the human genome project
1/9
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
10 Terms
sanger sequencing
DNA clones to be sequenced are generated by standard PCR reaction
The clones are then subject to a polymerase-mediated synthesis step
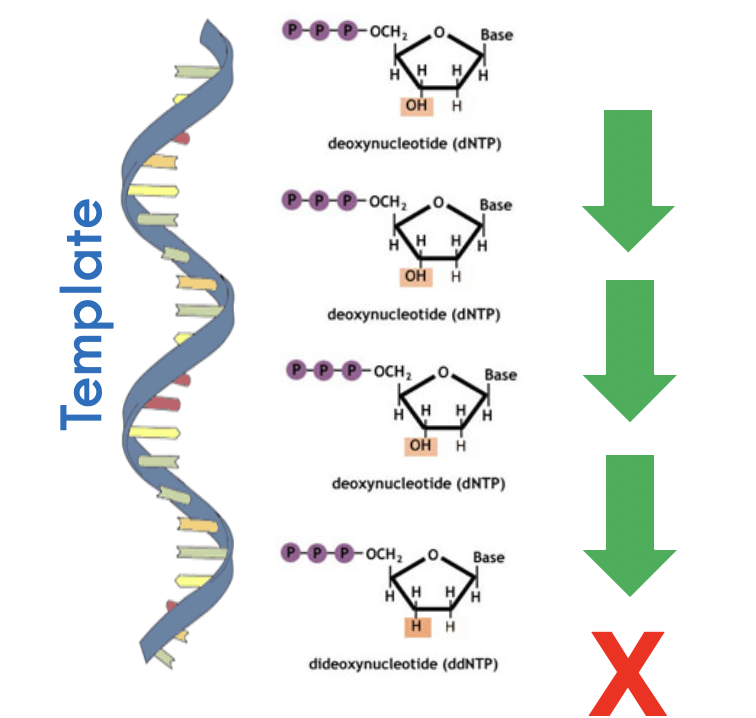
Critical is the random termination of extension at each nucleotide position
reaction mixture contains DNA template, primer, DNA polymerase, ddNTPs and dNTPs
The random termination results in DNA fragments of varying sizes which can be analysed to determine nucleotide sequence
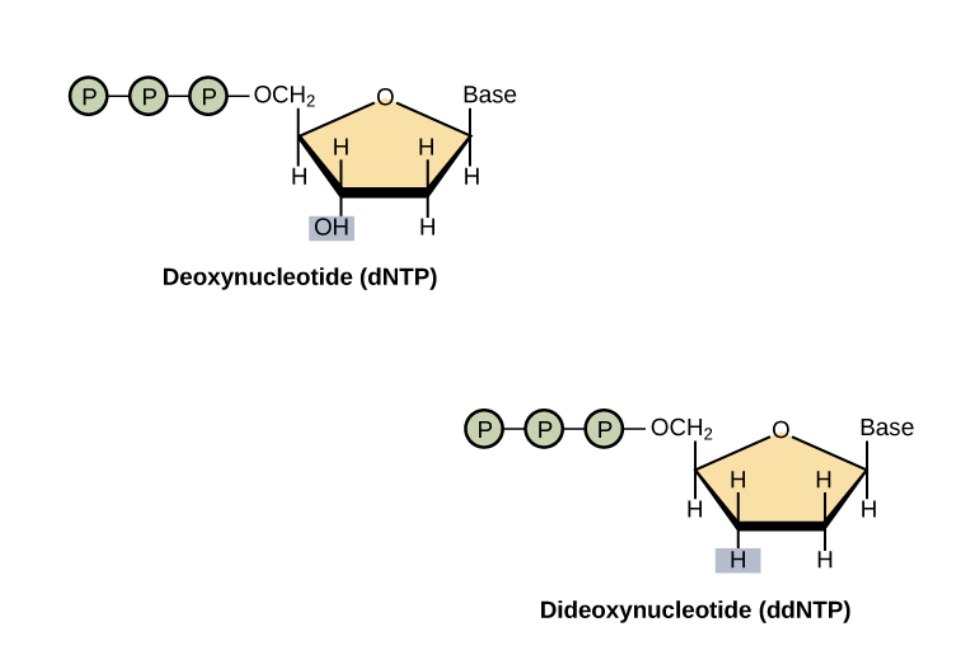
ddNTP
ddNTP have a hydrogen replaced from hydroxyl in dNTP
The hydroxyl is important for addition of next nucleotide
Thus next nucleotide is not able to be added
dNTPs are added in excess over ddNTPs in reaction mixture
Termination Is random
But will have termination at every single position
Provided there are enough clones
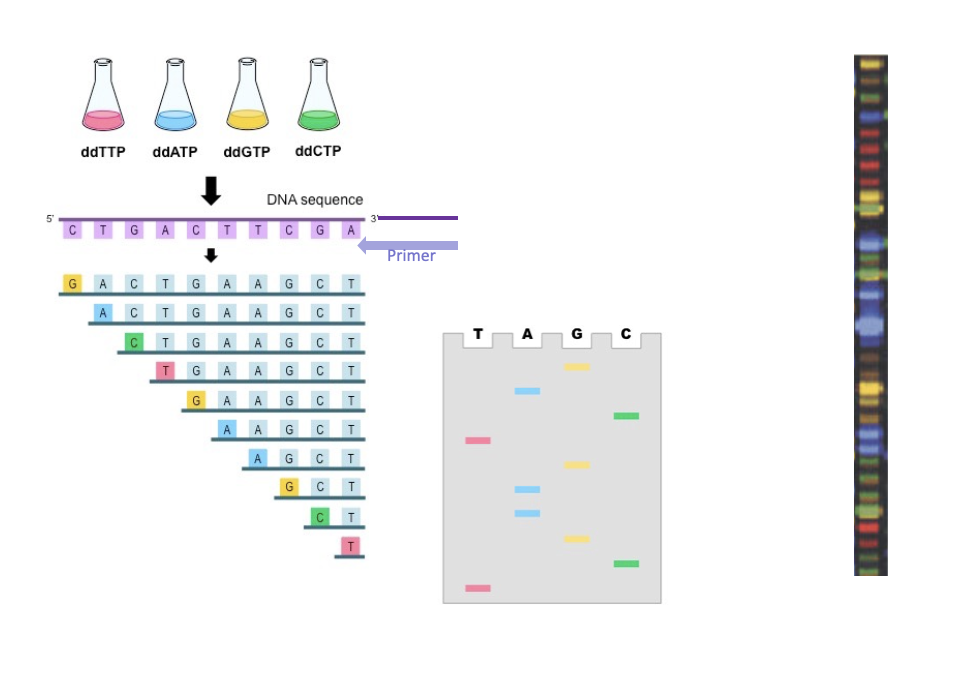
ddNTPs are labelled but dNTPs are not
with fluorophores
reading DNA
Need a primer
Run on a polyacrylamide gel
Shortest DNA fragments run faster
Will be at the bottom
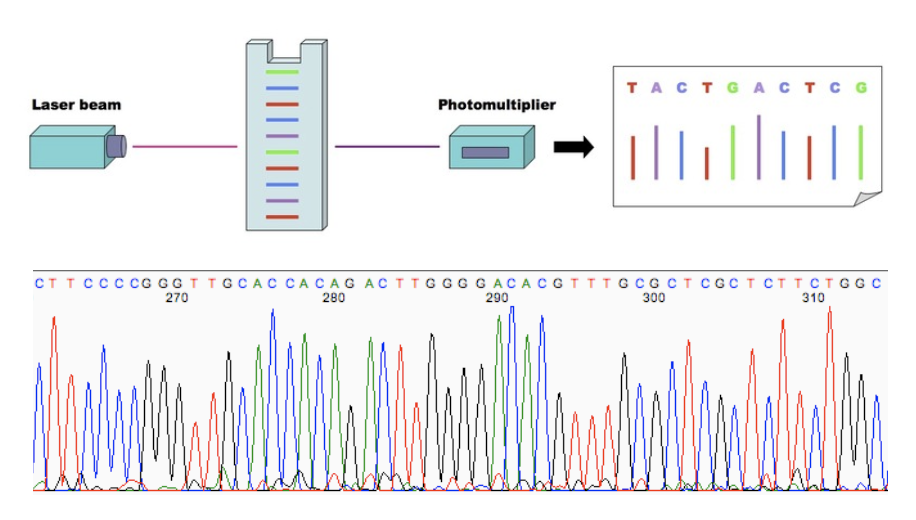
fluorescence detection systems
Fluorescence detection systems are used to image the fluorescence producing an electropherogram
Peaks is due to number of DNA strands with that sequence
Capillary electrophoresis allows the standard gel electrophoresis step to be bypassed
Further technical upgrades meant that 384 samples could be run in parallel
sequencing the human genome for the first time
Sanger sequencing yields only up to a maximum of 1000 nucleotides per reaction
Challenge for large scale projects
The sequencing strategy was shaped by 3 key limitations of sanger sequencing
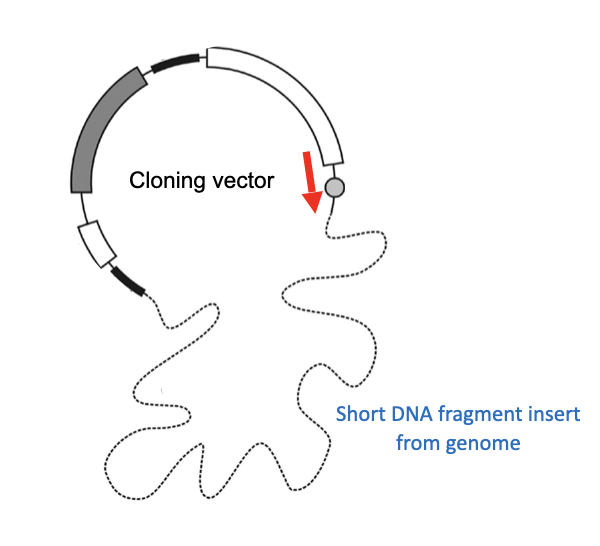
The necessity to have a clone of the DNA template (so that the levels of fluorescence emitted is able to be detected)
The requirement that at least some sequence information is known beforehand (so that primers can bind to the template)
The short sequencing read length
Due to using a vector, the primer can be designed to be complimentary to the vector
process
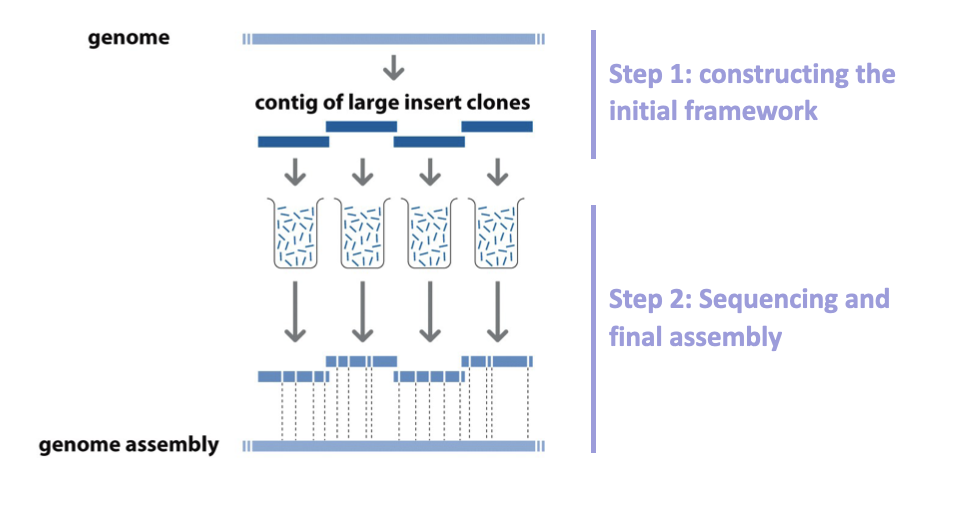
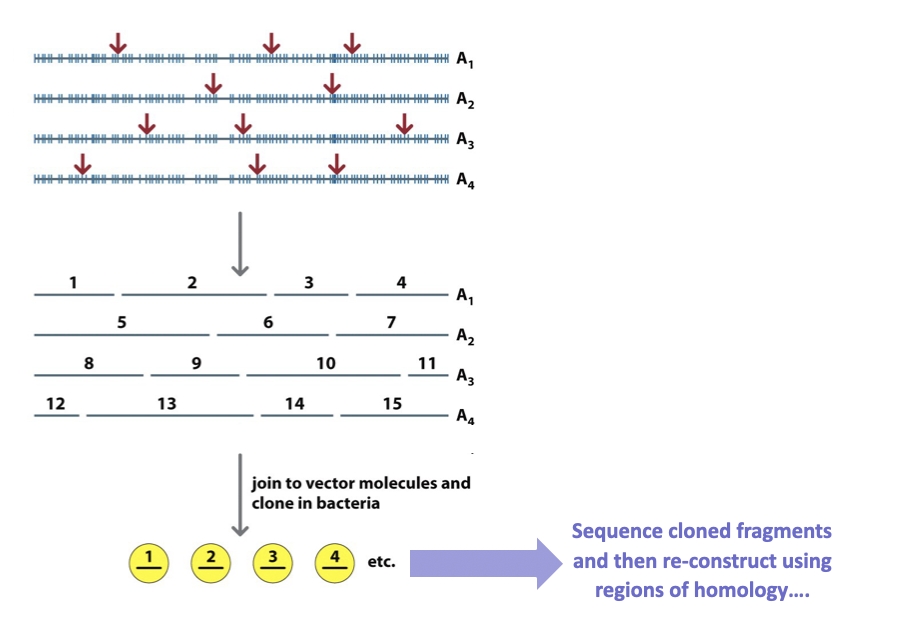
Fragment a chromosome
Order them
Fragment those fragments
Sequence
Reorder fragments
problem = fragmentation happens randomly
ordering larger fragments
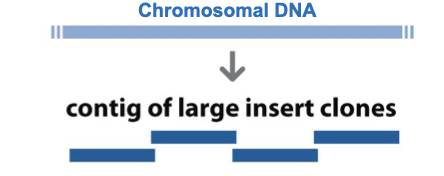
Chromosomal DNA was initially fragmented into large pieces and cloned into vectors known as YACs (yeast artificial chromosomes)
The clones were then mapped in terms of their original chromosomal location by 2 major techniques
FISH-type experiments
Label specific sequences
Can be detected on both chromosome and fragments
Can thus be reordered
PCR-based screening for STSs
Bits have already been sequenced
Called sequence tagged sites
Use sequence to design a complimentary primer
One that is positive thus has the same sequence and comes from the same location
A series of overlapping clones whose chromosomal location had been mapped was thus generated and these were referred to as clone contigs
No gaps
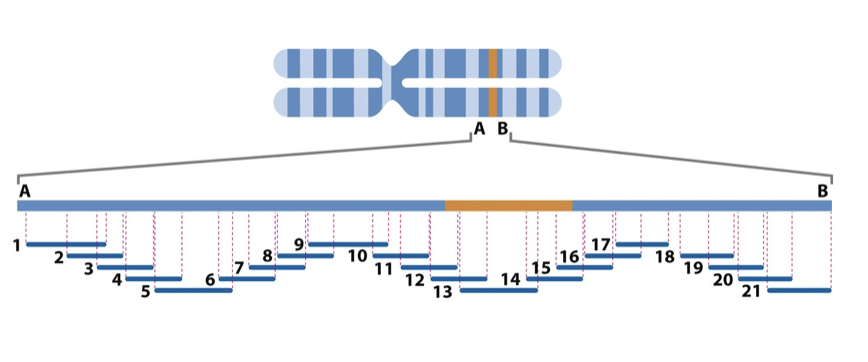
reordering smaller fragments
Do reaction so that fragmentation only happens at certain sites
Add restriction endonuclease at smaller concentrations
Random fragments
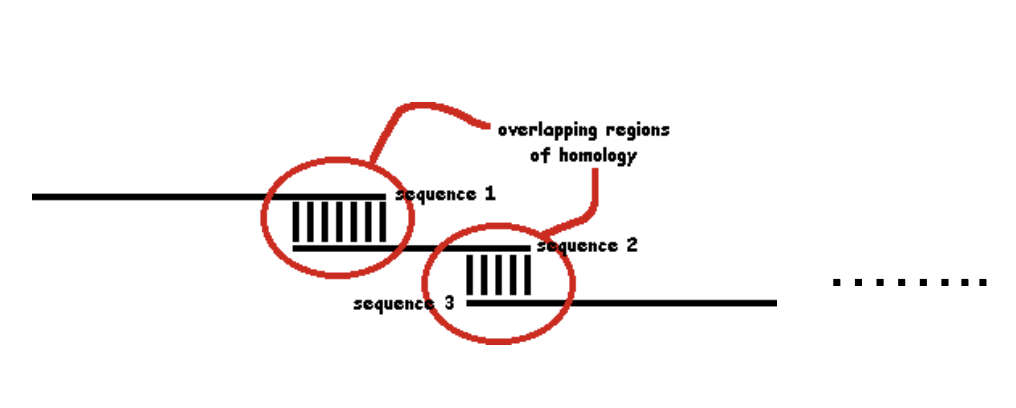
Overlap
And match overlapping ends (sequences of homology)
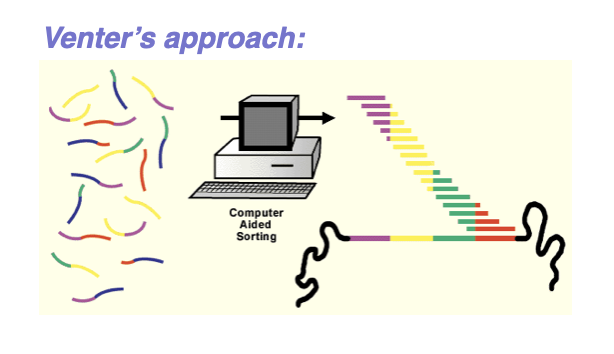
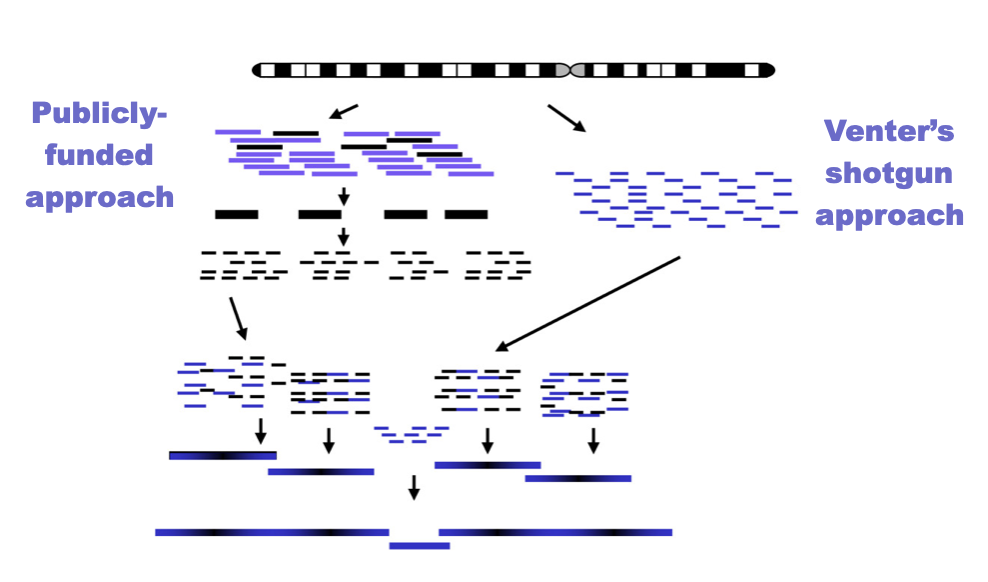
whole genome shotgun sequencing
Shotgun sequencing is used to determine the DNA sequence of an entire genome.
The genome is broken into many small, random fragments.
These fragments are cloned into vectors or prepared as sequencing libraries.
Each fragment is sequenced individually.
Sequencing produces many overlapping DNA reads.
Computers compare overlaps between reads.
Overlapping sequences are assembled into longer contigs.
Contigs are ordered and oriented to reconstruct the genome.
The randomness increases the chance that every region is sequenced.
Shotgun sequencing is faster and more efficient than sequencing DNA in order.
capillary sequencing / automated Sanger sequencing
Capillary sequencing is used to determine the nucleotide sequence of DNA fragments.
DNA is first amplified (often by PCR).
The sequencing reaction uses dideoxynucleotides (ddNTPs) to terminate DNA synthesis.
Each ddNTP is fluorescently labeled with a different color.
DNA fragments of varying lengths are produced.
The fragments are loaded into a thin capillary tube filled with polymer.
An electric field separates fragments by size during electrophoresis.
Shorter fragments move faster through the capillary.
A laser detects fluorescence as fragments pass a detector.
A computer reads the colors to determine the DNA sequence.