Module 2 (Labs 7-12)
1/144
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
145 Terms
What is the bacterial host used in this lab?
Streptococcus thermophilus
Circular and in chains
Gram+
Involved in acidification in milk and fermentation of dairy products (yogurts and cheese) → Probiotic
DGCC7710 strain
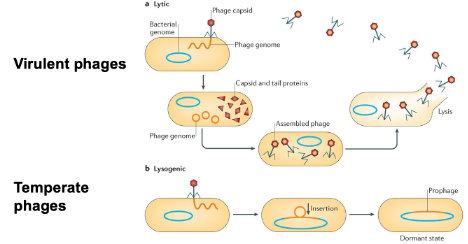
What are bacteriophages? Describe the life cycles.
Most abundant microorganisms on the planet
They are viruses that infect bacteria
Two life cycles
Lytic: phage infects and kills the infected host cells (virulent phages)
Lysogenic: phage genome integrates into the host DNA (prophage), stable for thousands of years so the phage can alter the phenotype of the bacterium (temperate phages → can go to lytic path)
Describe bacteriophages 2972 and 858.
Virulent phages isolated from yogurt samples
Infects S. thermophilus
Phages 2972 and 858 are 90% identical at the nucleotide level
Unclear if one phage produces more BIMs than the other
A BIM carrying a spacer from 2972 can be resistant to 858
Phage genome is ~35 kb
Siphoviridae family: dsDNA, nonenveloped head and noncontractile tail (lacks protein sheath around tail but is more flexible)
Contractile can physically release genome into bacteria, noncontractile needs enzymes
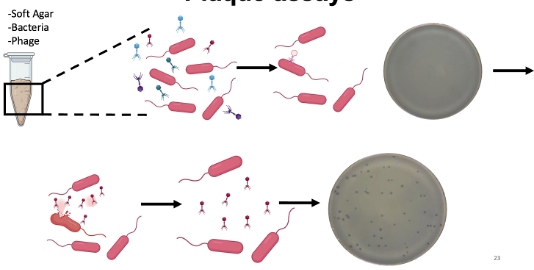
Describe the role of soft agar in plaque assays.
Soft agar has a lower percentage of agar and melts at 45deg but solid at RT → quickly add phage (make sure that phage and bacteria are in pipette)
Pour soft agar on plate and lytic cycle with take place → lyses → newly formed phages (plaques where bacteria lysed and released phages)
Creates a circle because the soft agar solidifies and the phages cannot go anywhere to infect bacteria
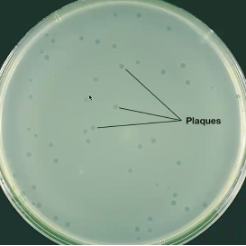
What is plaque forming units (PFU)?
Bacteriophages form plaques, which are clearings on a lawn of bacteria on the surface of agar
Each plaque corresponds to a single virus; can be expressed as a PFU
Plaque assays are used for:
Viral titers
Host susceptibility to phage infection
Phage virulence
Isolation of different phages
Generations of BIMs
How is viral titer using plaque assays calculated?
Phage titer is a PFU/mL which represents the number of infectious viral particles within a sample
This unit is used assuming that each plaque represents one infectious phage
Only plates with 30-300 plaques are counted and the following formula is used:
(# of plaques (PFU))/(volume plated in mL) × 1/Dilution (DF)
Rule for 30 is more flexible, but NEVER calculate PFU/mL for more than 300 plaques because of overlapping colonies or might lose track
I plated 100 ul of a phage solution that underwent a 1:10000 dilution. I then counted 100 plaques on my plate. What is the PFU/mL of this phage solution?
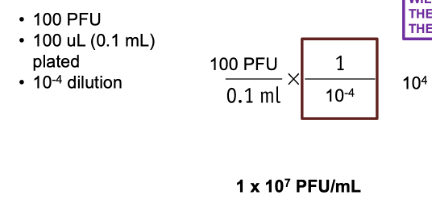
I plated 50 ul of phage solution that underwent a 10-5 dilution. I then counted 200 plaques on my plate. What is the PFU/mL of this phage solution?

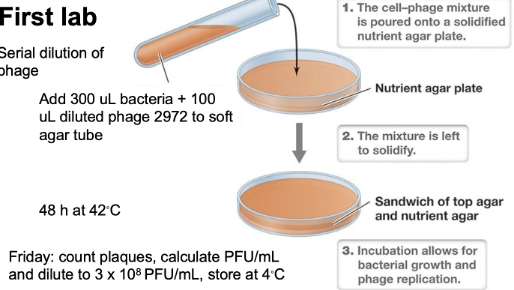
What are the steps for the first lab in this module? What are the plaque assay samples?
S. thermophilus uninfected (-) control
Bacteria growth without phage
Phage 2972 10-4
Phage 2972 10-5
Phage 2972 10-6 in triplicate
Phage 2972 10-7 in triplicate
9 in total
Triplicates to allow for more data points of the number of plaques
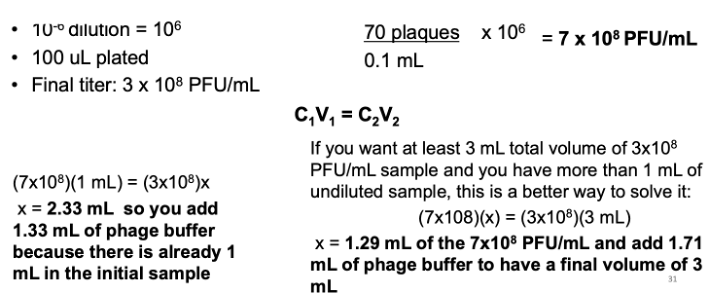
I counted 70 plaques on my 10-6 dilution plate. I plated 100 uL of my phage dilution. What is my PFU/mL and how much volume do I need to add to my 1 mL undiluted phage lysate to have a final concentration of 3 × 108 PFU/mL?
Which media is used?
LM17
Lactose
M17: Growth medium isolate from milk products
Highly nutrient and prone to contamination
Agar: different percentages in soft agar and petri dish
10 mM CaCl2 → needed for bacteriophage to attach to host
What are BIMs?
Bacteriophage Insensitive Mutants
Bacteria that have mutated to become resistant to one or more types of bacteriophages
What is plaque assay efficiency (plating efficiency)?
The number of plaque forming units is always lower than the actual counts of viral particles made microscopically
In other words, the viral titer has more viral particles than what is calculated since we assume 1 plaque = 1 bacteriophage, but two phages can infect 1 cell that leads to 1 plaque
Could have assembled incompletely during maturation, contain defective genomes, suffered spontaneous mutation that prevents from attaching or replicating
Phage could have infected a bacteria but did not complete the life cycle and no lysis
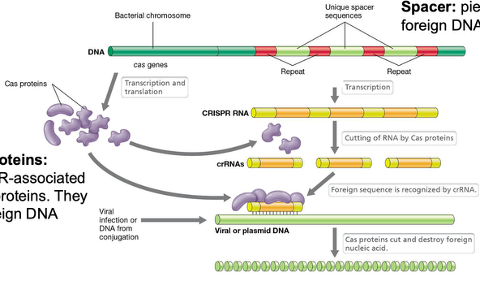
What does CRISPR stand for? Give a brief overview of what CRISPR is.
Clustered Regularly Interspaced Short Palindromic Repeats
CRISPR is a series of short 25 – 50 nucleotide repeats interspaced with variable yet similarly sized sequences known as spacers. These spacers are viral DNA or other foreign DNA that are inserted into the CRISPR locus. Protospacers are sequences in the plasmid or viral genome that will essentially be spacers in the CRISPR array. CRISPR is associated with a set of genes, known as Cas, and as discussed above, this system acts as an adaptive immune system for several bacteria and archaea
Why are BIMs resistant to phage infection?
Because of CRISPR
It is a major antiviral defence system in some bacteria and archaea
Adaptive immunity (memory) - specific to virus it recognizes
Seeks and destroys foreign/invading DNA
What are some possible outcomes that bacteria use to prevent viral attacks?
Some bacteria can alter the receptor that the bacteriophage uses to attach, so the phage can no longer attach to the host
Bacteria also use restriction enzymes to cut foreign nucleotides or DNA
Can also methylate the viral DNA or RNA so they can no longer replicate and transcribe
Bacteria can program cell death
There are different ways bacteria can prevent phage infection, but we will focus on CRISPR-Cas system, which specifically recognizes foreign DNA and degrades it
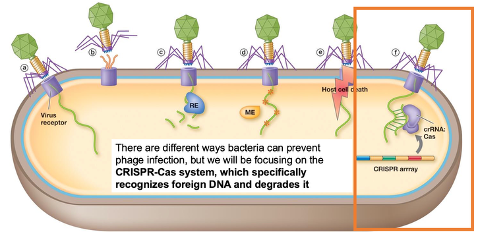
What components is the CRISPR-Cas system made up of? Explain how this system works.
Two sections: CRISPR array and Cas proteins
Interspaced repeats: short (30 nt), the same sequence but interspaced between unique spacers
Spacer: pieces of foreign DNA (the memory banks of foreign DNA that was injected)
CRISPR array is transcribed into CRISPR RNA
The Cas genes are transcribed and translated into Cas proteins
Cas proteins: CRISPR associated (Cas) proteins that cut foreign DNA
The long piece of CRISPR RNA is chopped up by Cas proteins so each piece has one of the unique spacers
Each piece forms a segment with the Cas protein and if there is any viral or invading DNA, the Cas protein with the spacers scan through the foreign DNA
If the viral or plasmid DNA is complementary to the spacer, the Cas protein will chop it up
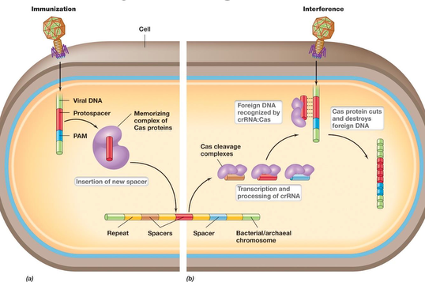
Explain the CRISPR defence system against viruses. What is immunization and interference?
PAM: protospacer adjacent motifs (5-6 nt) determines non-self, essential for cutting
If a Cas protein recognizes a PAM sequence, it knows it’s non-self
Protospacer: viral DNA near the PAM sequence
The Cas protein will take the protospacer, puts it into the CRISPR array and that is when it becomes a spacer (immunization)
If it gets infected again, it will recognize it and scan it (interference)
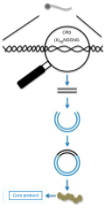
What are the steps of the CRISPR/Cas9 system?
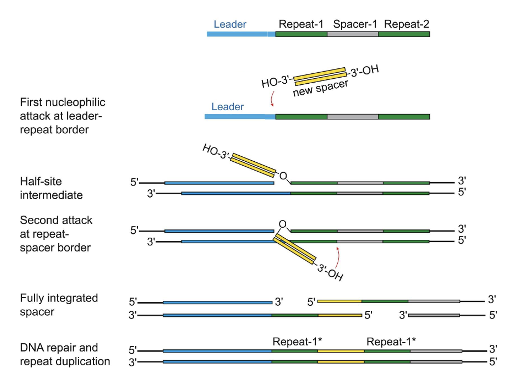
Cas proteins (1 and 2) recognize and integrate a piece of foreign DNA as a spacer into the CRISPR array at the 3’ end of the leader sequence
CRISPR array is transcribed into long pre-CRISPR RNA (pre-cRNA)
Trans-activating CRISPR RNAs (tracrRNA), crRNA and Cas9 form a complex that is cut → an antiviral complex (uses RNase III)
Scans dsDNA for the presence of PAM and complementary binding to crRNA
Cas9 cuts (silences) the invading DNA

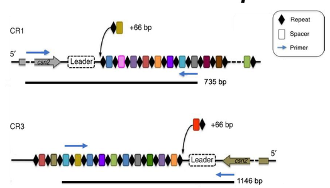
What are the CRISPR loci in S. thermophilus?
PAM sequence for CR1: NNAGAAW (N=nucleotide; W=A or T)
PAM sequence for CR3: NGGNG
S. thermophilus has mainly two active CRISPR-Cas sytems: loci 1 and 3 (cr1/3)
*A repeat sequence is always added with a spacer (60-66 nt added)

What is the difference between DGCC7710 vs DGCC7710 + pNZCR3?
DGCC7710 strain of S. thermophilus is wild type: found in its natural, non-mutated form (unchaged)
DGCC7710 + pNZCR3 contains a plasmid that increases BIM generation >400 fold
Plasmid contains a chloramphenicol resistance gene
How is CRISPR able to work in DGCC7710 + pNZCR3?
For CRISPR to work, spacer must already be present
Spacers can be acquired by:
Inactivated viruses (by UV to weaken lytic aspect of phages, so viral DNA is injected but bacteria can get immunized and not lysed)
Plasmids
pNZCR3 contains the protospacer and PAM sequence of phage 2972 that can be acquired into cr3 (plasmid is foreign but not dangerous, so CRISPR can still scan)
Cas proteins are responsible for:
a. Transcribing the CRISPR array into pre-crRNA
b. Finding PAM sequence and cutting the protospacer from foreign DNA
c. Programming cell death
d. All of the above
B
(a is polymerase, c is apoptosis mechanisms)
What is the overview for lab 8?
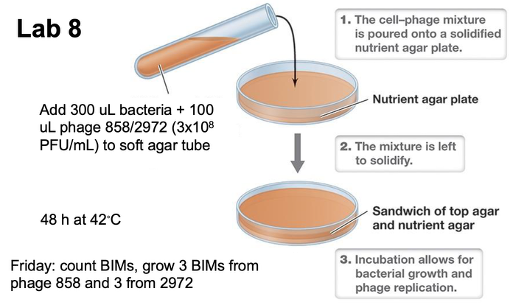
Which plaque assays will be done in lab 8? What are the expected results?
Negative control (no phage) S. thermophilus DGCC7710
Phage 2972 + S. thermophilus in triplicate
Phage 858 + S. thermophilus in triplicate
Keep in a sterile environment
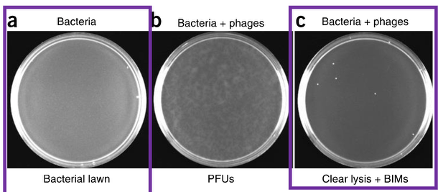
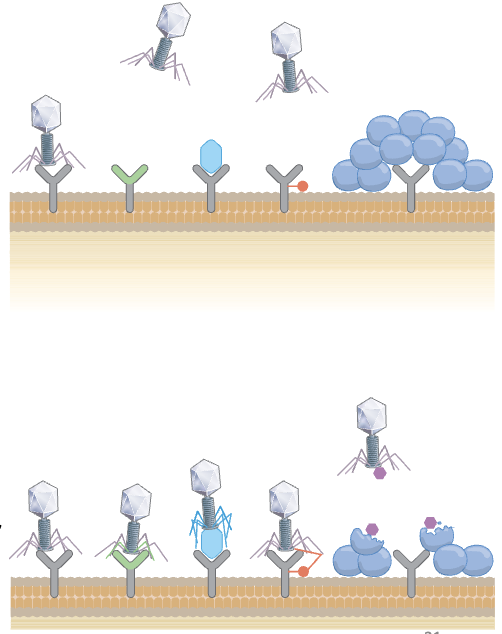
a is negative control (no phage); cloudy portions in b is bacteria so they did not get infected (that’s why high concentration and high PFU/mL to get clear lysing, and only the colonies that show up are BIMs)
b is inconclusive – not enough phage to infect bacteria
When the colonies are present it means BIMs because phage could not infect the bacteria
What forms the antiviral complex in the CRISPR-Cas9 systems?
a. Pre-crRNA and Cas 9
b. crRNA, tracrRNA, and Cas 9
c. The leader sequence
d. The CRISPR array
e. The protospacer and spacer
B
What is the difference between protospacer and spacer?
Protospacer is viral DNA near the PAM sequence in the viral genome vs spacer is a piece of foreign DNA in CRISPR array
Where is a new spacer added in the CRISPR loci in S. thermophilus?
New spacer is added to the 5’ end of the CRISPR array or leader-adjacent.
What are the 3 possible sources of selective pressure on spacers?
Some protospacers are easier to acquire by Cas proteins
Could be that they are located in a place where it is easier for the Cas protein to cut and take up
Protospacers that are more conserved in the viral population (more abundant)
Structural proteins, which can be taken up and usually more effective against a broad range of phages
Some spacers might be more effective than others at clearing viral infections (what the protospacer encodes for)
More effective at interference of phage infection, can encode for structural proteins or integrase or other important viral proteins
(The way in which the CRISPR acquisition mechanism selects between tens or hundreds of protospacers in the viral genome is not fully understood)
Explain the steps for the integration of new spacers into the CRISPR array/locus.
Nucleophilic attack by 3’OH of protospacer at the leader end of the repeat
Nucleophilic attack by 3’OH of the spacer at the spacer end of the repeat
The spacer is acquired but there are ss repeat sequences where the spacer is sandwiched between the repeats → DNA repair and ligation
How can the effectiveness of CRISPR differ based on the number of spacers?
Cas protein complexes are more spread out with diverse target sites when there are multiple spacers → reduced immunity but diverse
With less spacers, the Cas complexes (antiviral complexes) are concentrated on targeting fewer sites → highly effective and efficient but less diverse immunity
Large acquisition of spacers leads to a broader spacer distribution, whereas smaller acquisition leads to a focused and more effective spacer
(In S. thermophilus, the most effective immunity was achieved when all Cas protein sequences were focused on a single spacer. Most CRISPR array evolved to have a length of 20-30 spacers (max is 100))
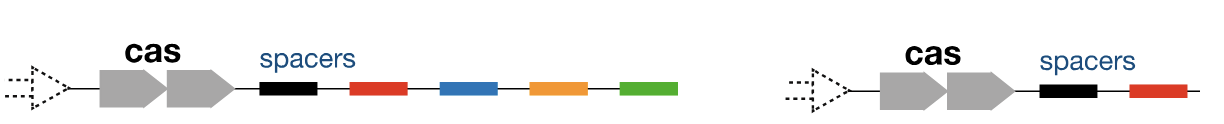
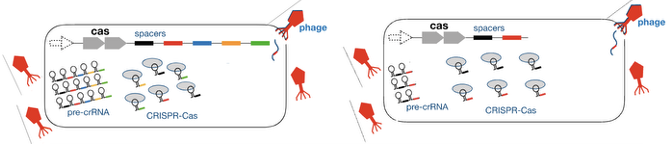
Which one would more effectively interfere with this viral infection, A or B? Why?
B. Faster to make crRNA and antiviral complexes and more antiviral complexes focused on the spacer
There is a trade-off between effectiveness of the defense and broader immunity
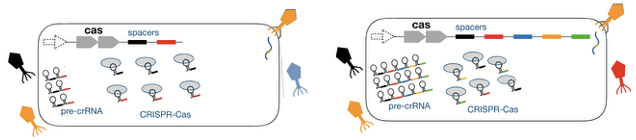
Which one would give you an effective interference of the phage infection, A or B? Why?
B. More diverse
There is a trade-off between effectiveness of the defense and broader immunity
Where does the second 3’-OH nucleophilic attack of the protospacer occur on the CRISPR array?
a. Between the leader sequence and the previous spacer
b. Between the leader sequence and the adjacent repeat sequence
c. Between the leader-adjacent repeat sequence and the previous spacer
d. Between two spacer sequences
C
Which statement is true?
a. Protospacers that are conserved in certain viral populations are not favoured in spacer acquisition
b. Spacers are pieces of foreign DNA that contain PAM sequences
c. More spacers in a CRISPR array provide more diverse immunity
d. The only difference between pre-crRNA and crRNA is that crRNA is bound by tracrRNA
C
What are the 8 PCR reactions that will be performed to characterize the BIMs?
1 BIM from 2972 infection
1 BIM from 858 infection
1 positive control (DGCC7710)
1 negative control (no bacteria)
*4 reactions with CRISPR1 primers, 4 reactions with CRISPR3 primers
*Add all PCR reagents + primers for CRISPR1/3 and add master mix to (-) control last
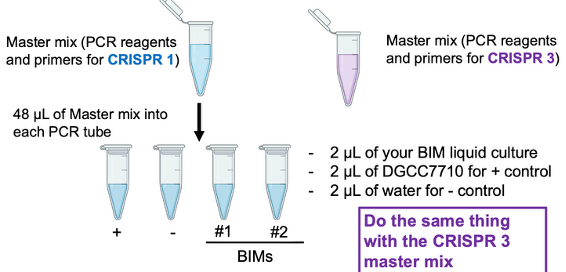
Name the PCR reagents.
dNTPs
Taq buffer
Taq pol
Forward primer
Reverse primer
MgCl2 (ensures pH is good for pol to work)
Why is 95 degrees C for 5 min needed before the PCR reaction?
To lyse the bacteria to release its genome
Why are different annealing temperatures needed?
Depends on the primers’ melting temp and the buffers used (GC content differs)
CR1 requires a higher annealing temp
Why do the BIMs need to be streaked?
Ensures that the BIMs are not contaminated
Should be small, circular, golden yellow
What are the expected PCR results? What are possible explanations if there is no spacer acquired?
The BIM might show a higher band for CR1 or Cr3 – new spacer acquired +66 bp
WT products should be ~730 bp for CR1 and ~1200-1500 bp for CR3
If your BIM acquires a spacer in CR1 it will not acquire a spacer in CR3 and vice versa – only acquired once in one of the loci
If you have BIMs that may have acquired spacers → send for sequencing
No height differences?
Maybe it acquired a spacer in one of the other 4 CRISPR-Cas systems in S. thermophilus
Other ways a bacteria can interfere with phage infection

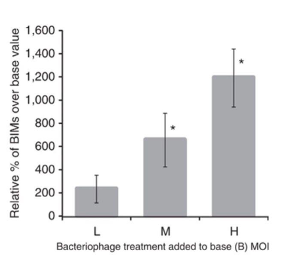
This graph shows how many BIMs were generated when challenging bacteria with 2972 after exposing the bacteria to different MOI (multiplicity of infection – ratio of phage to host) of defective phage. What do you observe on this graph?
Low, medium, and high MOI meaning the amount of defective phage added to a number of S. thermophilus.
When a lot of defective phages is added (phage is not lytic, it does not kill bacteria, but there are enough time/opportunities for the phage to infect and the bacteria can acquire the spacer) and the bacteria is washed up and WT phage is added, a lot of the bacteria has already acquired a spacer from the defective phage.
Low MOI means adding less phage per number of bacterial cells - not all bacterial cells are exposed to defective phage to acquire a spacer
Can a spacer from 2972 be effective against 858?
Yes
Spacers do not have to be 100% identical to protospacers to undergo phage interference
A spacer from 2972 can be effective against 858 (90% identity at the nucleotide level)
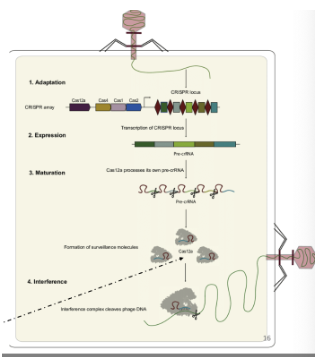
What is the 3-step CRISPR-Cas system?
Adaptation: Foreign DNA is added to the CRISPR array as a spacer
Cas proteins recognize and integrate a piece of viral RNA into the CRISPR array (at the 3’ end of leader, but the 5’ end of entire array/leader adjacent)
Maturation: CRISPR array is transcribed and cleaved into crRNAs
CRISPR array is transcribed producing pre-crRNA → tracrRNA, Cas9 proteins, and Rnase III bind to pre-crRNA → pre-crRNA processed into gRNA
Interference: Antiviral complexes cleave foreign DNA that is complementary to crRNA
Antiviral complex scans dsDNA for PAMs + a complementary sequence to gRNA
(for S. thermophilus, Cas9 is part of the antiviral complex)
In which locus are spacer acquisitions mostly found in? What could be a reason for this?
In DGCC7710, >90% of acquisitions are observed at the CR1 locus, and <10% are observed at the CR3 locus
CRISPR2 has been shown to incorporate new spacers but to a lesser degree
CRISPR1-Cas system is more active than CRISPR3-Cas system
Not due to the number of PAMs in phage genome (more CR3 PAMs in 2972)
CR1 found in most S. thermophilus strains (more active earlier on), CR3 is less frequent
What is primed adaptation/acquisition?
Efficient adaptation can occur at the locus with pre-existing spacers: primed adaptation/acquisition
The presence of pre-existing CRISPR spacers that partially match invader sequences dramatically increases spacer acquisition from the same invader (like when a friend goes to a party and you are more inclined to go)
Only shown to occur in Type I subtype of CRISPR-Cas systems
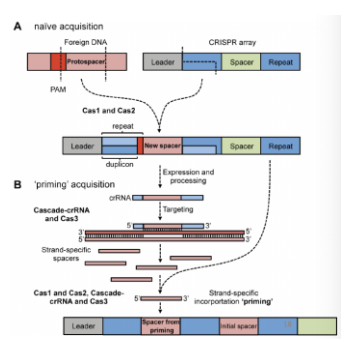
What is naive acquisition vs primed spacer acquisition? What is cascade?
Naive acquisition: no spacers in the CRISPR array have homology to the target genome
No spacers in CRISPR array that are homologous to newly acquired spacer
Primed spacer acquisition: presence of spacers with a full or partial match to the target DNA → increasing the frequency of acquiring another spacer
‘old’ spacers within CRISPR array are full/partial match to the target phage sequence DNA → resulting in the acquisition of another spacer that is similar to one that already exists in the array (type I subtype)
One part that is chopped up can then be acquired as a spacer if it still has its PAM → increases chance of getting another spacer that has homology to a pre-existing one
Cascade: CRISPR-associated complex for antiviral defense (antiviral complex)
How can bacteria and phages evolve regarding the receptors?
Bacteria: altering (starts at genetic level), disguising (sits on receptor), modifying, masking
Phages: mutations, production of extracellular matrix degrading enzymes, some phages have more than one receptor binding protein → enables recognition of multiple receptors
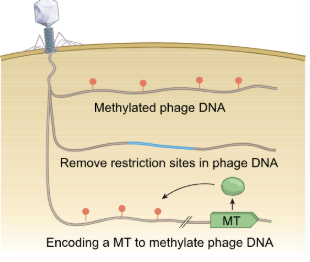
What are restriction-modification (RM) systems?
RM systems present in ~90% of bacterial genomes
Distinguish self from non-self DNA to recognize and destroy phage DNA after injection → DNA modifications at specific sequences
Involves methyltransferase (methylates DNA) and a restriction endonuclease (cleaves unmethylated sequence)
Phage DNA can
Become methylated by the host (at random)
Remove restriction sites (target region)
Encode its own methyltransferase and methylate its genome
How can phages overcome the CRISPR-Cas system?
Mutation of PAM or protospacer
DNA modifications or deletions = not bound by Cas proteins
Production of Cas protein inhibitors (anti-CRISPRs - Acr) -> most interact with the Cas proteins to block activity
Jumbo/giant phage or phage-like viruses: create this nucleus-like structure so the anti-viral complex cannot get to it and they carry machinery so they do not need to use the host’s
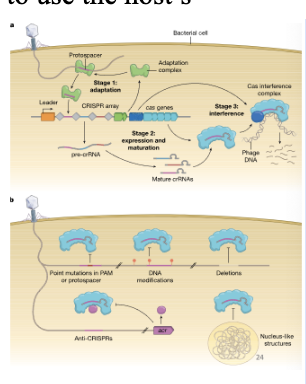
Why is primed spacer acquisition advantageous to bacteria?
Allows bacteria to defend themselves against rapidly evolving phage populations or against related phages with conserved but not identical targets
How can phages evade primed strains with multiple spacers?
By deleting the target region
Why is the ecological context important in defense systems?
The differences in defense systems can be due to reduced nutrients → importance of considering the ecological context
Which statement is true?
a. Primed spacer acquisition is when no spacers in the CRISPR array are homologous to the target genome
b. Naive acquisition is when there is presence of spacers with full or partial match to the target genome
c. 10% of spacers acquired in S. thermophilus are in CR1
d. Bacteria modifying their receptors is one way they can resist phage infection
D
How do phages avoid the CRISPR-Cas system? Name one method.
Mutation of PAM/modifying PAM
Deletion of PAM or protospacer
Cas protein inhibitors
Nucleus-like vesicles
What is a spot test?
Spot tests are simply done to determine if the phage can infect the host
Can be treated like a yes/no test: yes it is infectious = clearance; no it is not infectious = no clearance
A small amount of phage is placed onto a plate that is already inoculated with the host bacteria. If there is a plaque that forms where the spot of phage was placed, that suggests that 1) there is phage present in your sample and 2) the phage infects the bacteria on the plate
Describe lab 10 (Wed).
Serial dilute your phage (858 and 2972)
1 selected BIM from 2972 infection and 1 BIM from 858
(+) control, DGCC7710 with 858 and 2972
Spot tests
Plaque assays with your phage dilutions
BIM: 10-1, 10-2, 10-3
(+) control, DGCC7710: 10-4, 10-5, 10-6
More concentrated phage for BIMs and less for control because the BIMs are more resistant so you want to add more phage, whereas more phage for control will result in full lysis
(-) control, BIM with no phage
(-) control, DGCC7710 with no phage
15 total plates: 6 2972, 6 858, 3 (+) controls
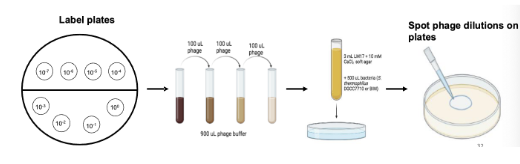
What is efficiency of plaques (EOP)? How is it calculated?
Used to determine both host range and relative phage killing - virulence of phage
Calculated by dividing the titer of the phage on the resistance strain by the titer of the same phage on the susceptible strain
Phage titer in BIM plates / Phage titer of WT plates
What are the ranges of virulence in EOP?
Highly virulent: 0.1 < EOP >1.00
Moderately virulent: 0.001 < EOP < 0.099
Avirulent: Less than 0.001
What is the spot test follow up in lab 10?
Count number of plaques
Calculate PFU/mL (viral titer). Difficult to do with spot test as not many plaques will be seen. In this case, it is okay to consider less than 30 plaques in the calculations
Calculate EOP of the phage on the BIM (resistant strain)
Compare results to the plaque assays
A positive spot test is not 1 plaque, it is many plaques that are fused together
What is the plaque assay follow up in lab 10?
Count number of plaques
Calculate PFU/mL (viral titer)
Calculate EOP of the phage on the BIM (resistant strain)
Calculated by dividing the titer of the phage on the test strain by the titer of the same phage on its isolation strain
Pick BIM infectious phage(s) from a plaque from your plaque assays to amplify (plaque on BIM plate means phage have evolved to kill it)
On your WT spot test, the last dilution that showed plaques was 10-6, with 6 plaques. On the other hand, 20 plaques were seen in the 10-4 dilution marked area on your BIM plate. With 5 uL of phage dilutions deposited on each plate, what is the EOP?
3.33 x 10-2 (0.033) meaning it is moderately virulent
WT titer is 1.2 x 109 PFU/mL
BIM titer is 4 x 107 PFU/mL
BIM/WT
What is CRISPR-Cas9 genome editing used for?
Use of this system to alter eukaryotic genomes in living cells
What does CRISPR-Cas9 genome editing require? What is designed?
A PAM sequence on target DNA for complete endonuclease activity is required
Synthetic RNA (sgRNA - synthetic guide DNA) can be designed to recruit Cas9 and bind to target DNA, enabling cutting in genome of almost any cell
sgRNA is the tracr-RNA + crRNA
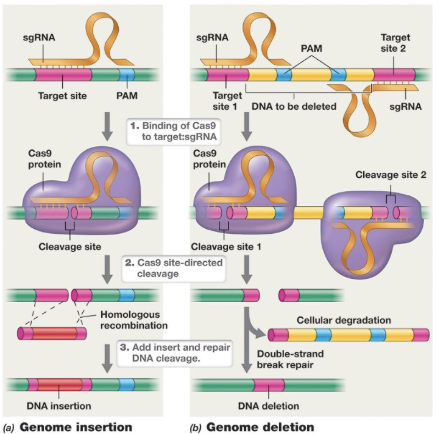
At the cut site in CRISPR-Cas9 genome editing, there can be an insertion or deletion. How is it incorporated?
At cut site, DNA can be ligated or used to insert new DNA
Homologous recombination can be used to incorporate new DNA (insertion) (Non homologous is more error prone)
Two separate target sites flanking the DNA to be deleted are selected
dsDNA break repair pathway can ligate after deletion
In deletion, it cannot be cut on one site, it must be on both so the DNA repair mechanism does not fix it so that degradation and repair can take place
To insert → homologous recombination
To delete → two target sites
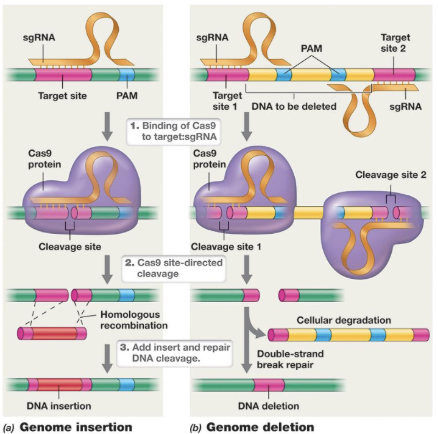
How can a CRISPR system be delivered to target cells?
Using a plasmid
Genes corresponding to sgRNA and Cas9 proteins are often cloned into a plasmid
GFP to see expression of plasmid

If I want to insert a piece of DNA into an eukaryotic cell using the CRISPR-CAS 9 system. What do I need?
A plasmid/vector with Cas9 and sgRNA
PAM sequence next to target site
The sequence we want to insert, so ends must be flanked with a similar or identical sequence to where the double break is
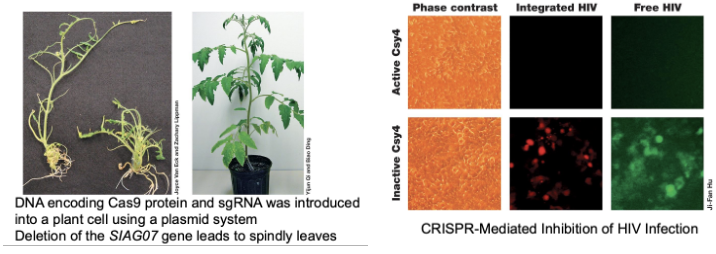
How can CRISPR editing be used in practice?
Targeting tomato mutation leads to needle-like or wiry leaves
Targeting HIV or other viral RNAs, acting as diagnostic tools and targeting viruses and cells
Removal of 62 copies of porcine endogenous retrovirus from swine cells to facilitate organ transplants
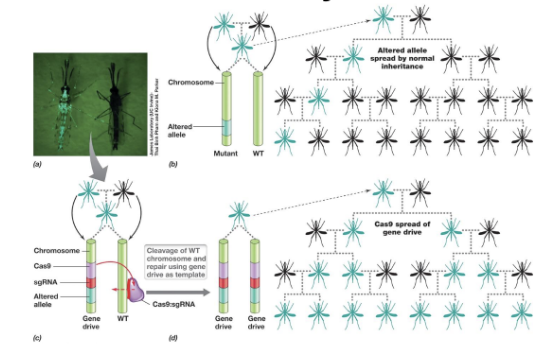
What is gene drive? How is used with mosquitoes?
Cas system can be used as gene drive to spread mutations throughout generations of populations of sexually reproducing organisms
Occur naturally via transposons but difficult to control
CRISPR and Cas9 allow inheritance to be controlled
Heterozygotes become homozygous due to ds breaks in WT copy
Modification of mosquitoes to prevent diseases like malaria
What is the expected result of amplified phage? What should it contain?
Clear tube
Should contain:
Phage
Chemicals from broth and phage buffer
Bacteria debris
Must be filtered before extracting DNA
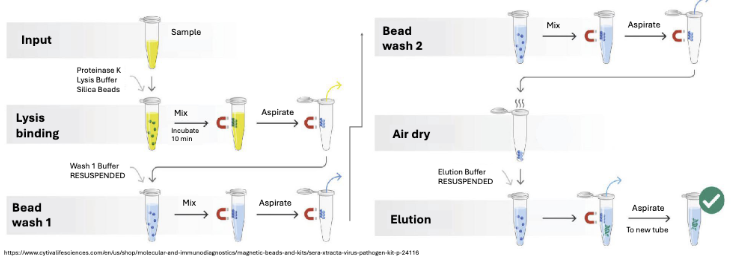
Describe the phage DNA extraction process.
DNA extraction using positively charged magnetic beads
Add proteinase K and lysis buffer and binding buffer
Lysis buffer opens up the phage (extract DNA and free from proteins)
Lysis buffer also precipitates DNA and promotes binding to beads (- and + binding)
Tubes go on magnetic rack
Remove supernatant - phage DNA is attached to beads
Wash beads twice (remove impurities) and air dry
Add eluant to disturb beads and DNA binding so DNA can be taken up
Nuclease free water or Tris solution reduces the interaction between the beads and genomic material -> elutant of viral DNA
What should the concentration of extracted DNA be?
Anything higher than 12 ng/uL
What should the ratio of A260:230 be and what does it measure? What does it mean when the ratio is off?
2-2.2 → DNA purity
If the ratio is too low or high, the sample is contaminated with chemicals
Using our kit, it is very possible the ratio will be low because of the chemicals we use but the sequencing will work
What should the ratio of A260:280 be and what does it measure? What does it mean when the ratio is off?
1.8 → DNA contamination
If ratio is too low or high, the sample is contaminated with protein and/or RNA
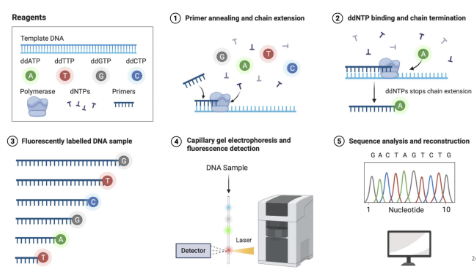
What is Sanger DNA sequencing?
Di-dieoxy nucleotides (ddNTPs) that stop chain extensions
A fluorophore will be at every positions and do capillary gel electrophoresis and get sequence
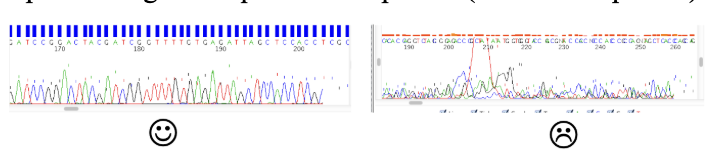
What are chromatograms?
Visually examine the chromatogram to get an idea of the sequence quality. Usually the first ~50 bases are unreliable and the sequence quality will trail off after a while
Optimal range of sequence after primer (50 nts after primer) and lasts 300-500 nts
What is nanopore sequencing?
An adaptor is added to the DNA strands to allow them to get anchored to the nanopores
As nucleotides pass through the nanopore, a characteristic current change is measured, each nucleotide has a different size and shape
Nanopore sequencing allows for more nucleotides to be sequenced more quickly and accurately and requires less sample
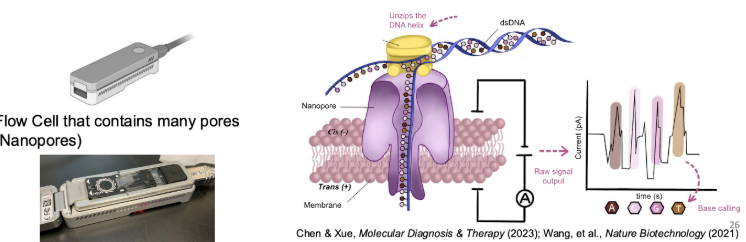
How does nanopore sequencing differ from Sanger sequencing or PCR?
98% accuracy while accuracy is ~70% for Sanger
Nanopore is faster, more accurate, requires less material and longer reads
Smaller sample size than PCR and some PCRs still failed
Which statement is false?
a. Nanopore is more accurate than Sanger Sequencing
b. If we don't filter our amplified phage, the final extracted eluant might not be pure viral DNA
c. Genome editing can be used as a detection method of certain genes
d. sgRNA stands for synthesis of generic RNA
e. To insert a DNA fragment, the process of homologous recombination can be use
D
What are the goals (3) of lab 12 bioinformatics?
Identify components of the BIMs and phage through sequence analysis
Identify the presence and location/position of new spacer acquisition
Identify mutations in the BIM infectious phage
Describe the steps in lab 12 bioinformatics.
Given sequences:
CR1 BIM sequences
The names of those whose BIM is listed above the sequence
Mentioned which BIMs did acquire a spacer
There are also some of profs BIMs at the end that can be used if the BIM was not successfully sequenced
DGCC7710 sequence on MyCourses
BIM infectious phage sequences
Goal:
Identify components of the BIMs and phage through sequence analysis
Identify the presence and location/position of new spacer acquisition
Identify mutations in the BIM infectious phage
Identify:
Repeat sequences
Newly acquired spacer and which phage -> was the phage against 2972 or 858
The position of the protospacer and the function in the phage (what does it encode for)
CRISPR Finder: DGCC7710 sequence to determine where the spacers and repeats are. Identified the sequence of the leader, repeat and already acquired spacers’ sequences
CR1=CRISPR1 but CR3=CRISPR4
Compare the BIM sequences with the phage genome (accession # in lab manual)
Leader sequence -> First repeat -> Newly acquired spacer
Leader sequence in lab manual intro
Query is BIM sequence an Sbjct is phage
Protospacer is located between 31582 and 31611 (in the example)
Determine what protein that gene encodes for (ORF, unknown, etc)
Compare phage sequence to WT 2972 or 858 with BLAST
There will be a few areas that are different between mutated and WT phage, only pick one (query is BIM infectious phage and Sbjct is WT)
Gaps can lead to frameshift mutation
What is percent identity?
How good of a match your sequence is to the database
What is the E value?
The number of expected hits of similar quality that could be found by chance, in other words, random hits or background noise (closer to 0 is better)
What is query cover?
The length of your sequence used to compare to the database
What is the total score (in BLAST)?
Sum of alignment scores for all the sequence segments
What is the max score (in BLAST)?
Highest alignment score calculated from the sum of matches (rewards) and mismatches/gaps (penalties)
You have a phage sample and saw that the concentration written on the side is 3 × 1010 PFU/mL and decided to make a stock of 5 mL of 3 × 108 PFU/mL working solution. After making your working solution, you looked at your sample phage and realized you made a mistake! The concentration of the sample is not 3 × 1010 PFU/mL but rather 3 × 1011 PFU/mL. Based on this, what is the concentration of your working sample?
Step 1:
C1V1=C2V2 → (3×1010 PFU/mL)V1=(3×108 PFU/mL)(5 mL)
V1=0.05 mL = 50 uL
Step 2:
C1V1=C2V2 → (3×1011 PFU/mL)(0.05 mL)=C2(5mL)
C2=3×109 PFU/mL (10x more concentrated)
You are mentoring a student who is doing this module of MIMM 384. They start with a plaque assay to generate BIMs. On their control plate, they see a bacterial lawn with no plaques but they see the exact same thing on their plate with phage. What do you propose should be the next step? Explain why the following experiments are or are not appropriate follow-up experiments.
a) Spot test with the phage and bacteria
b) Redo the plaque assay to generate BIMs
c) Amplify the CR1 and CR3 of the bacterial host
a) Spot test with the phage and bacteria
It is an appropriate step because it will also tell us if the phage is actually virulent and if it susceptible to the host, maybe it is a temperate phage
Not generating BIMs, but to see why the plaque assay is not working
b) Redo the plaque assay to generate BIMs
Appropriate, can do in parallel to spot test
Maybe there was human error
c) Amplify the CR1 and CR3 of the bacterial host
Maybe the host is a BIM but then a spot test would answer that question
Not an appropriate step because there is not a control that can be used
Maybe it can be used to see that the host we think we are using is actually it
Why is studying the CRISPR-Cas system in S. thermophilus useful?
From an industrial perspective, generating BIMs of S. thermophilus provides a direct mechanism for producing strains with the capacity to evade phage attack
Used often in food industry because they want BIMs
Having a CRISPR-Cas system that is resistant to phage infection is more effective in obtaining robust resistance compared to other defense methods
Understand phage-bacteria evolution
Understand the application of the CRISPR-Cas system
Bacterial defense:
Can restore antibiotic sensitivity in Pseudomonas aeruginosa (regarding other bacterial defense mechanisms against phages) (trying to be resistant to phage so it ends up being susceptible to antibiotics)
Describe co-evolution of bacteria and phages and how studying it is useful.
Co-cultures of bacteria and phages enable assessment of changes in phage resistance and susceptibility → can study co-evolutionary dynamics
In soil, co-evolution would occur but in a different manner
Different nutrients
Different communities of microorganisms (not all can be cultured in lab setting)
Evolutionary studies provide insights into conditions in which bacteria might favour different resistance mechanisms
Example: CRISPR–Cas used when grown under nutrient-limiting conditions, whereas excess nutrients typically led to surface mutations
Metagenomics – one way to examine co-evolution in complex environment
Describe the significance of studying CRISPR array and dark matter.
Provides insight into genetic material encountered and selected against during the course of bacterial evolution
Molecular fossil record
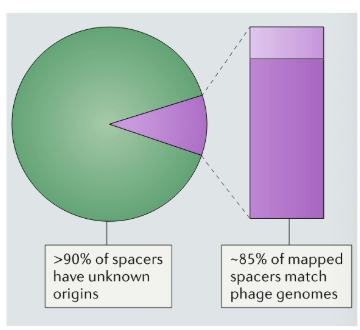
8.5% of the mapped spacers match phage genome (majority of spacer sequences are considered dark matter - no idea what the sequences are)
Can give clues to new phage discoveries
90% of the spacer sequences originate from unknown sources -> CRISPR “dark matter”
Learning a technique used in many different fields
Plaque assays: find viral titer of sample, then wanted to generate BIMs, then wanted to find BIM infectious phages
True or False: Plaques are clearings on a lawn of bacteria on the surface of agar.
True
True or False: In this week’s lab (7) exercise, you are generating BIMs.
False
True or False: This lab module does NOT require you to work in a sterile environment.
False
True or False: BIM stands for bacteriophage insensitive mechanism.
False
You plated 200 uL of your 1:1000 dilution and you noticed that you had around 500 plaques. You’re then asked to calculate the PFU/mL of this. What is your response?
The PFU/mL cannot be calculated as the number of plaques exceeds the 30-300 plaque range. Over 300 plaques does not work well as there is overlapping of plaques and it is easy to lose track when counting.
You have a 6 mL phage sample with an unknown concentration. You want to make a working phage sample that has a final concentration of 3×108 PFU/mL and a total volume of 10 mL. To determine the concentration of your phage sample, you do serial dilutions and plaque assays. You plated 100 uL of your 10-6 phage dilution and saw 50 plaques. What is the viral titer of your sample? How much phage buffer and undiluted phage do you need to make your working phage sample?
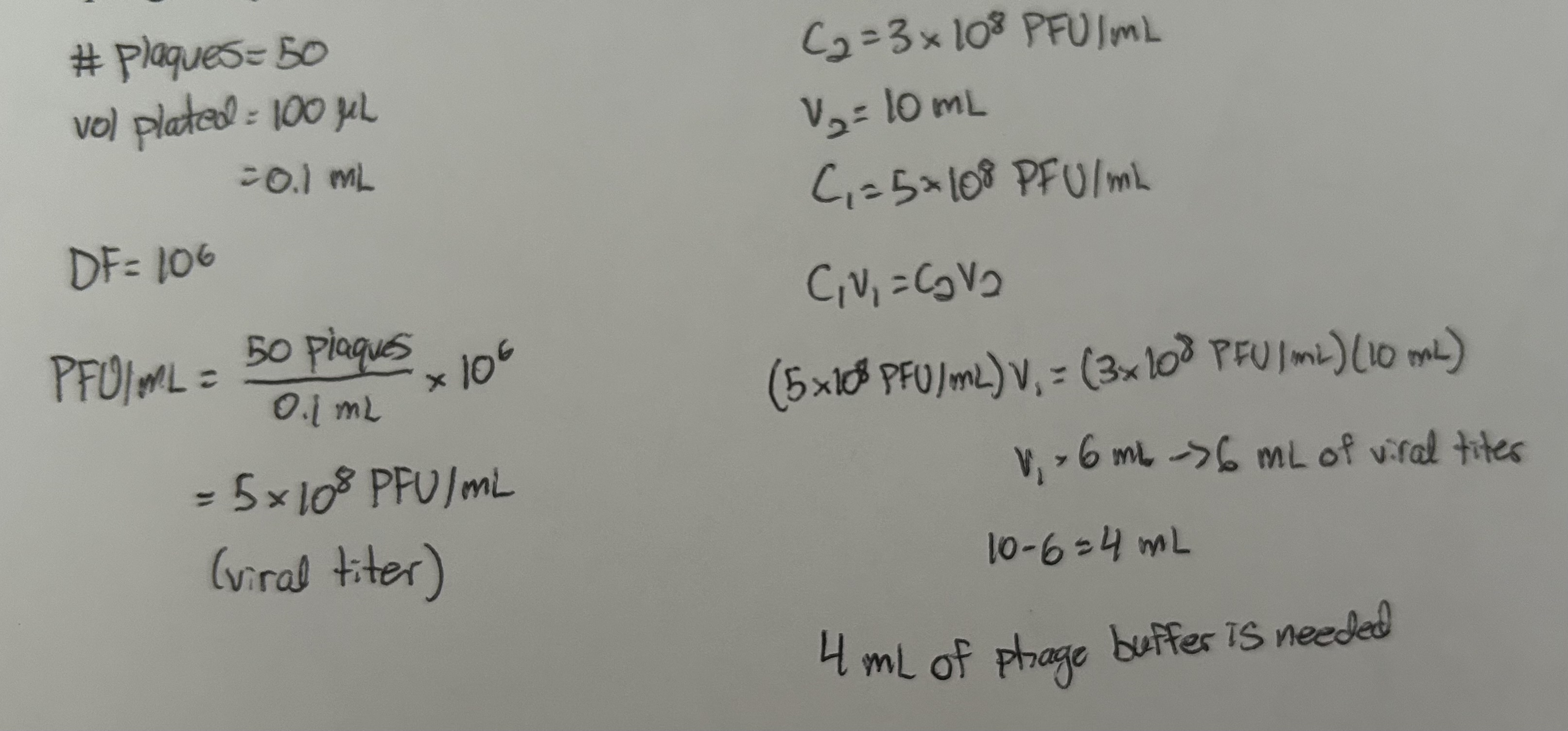
True or False: In this lab exercise (lab 8), if your plaque assay has clear lysis and only a few colonies, then those colonies are BIMs.
True
True or False: The ~66 bp addition in the CRISPR array is due to the length of the integrated spacer.
False
What is the difference between a protospacer and a spacer?
A protospacer is present in the foreign DNA, whereas the spacer is part of the CRISPR array.