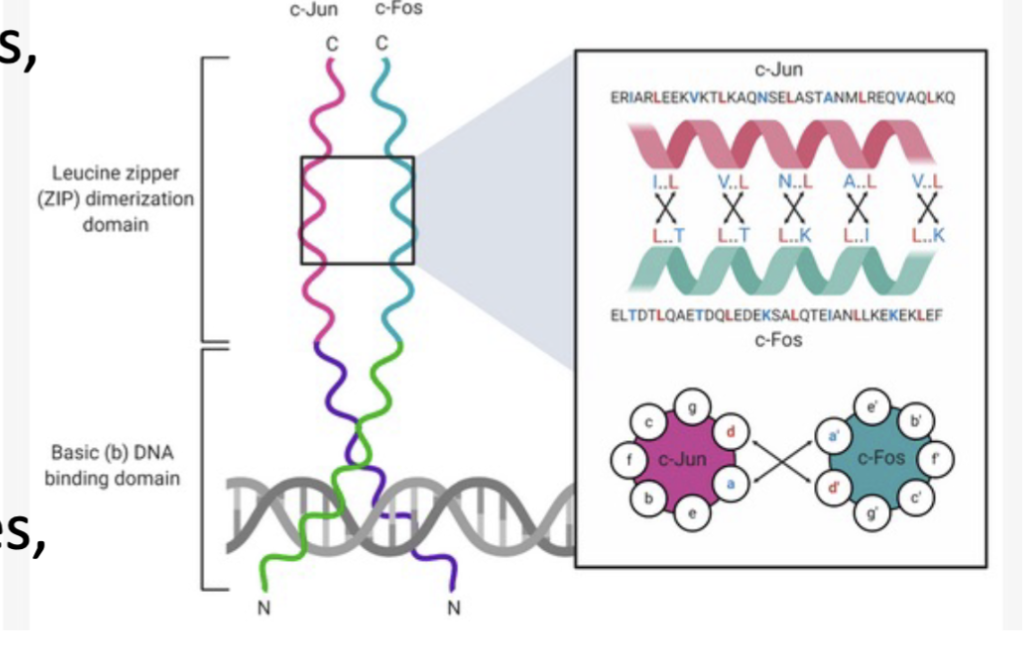
bZIP protein like c-Jun
c-Jun
another bZIP protein, is also directly activated by the ST pathway and is phosphorylated by MAPK and JNK protein kinase.
Ras→ Raf → Mek → Mapk → Elk1 and Jun
Ap1
Heterodimer between Fos + Jun that work as a TF
AP1 turns on genes involved in cell proliferation, such as Cyclin D1.
bZIP Proteins
Basic region (“b”):
Rich in positively charged amino acids (e.g., lysine, arginine).
Binds to specific DNA sequences, often with a consensus like TGACGTCA (the CRE motif).
has a Leucine Zipper region
A stretch of amino acids where leucine appears every 7 residues, forming a coiled-coil structure.
Allows dimerization (either homodimers or heterodimers).
Acts like a molecular Velcro, zipping two proteins together.
JNK
stress-activated pathway is named after c-Jun.
c-Jun stabilization
by phosphorylation, increasing its concentration in the nucleus.
Phosphorylation also increases its transcriptional activity.
c-Fos concentration
increased by the action of TCF/SRF on its promoter.
High levels of both Jun and Fos allow for the formation of the Jun-Fos heterodimer, AP-1.
The Fos/Jun heterodimer is the most stable and has the highest affinity for DNA, higher than Fos or Jun homodimers.
Fos/Jun heterodimer
most stable and has the highest affinity for DNA, higher than Fos or Jun homodimers.
Ras/MAPK - Jun/Fos Connection
Ras activates MAPK and JNK in response to GFs and other signals.
Both phosphorylate c-Jun; MAPK phosphorylates c-Fos.
Phosphorylation of c-Jun stabilizes it, increasing the concentration of c-Jun.
MAPK phosphorylates Elk-1, which turns on the expression of the c-Fos gene.
c-Jun heterodimerizes with c-Fos, forming the AP1 complex.
AP1 turns on genes involved in cell proliferation, such as Cyclin D1.
Fibroblasts lacking c-Jun cannot be transformed by Ras, indicating that c-Jun is required for the transformation of cells by Ras!
Grb2 and SOS
Right next to RTK
Growth factor receptor-bound protein 2 (adaptor protein) which is an adaptor protein (has no enzymatic activity).
ensures specificity
SOS: Son of Sevenless (named from Drosophila genetics) which is a GEF (Guanine nucleotide Exchange Factor) for Ras.
Cyclin D1
increases after exposure of serum-starved cells to a mitogen.
cells grown in a medium without serum cannot get mitogenic cells
so no cyclin D1
if a mitogen is added → initiates proliferation