Module 1: Genetic Variation in Populations
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
28 Terms
How can we describe Mendelian variation?
Diploid individuals can be one of 3 genotypes: homo dom. homo rec. or hetero. Genetic variation occurs when all individuals are not homozygotes
How to quantify/compare Mendelian variation?
Allele frequency: count total number of one type of allele and divide by the total number of alleles
Heterozygosity: count up the total number of heterozygotes in a population and divide it by the total number of individuals.
Nucleotide diversity: gene diversity averaged over all nucleotide sites in a gene
what is allelic richness?
the total number of alleles at a locus in a population
what is gene diversity?
“expected” heterozygosity within a population
what are the assumptions for Hardy-Weinberg equilibrium?
mutation is absent
large populations (no genetic drift)
no selection
no gene flow/migration
random mating
what are the genotype frequencies in HWE?
homo dom: p²
hetero: 2pq
homo rec: q²
when the allele is rare where is most of it found?
most of it is in the heterozygote because when two rare alleles show the phenotype (homo rec) it is often deleterious
examples of HWE in the real world
estimating allele frequencies from genotype frequencies
estimating “carriers” (heterozygotes)
DNA fingerprinting
finding deviations from HWE to infer selection or population structure
how to get allele frequencies from genotype frequencies
p = f(AA) + ½ f(Aa)
q = 1-p = f(aa) + ½ f(Aa)
what is inbreeding?
The probability that the same allele in two diploids came from the same individual. Inbreeding increases the probability that two alleles at a locus will be copies of an allele present in an ancestor, and are considered to be identical by descent (IBD).
what is inbreeding coefficient (F)?
the probability that 2 alleles are identical by descent when there is one common ancestor. F = X (1/2)n (X = # of alleles)
what does inbreeding do in terms of genotype frequencies?
It increases the number of homozygotes (decreases heterozygotes)
How to calculate genotype frequencies when it is known that the 2 alleles are IBD?
AA = p, (no heterozygotes),
aa = q
fAA = p² + pqF
faa = q² + pq
does inbreeding change allele frequencies?
No, only genotype frequencies
how to get inbreeding coefficient of a population?
compare the number of heterozygotes expected under HWE and compare to observed number (F = (Hetexp-Hetobs)/Hetexp)
what is inbreeding depression
reduction in fitness in offspring that are inbred, likely due to effects from recessive alleles that are not masked (homozygous recessive).
what is genetic rescue?
introducing genetic variation to a population that has a large inbreeding coefficient in an attempt to decrease inbreeding.
what are 4 facctors that influence allele frequencies?
mutation
migration
genetic drift
natural selection
how does natural selection favour alleles?
It selects for the form with the highest fitness (where relative fitness = 1)
what is heterozygote advantage?
when both homozygotes have different deleterious phenotypes but the intermediate heterozygote has an advantage (overdominance for fitness)
what are s and u? and why is it important?
s = selection coeficient
u = mutation rate
q = (u/s)1/2 => what is q when there is selection and mutation
what is directional selection?
when selection favours one genotype over another, and changes the allele frequencies at a locus.
what is negative frequency-dependent selection?
when a rare allele is beneficial because there is more access to resources (high frequency = low fitness)
in negative frequency-dependent selection, when do heterozygotes have an advantage?
Never, they will always be below either type of homozygote
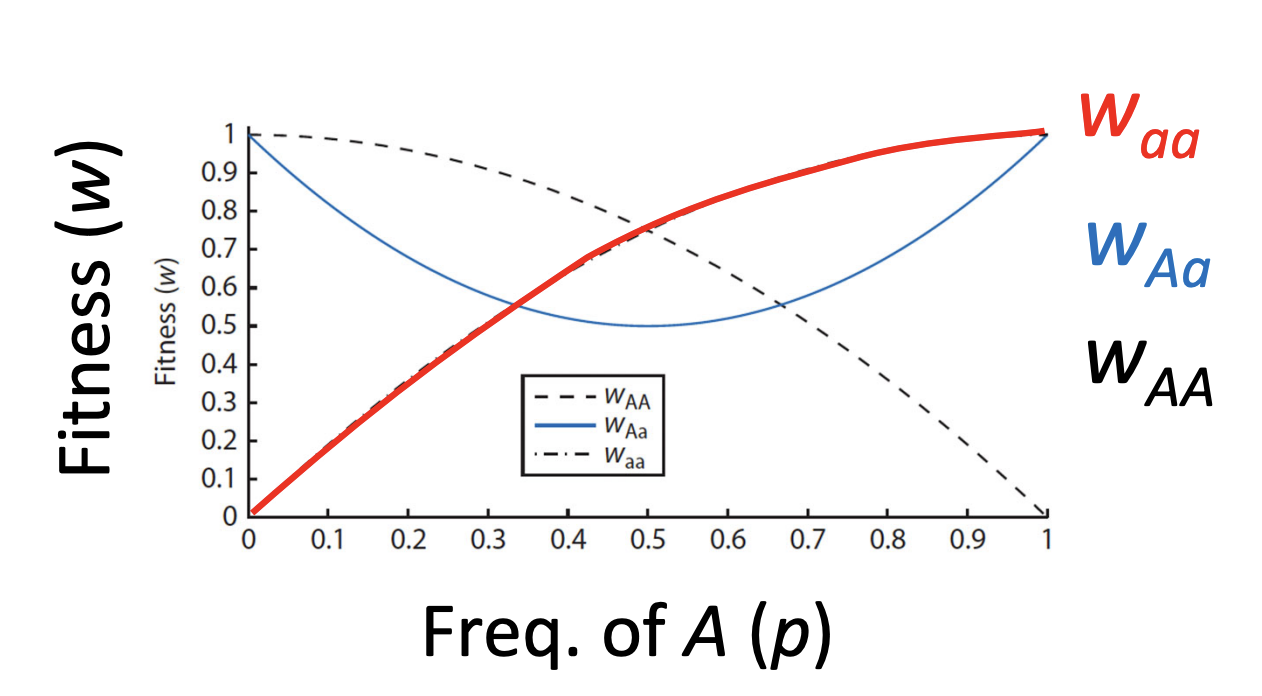
what is spatially-varying selection
multiple niche spaces, coexistence and different niche spaces favour different alleles in the same inter-mating population
how are three ways to maintain genetic variation?
heterozygote advantage
negative frequency-dependent selection
spatially-varying selection
how does genetic drift affect allele frequencies?
in small populations with random mating events there is the chance that one allele will fixate in a population and the other will die off simply due to chance
what is the founder effect?
the introduction of a new allele to a population that is higher than that allele’s frequency that already exists in the population