Components of DNA
1/63
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
64 Terms
Nucleic acids where discovered WHEN
1869
Nucleic acids consists of qual parts of WHAT
Pentose (5) sugar
Nitrogenous base
Phosphate
Pentose sugar
WHAT in RNA
WHAT in DNA
Pentose sugar
RIBOSE (C5H10O5) - used in RIBONUCLEIC ACIDS in RNA
DEOXYRIBOSE (C5H10O5) - used in DEOXYRIBONUCLEIC ACIDS in DNA
Nitrogenous base
WHAT
WHAT
Nitrogenous base
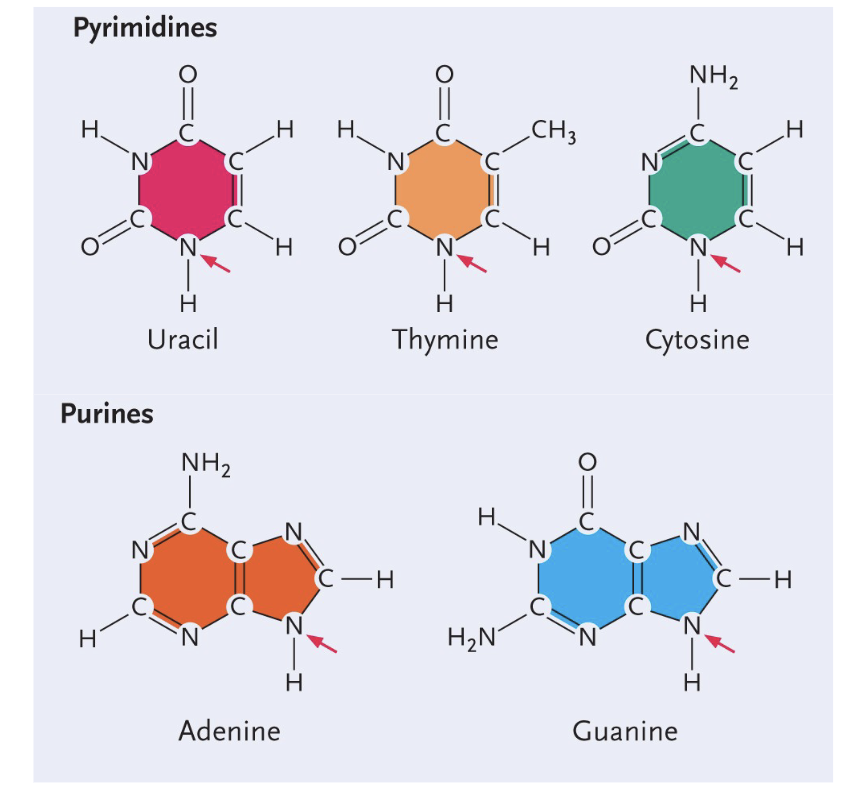
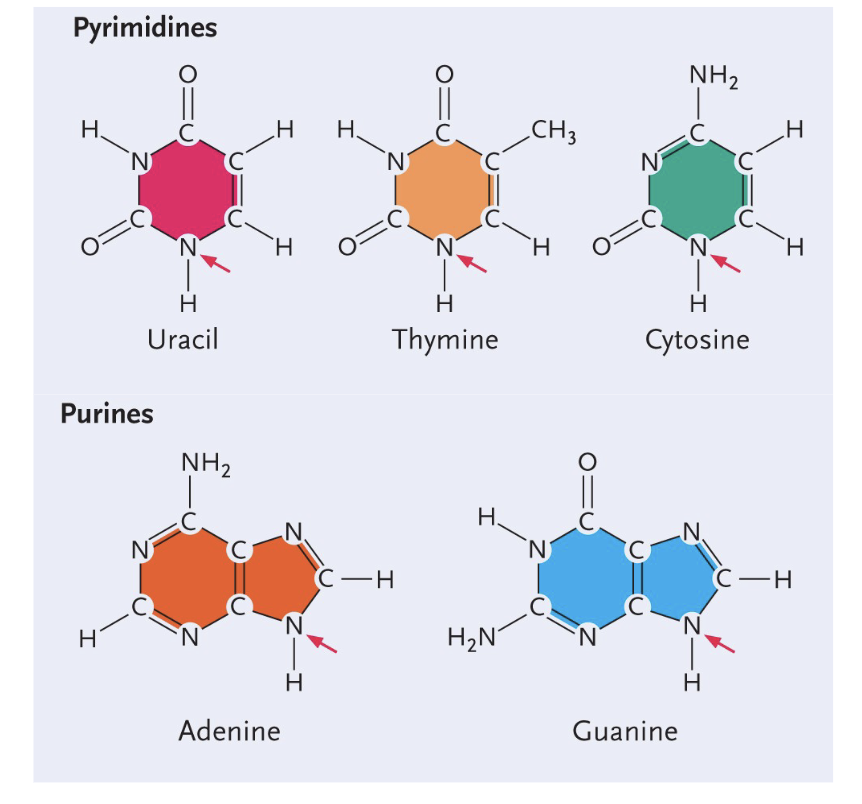
Purines (DOUBLE ring) - Guanine and adenine
Pyramides (SINGLE ring) - Uracil, thymine, cytosine
Phosphate (PO4) is found in WHAT
All nucleic acids
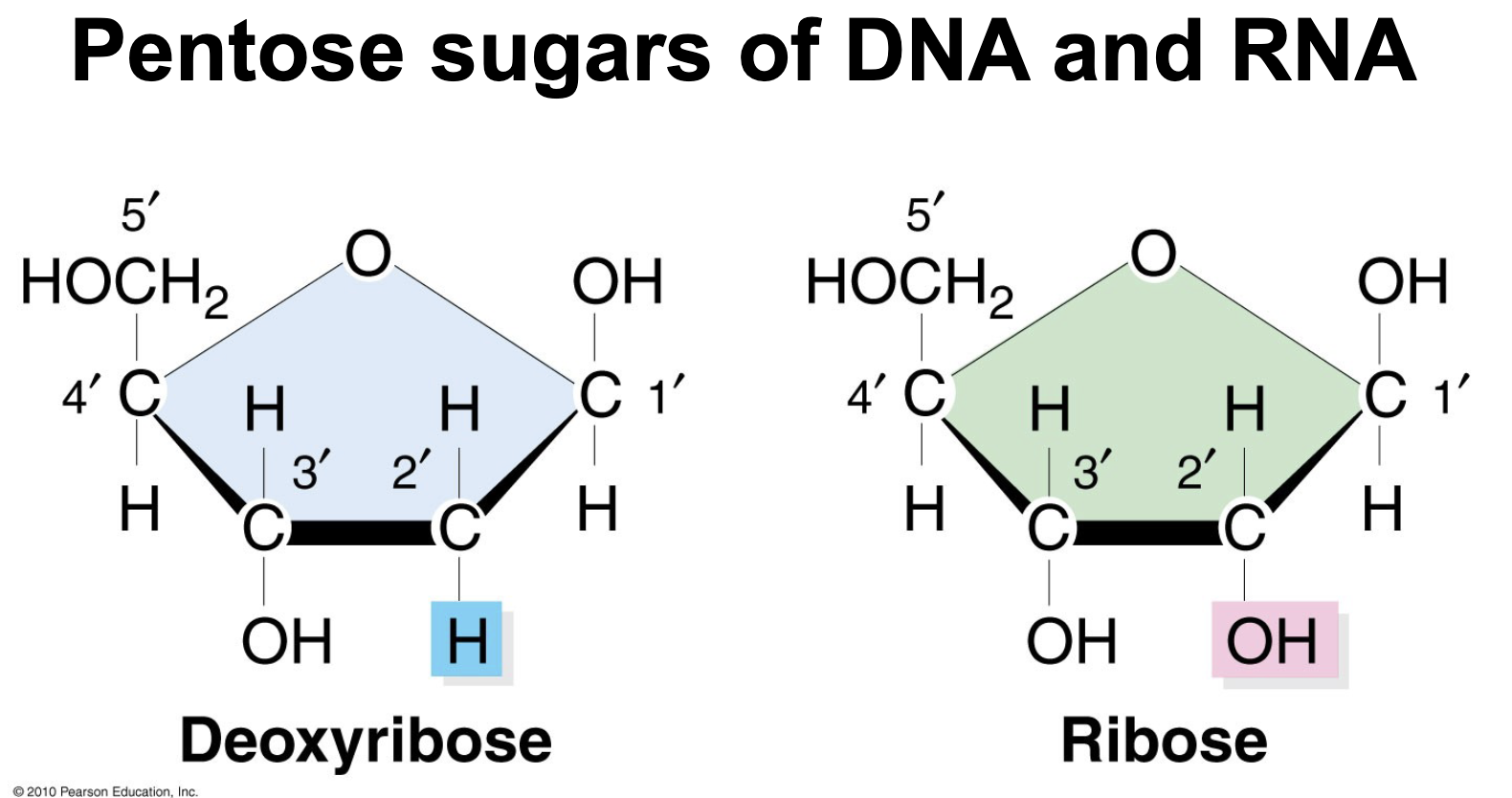
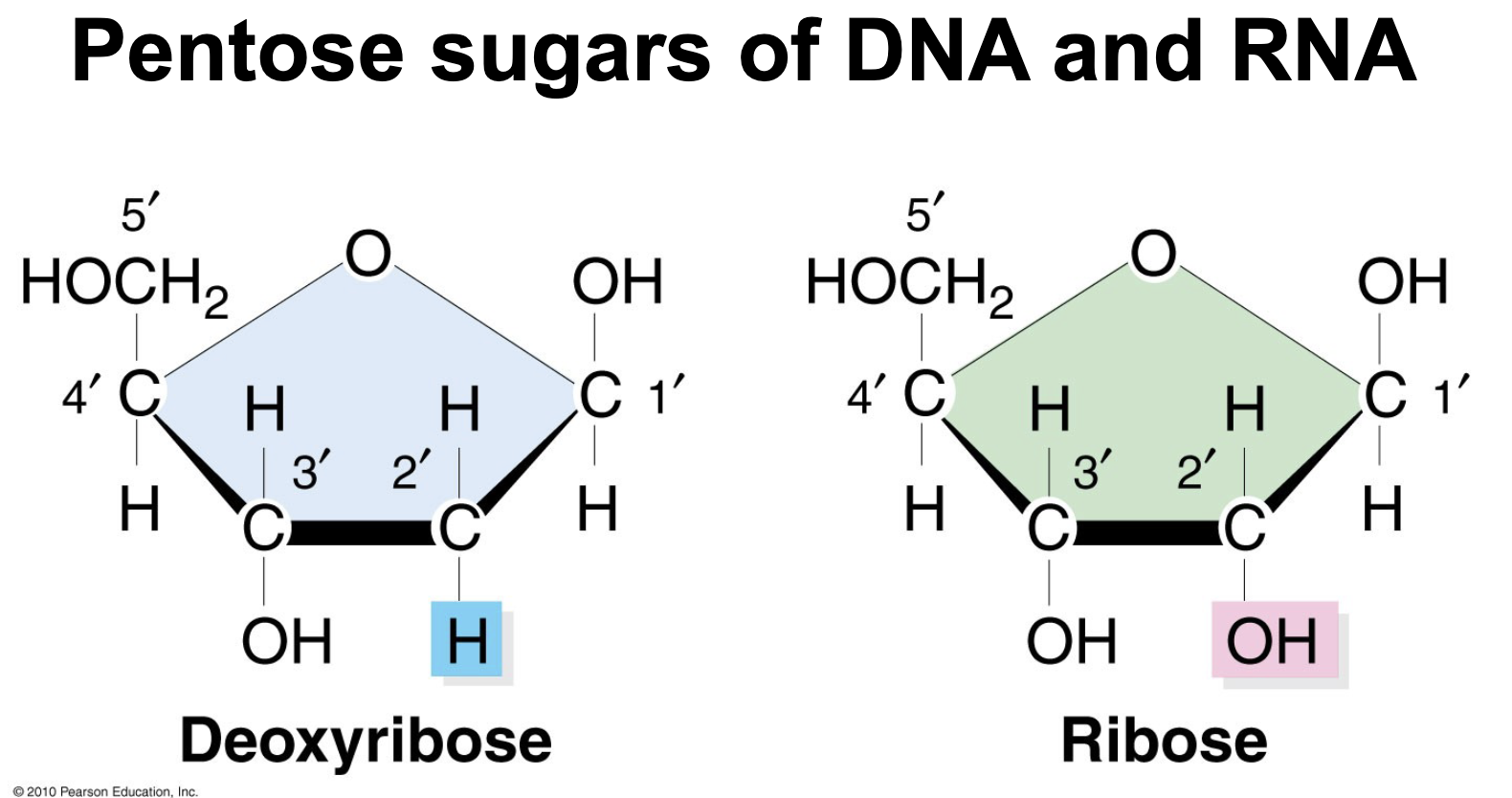
In the pentose sugar of DNA and RNA the two sugars only differ in one type of chemical group attached to WHAT carbon
In the pentose sugar of DNA and RNA the two sugars only differ in one type of chemical group attached to 2’ carbon
Nitrogenous bases of DNA and RNA:
DNA uses WHAT
RNA uses WHAT
Nitrogenous bases of DNA and RNA:
DNA uses Adenine, guanine, cytosine, thymine
RNA uses Adenine, guanine, cytosine, uracil
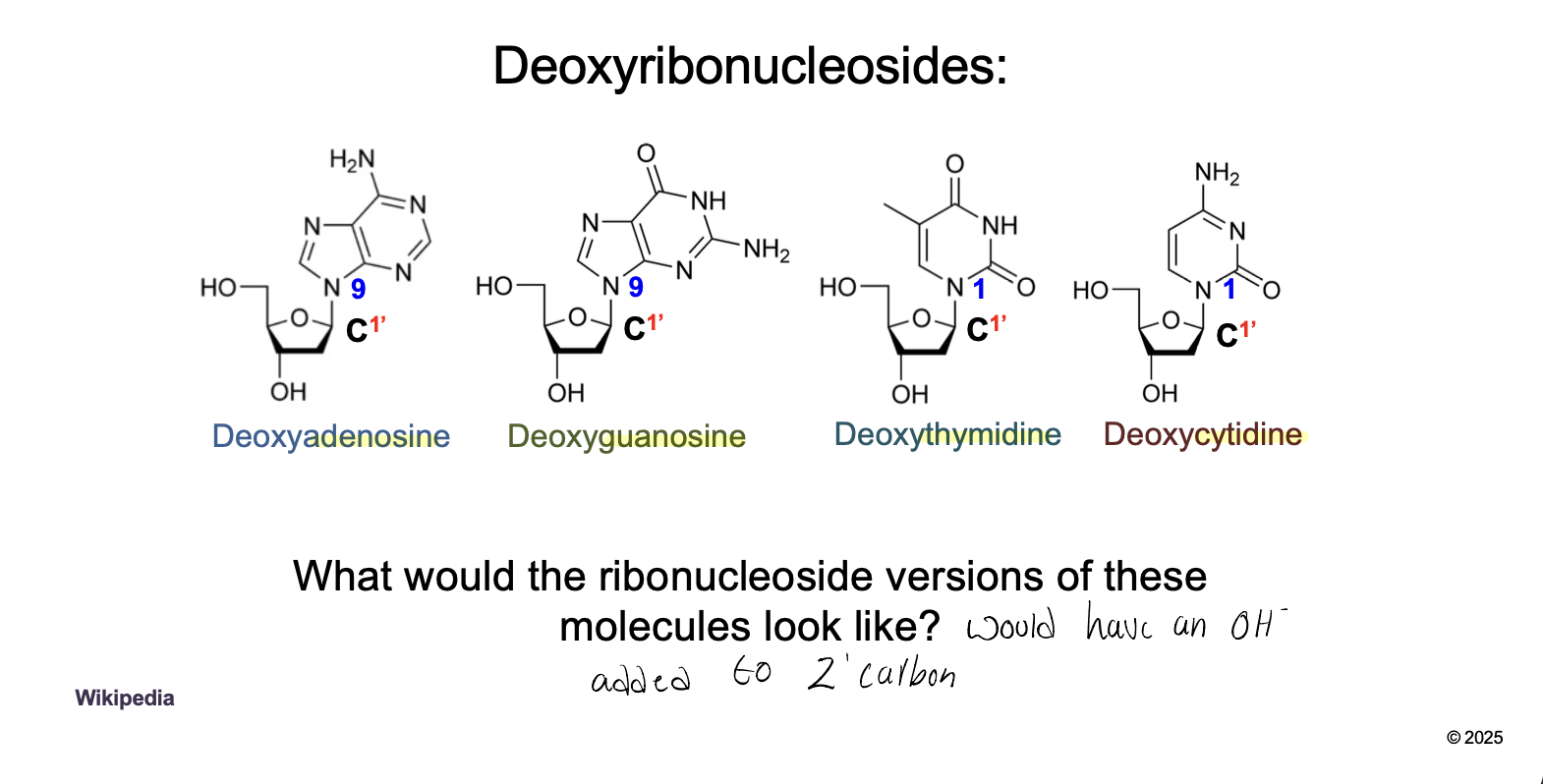
NucleoSIDES: WHAT + WHAT
The base are bound to the WHAT carbon of sugar
WHAT are all the new names
NucleoSIDES: Pentose sugar + nitrogenous base
The base are bound to the 1’ carbon of sugar
DeoxyADENOSINE, DeoxyGUANOSINE, DeoxyTHYMIDINE, DeoxyCYTIDINE
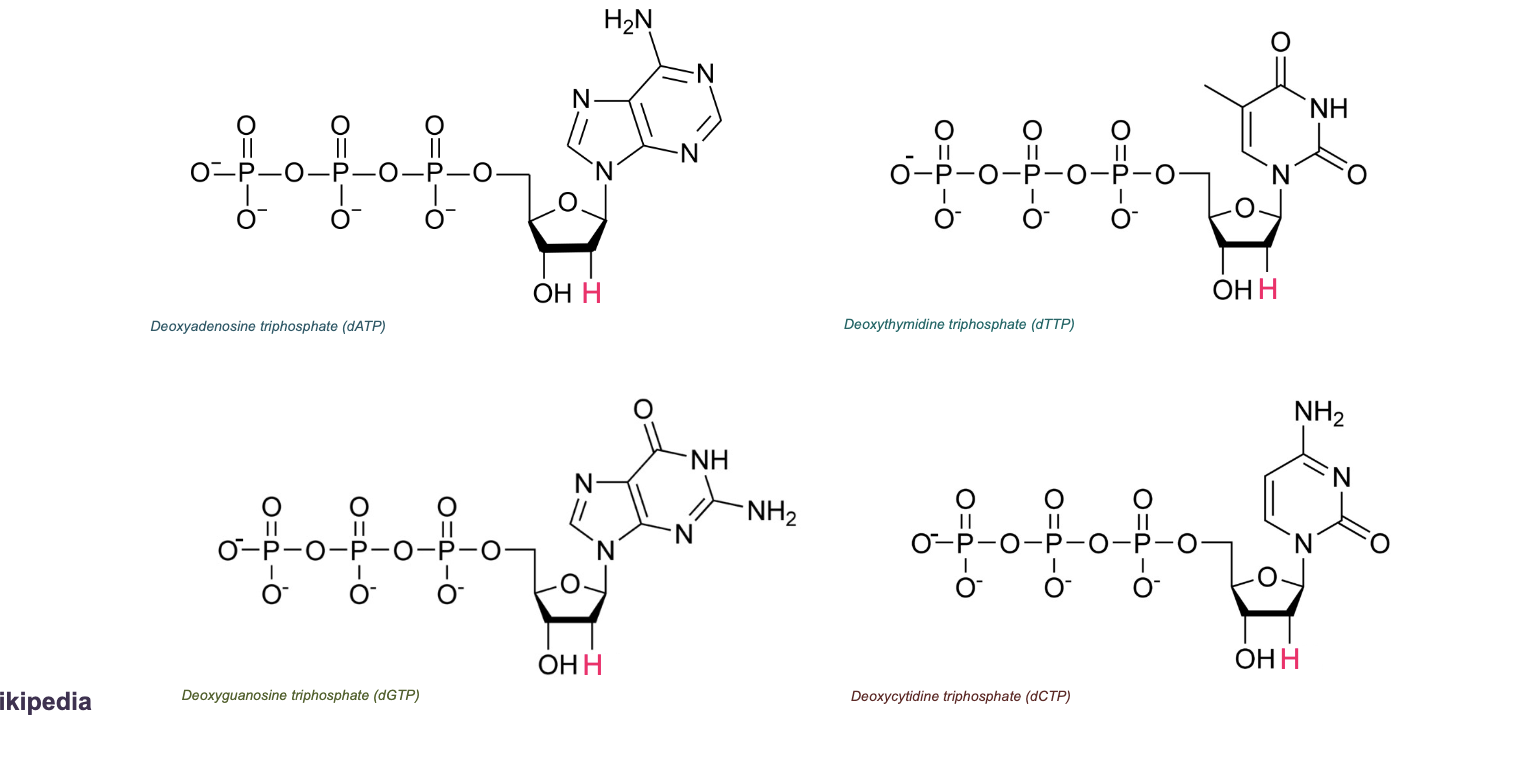
NucleoTIDES: WHAT + WHAT
The phosphates are bound to the WHAT of the sugar
Deoxyribonucleotides (dNTP)
NucleoTIDES: NucleoSIDE + Phosphate(s)
The phosphates are bound to the 5’ of the sugar
Deoxyribonucleotides (dNTP)
DNA: A polymer of WHAT
DNA: A polymer of DEOXIRIBONUCLEOTIDES
Nucleotide monomers polymerize (combine) via WHAT
Nucleotide monomers polymerize via PHOSPHODIESTERS
Ester = WHAT
Phosphoester = WHAT
Phosphodiester = WHAT
Ester = C-O-C
Phosphoester = C-O-P
Phosphodiester = C-O-P-O-C
The covalent bonds form between the WHAT
The covalent bonds form between the 3’ carbon and this 5’ of adjacent nucleotides
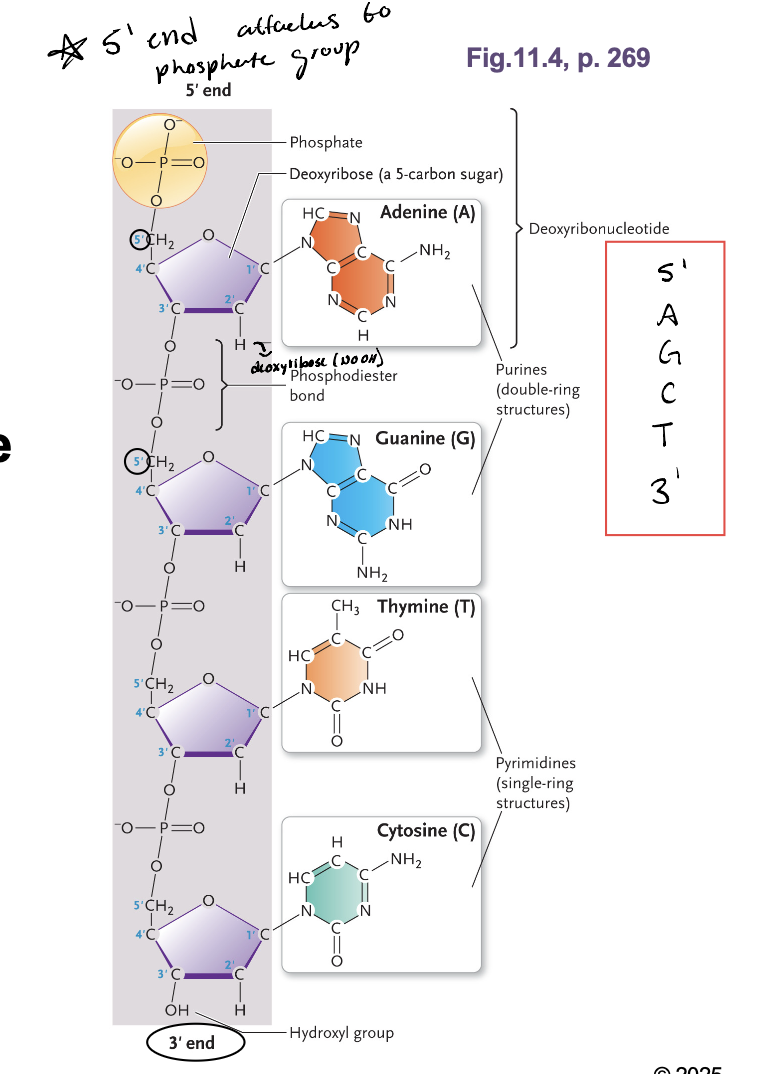
These covalent bonds form the WHAT
These covalent bonds form the PENTOSE-PHOSPHATE BACKBONE
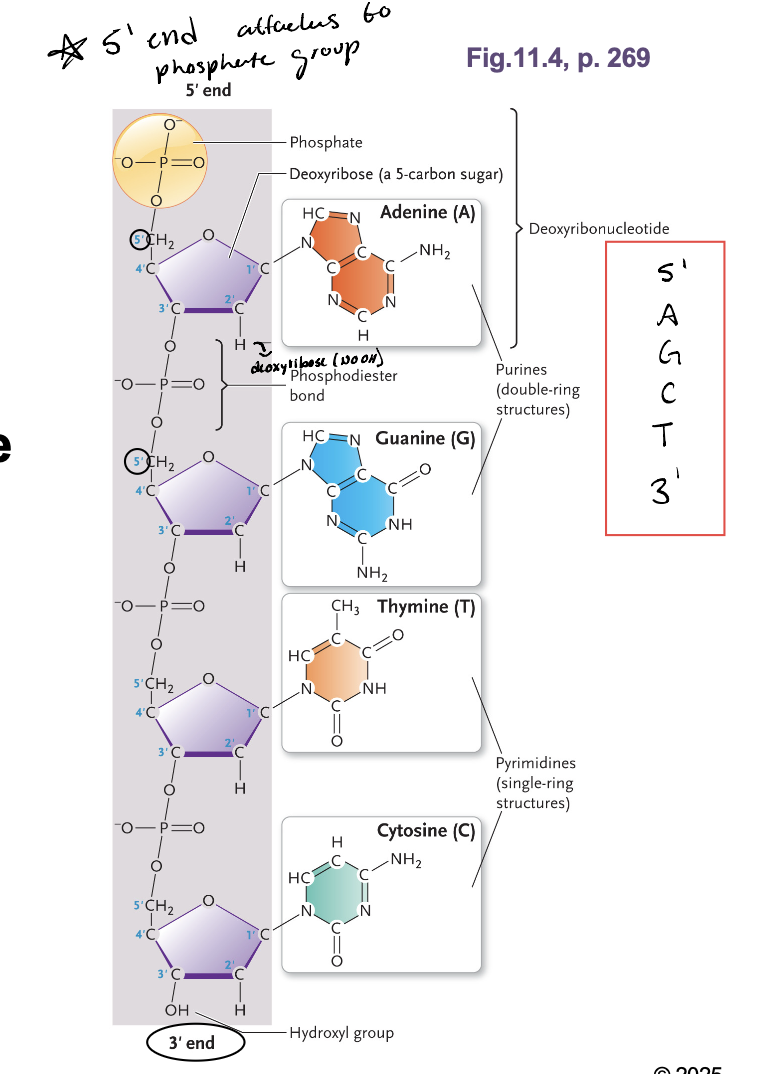
The polynucleotide has WHAT with a WHAT end and a WHAT end
The polynucleotide has POLARITY with a 5’ end and a 3’ end
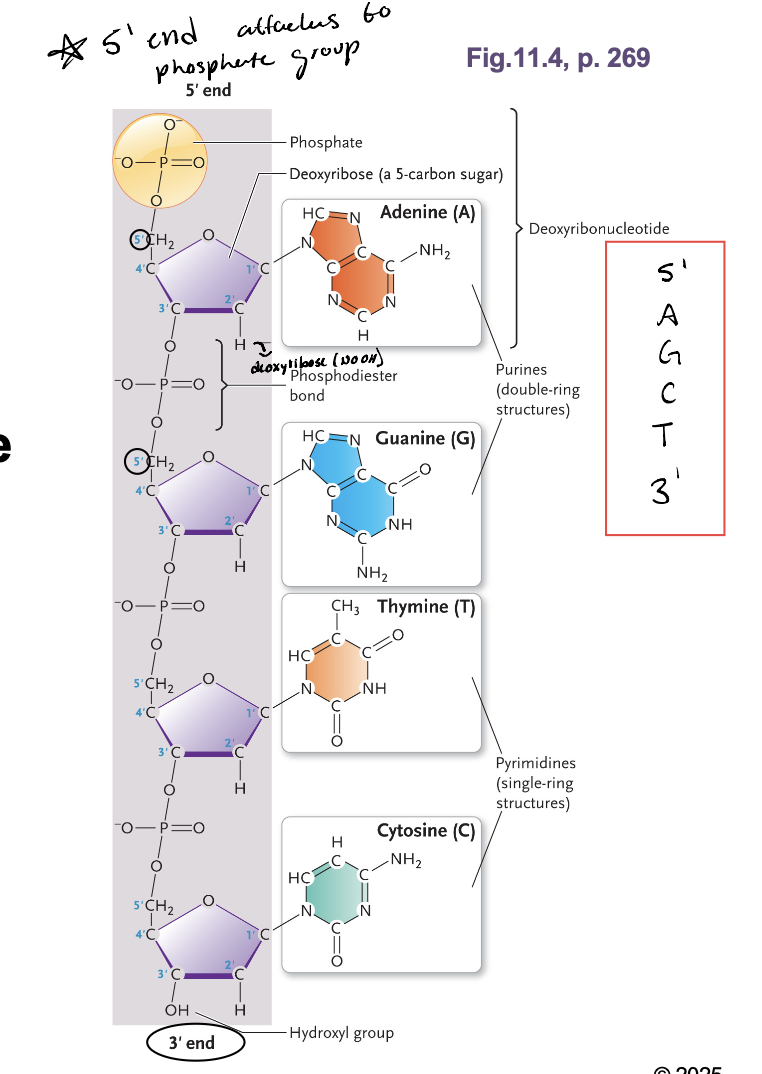
Type of nucleic acid depends on the WHAT in the WHAT
Type of nucleic acid depends on the SUGAR (DNA-deoxyribonucleic acid or RNA - ribose) in the PENTOSE_PHOSPHATE BACKBONE
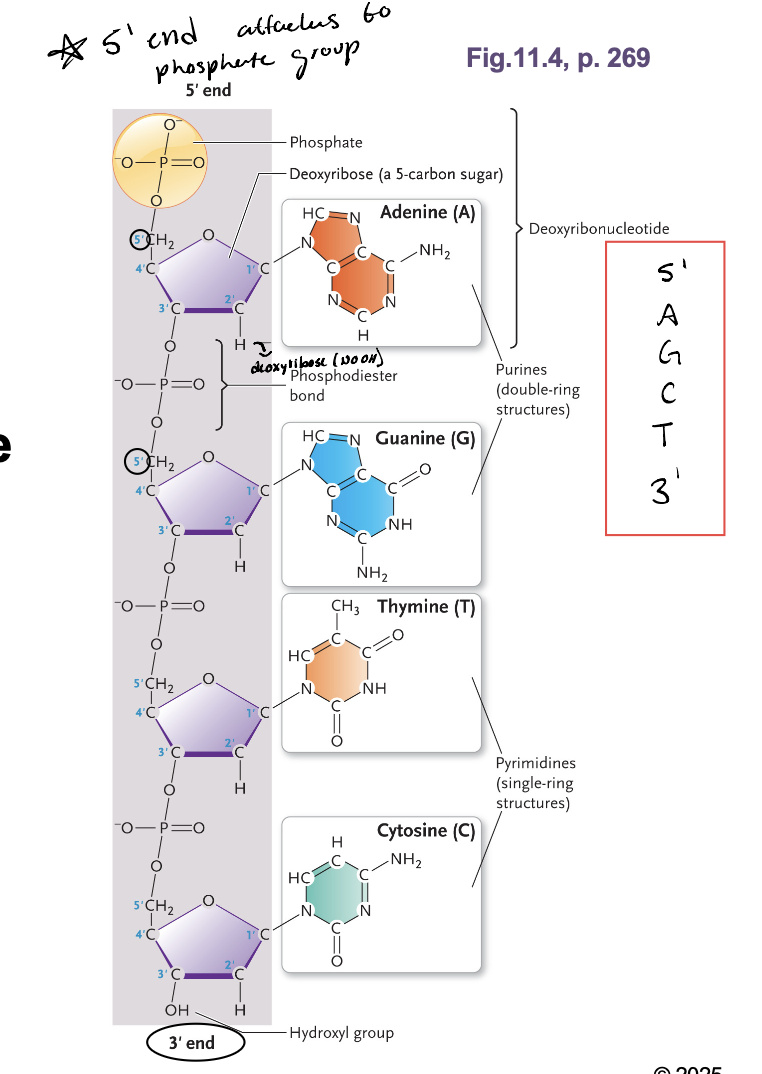
What charge is aa DNA molecule
Negative
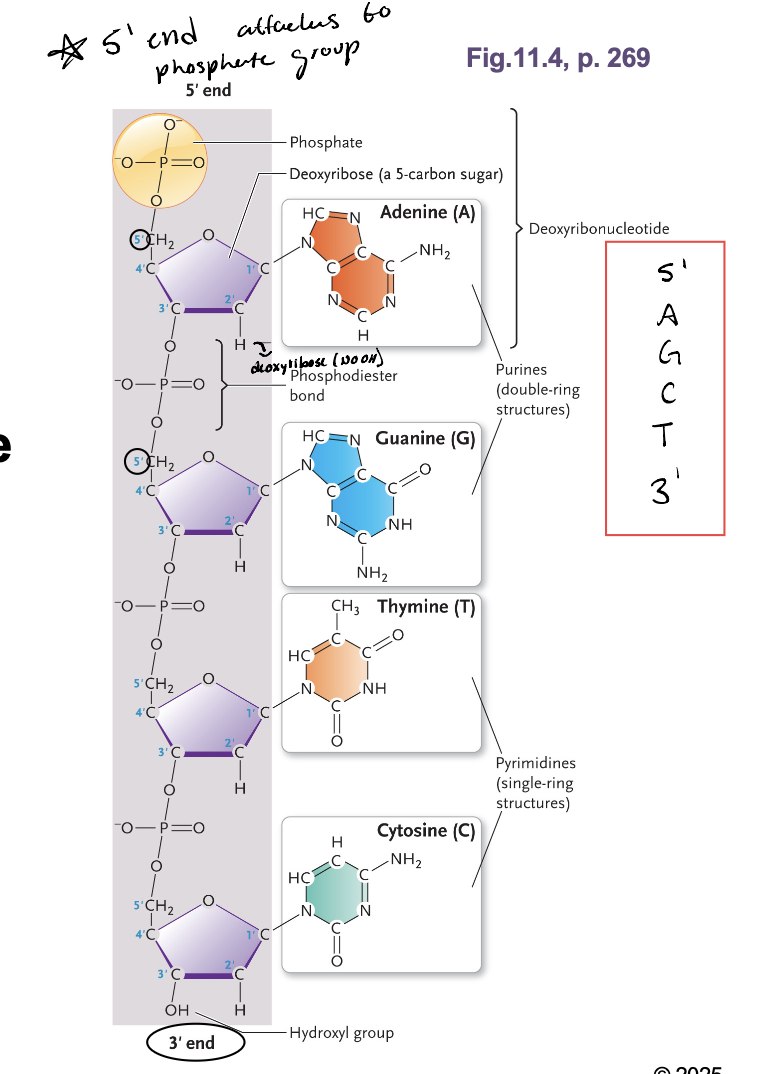
The 5’ end is always attached to WHAT
Phosphate group
The 3-D structure of DNA is based on the three chemical components of DNA, scientists knew that DNA:
Was WHAT
Had a WHAT
The WHAT in the nucleotide “WHAT”
The 3-D structure of DNA is based on the three chemical components of DNA, scientists knew that DNA:
Was RELATIVELY LINEAR
Had a PENTOSE PHOSPHATE BACKBONE
The NITROGENOUS BASES in the nucleotide “HELD THE CODE”
Chargaff analyzed overall WHAT of the four WHAT in various species
Chargaff analyzed overall QUANTITIES of the four NITROGENOUS BASES in various species
What is chargaff rule: WHAT
other conclusion:
WHAT
WHAT
WHAT
What is chargaff rule: %A = %T and %C = %G
other conclusion:
%Purines (A+G) = % Pyrimidine (C+D)
C+G wont = A+T
A, C, G, and T are NOT present in EQUAL amounts
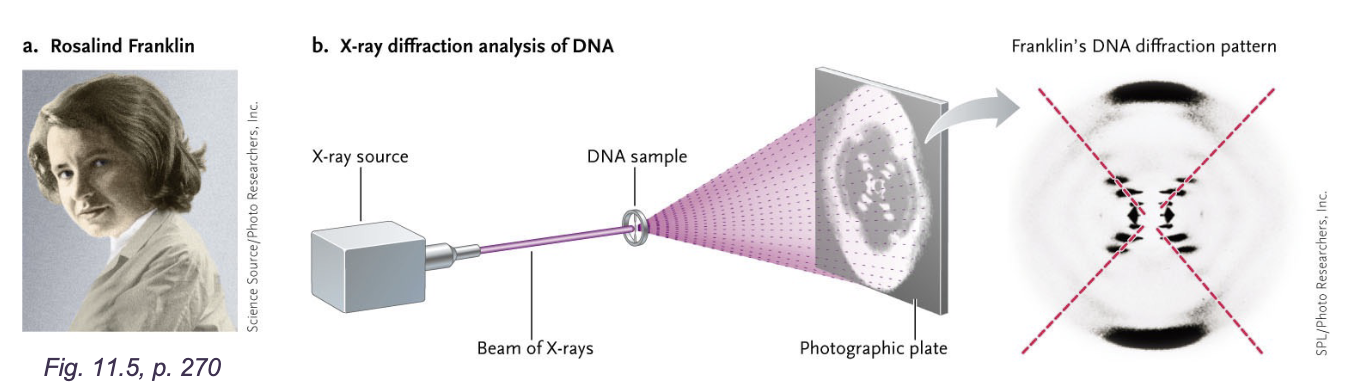
X-ray diffraction studies:
DNA molecules were WHAT and about WHAT in diameter
0.34nm periodicity suggested that bases were stacked like pennies on top of one another
WHAT pattern suggested WHAT
Franklin did not propose a WHAT
Wilikins (franklins colleague) shoes images Watson
X-ray diffraction studies:
DNA molecules were CYLINDRICAL and about 2nm in diameter
0.34nm periodicity suggested that bases were stacked like pennies on top of one another
X-SHAPES pattern suggested HELIX
Franklin did not propose a DEFINITIVE MODEL
Wilikins (franklins colleague) shoes images Watson
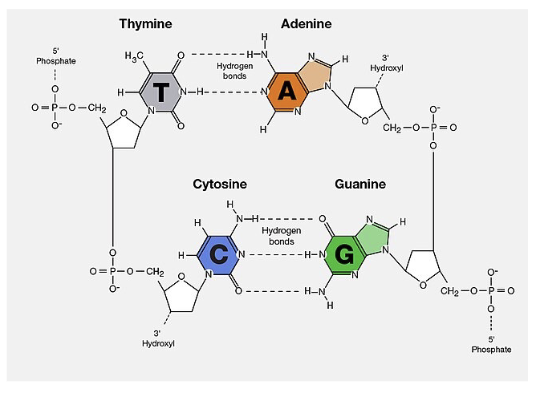
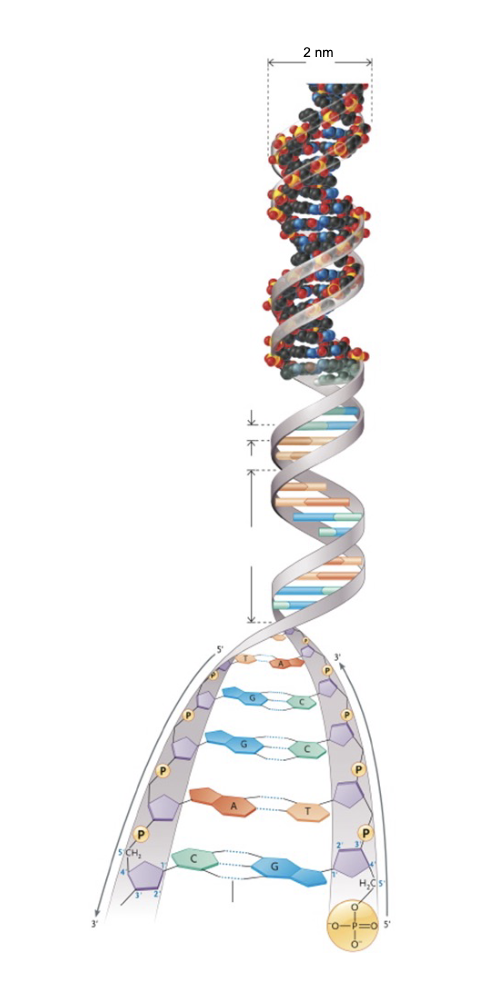
DNA double helix:
Base pairing is WHAT and therefore, base pair sequence on one strand can be used to specify the WHAT of the other strand
Base pairs are stacked WHAT to the axis and contributes to WHAT of the double helix
WHAT between bases keeps two strands together:
— A-T pairs share HOW MANY H-bonds
— G-C pairs share HOW MANY H-bonds
DNA double helix:
Base pairing is COMPLEMENTARY and therefore, base pair sequence on one strand can be used to specify the SEQUENCE of the other strand
Base pairs are stacked FLAT LYING PERPENDICULAR to the axis and contributes to STABILITY of the double helix
WHAT between bases keeps two strands together:
— A-T pairs share 2 H-bonds
— G-C pairs share 3 H-bonds
The double helix can be WHAT (separated or change shape of DNA)
The double helix can be DENATURES (separated or change shape of DNA)
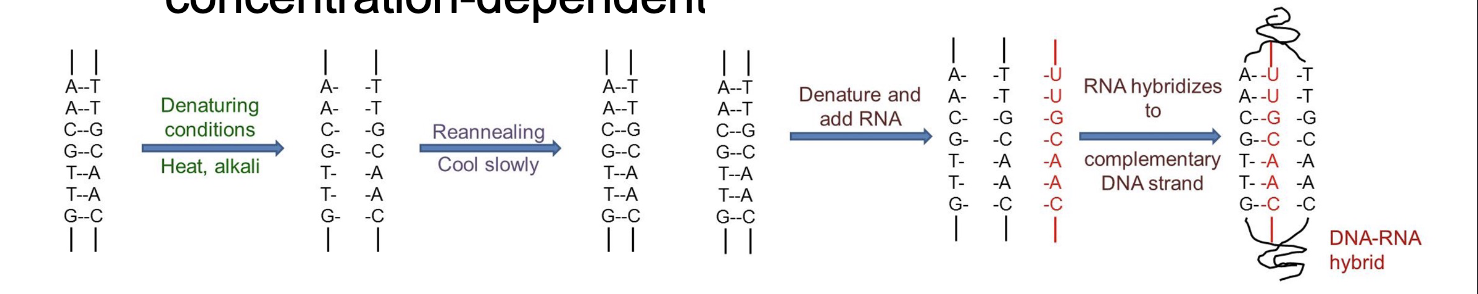
WHAT of single strands of DNA and RNA by forming H-bonds
WHAT-WHAT
WHAT-WHAT hybrids are possible
ANNEALING of single strands of DNA and RNA by forming H-bonds
DNA-DNA
DNA-RNA hybrids are possible
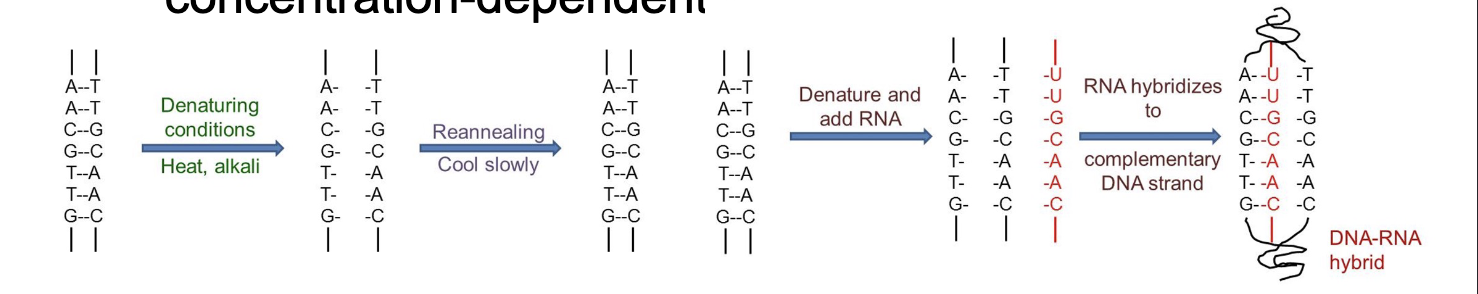
Nucleic acid hybridization highly-specific (two strands must be WHAT in sequence), WHAT-driven and WHAT-dependent
Nucleic acid hybridization highly-specific (two strands must be COMPLEMENTARY in sequence), TEMPERATURE-driven and CONCENTRATION-dependent
DNA-RNA hybrids are important in WHAT, WHAT and reproduction of some WHAT
DNA-RNA hybrids are important in DNA REPLICATION, TRANSCRIPTION and reproduction of some RNA VIRUSES
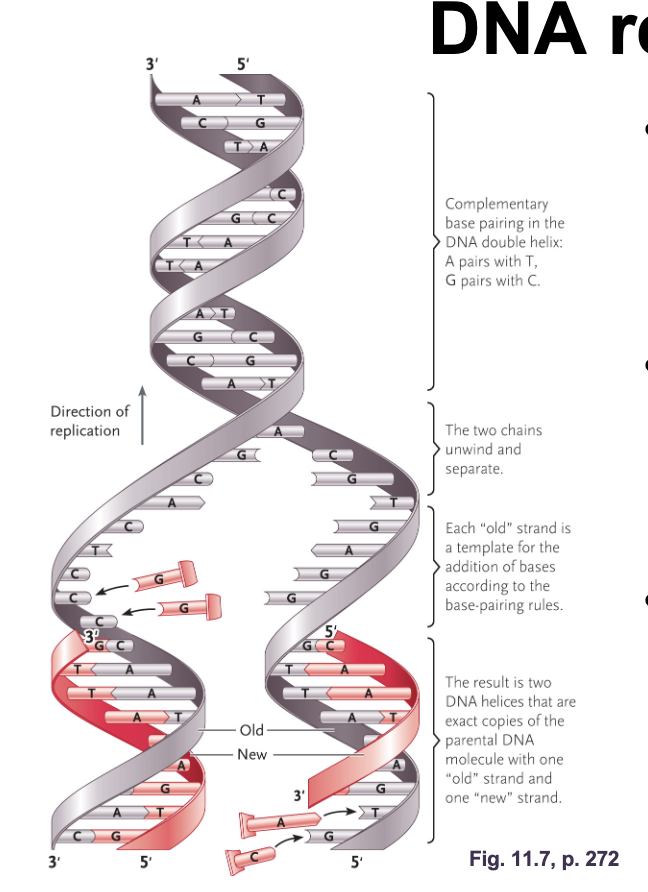
Watson and Cricks model of DNA replication:
WHAT allows WHAT (original strand) to act as WHAT
Parental strands can unwind by breaking the WHAT between bases
WHAT (conserved since half of the original strand is still present) where the double helix will contain. aWHAT and a WHAT (daughter) strand
Watson and Cricks model of DNA replication:
COMPLEMENTARY BASE PAIRING allows PARENTAL STRANDS (original strand) to act as TEMPLATE for DNA REPLICATION
Parental strands can unwind by breaking the HYDROGEN BONDS between bases
SEMICONSERVATIVE REPLICATION (conserved since half of the original strand is still present) where the double helix will contain. a PARENTAL STRAND and a NEWLY SYNTHESIZED (daughter) strand
Eukaryotes have WHAT enclosed in a nucleus
Eukaryotes have MULTIPLE LINEAR DNA MOLECULES enclosed in a nucleus
Organization in Eukaryotes:
To keep the DNA organized, and help regulate gene expression, DNA is condensed into WHAT
To keep the DNA organized, and help regulate gene expression, DNA is condensed into CHROMATIN (normal state of DNA)
Organization in Eukaryotes:
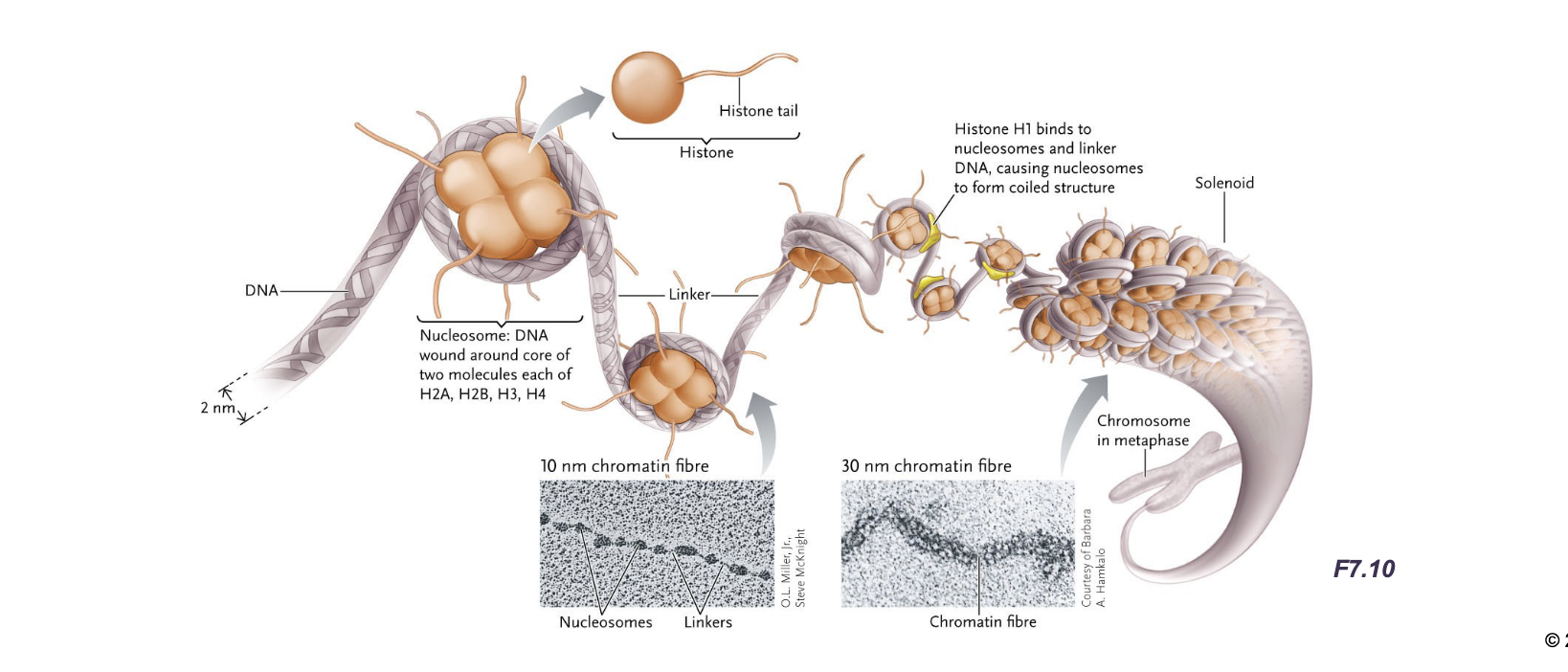
Before DNA is in chromatin form, the DNA double helix if first wrapped TWICE(ish) around WHAT to form a WHAT
Before DNA is in chromatin form, the DNA double helix if first wrapped TWICE(ish) around HISTONE PROTEIN to form a NUCLEOSOME (to make chromatin)
Organization in Eukaryotes:
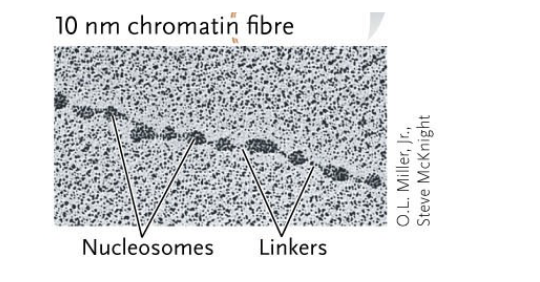
Each DNA molecule is a repeating series of WHAT, called WHAT, that looks like “beads on a string” under an electron microscope
Organization in Eukaryotes:
Each DNA molecule is a repeating series of NUCLEOSOMES, called 10nm CHROMATIN FIBRES, that looks like “beads on a string” under an electron microscope
Organization in Eukaryotes:
Additionally histones (H1) cause the chromatin to coil further into WHAT
Organization in Eukaryotes:
Additionally histones (H1) cause the chromatin to coil further into 30nm CHROMATIN FIBRES (solenoids)
Organization in Eukaryotes:
Chromatin is the “WHAT” state if our DNA molecules
Organization in Eukaryotes:
Chromatin is the “NORMAL” state if our DNA molecules
Organization in Eukaryotes:
The chromatin unwinds (expresses the double helix) during WHAT and WHAT
Organization in Eukaryotes:
The chromatin unwinds (expresses the double helix) during DNA REPLICATION and TRANSCRIPTION
Organization in Eukaryotes:
The chromatin than condenses further into WHAT - during WHAT and WHAT
Organization in Eukaryotes:
The chromatin than condenses further into CHROMOSOMES (Highly condensed state of DNA molecules) - during MITOSIS and MEIOSIS
Histones are basic, WHAT charged proteins
Histones are basic, POSITIVELY charged proteins
The nucleosome is a WHAT with about HOW MANY base pairs of DNA wrapped around it
The nucleosome is a HISTONE OCTAMER with about 147 base pairs of DNA wrapped around it
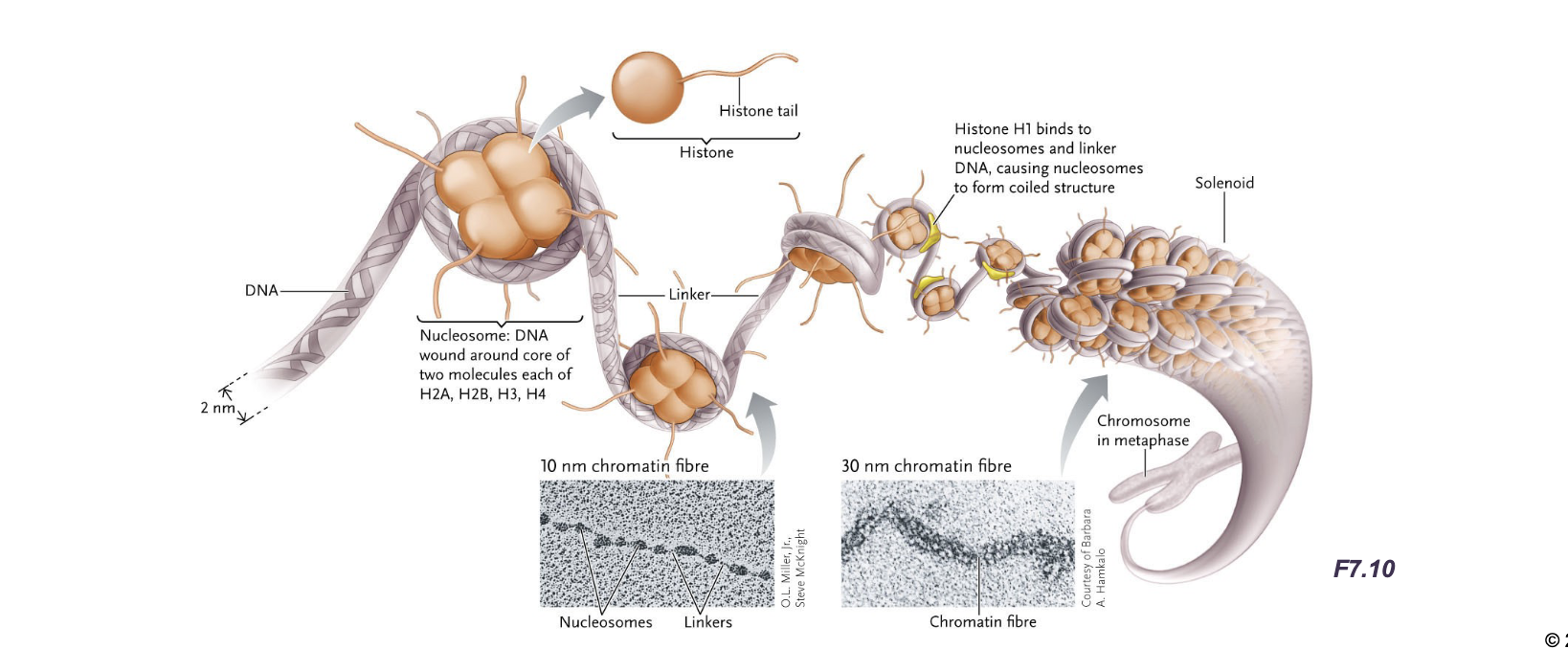
Histone (H1) binds WHAT and coils nucleosomes to from WHAT
Histone (H1) binds LINKER DNA and coils nucleosomes to from 30nm CHROMATIN FIBRE
DNA packing along the double helix is not WHAT
DNA packing along the double helix is not UNIFROM
Euchromatin regions have lower WHAT and genes are WHAT
Euchromatin regions have lower DNA COMPACTION and genes are ACTIVELY EXPRESSED
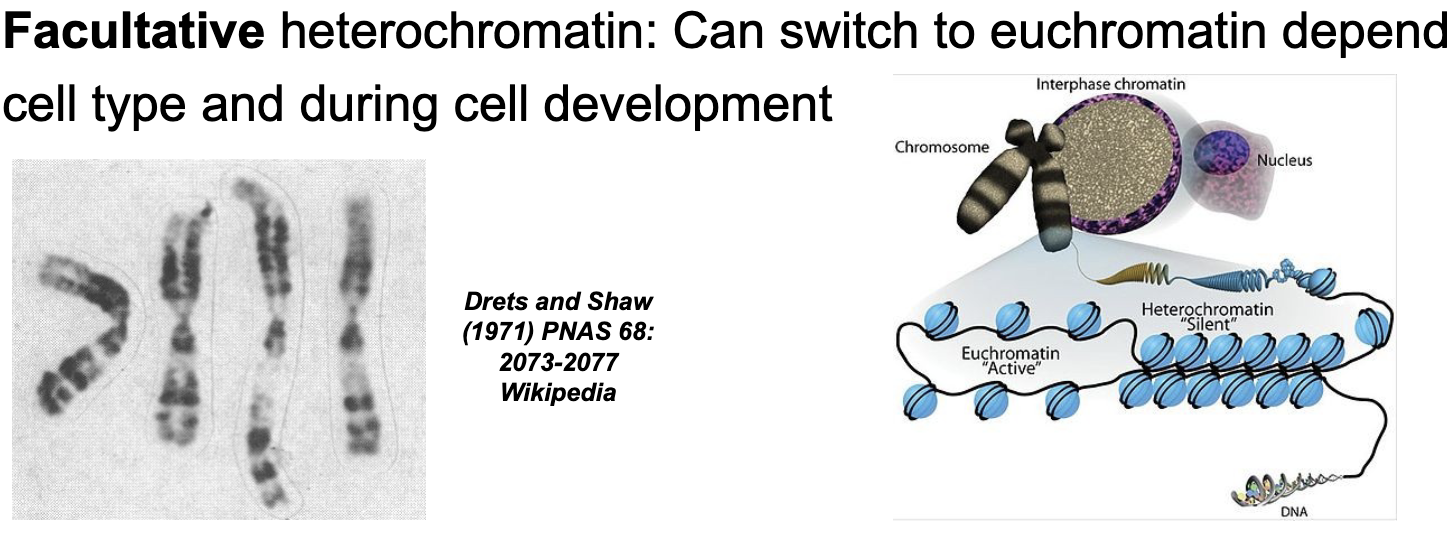
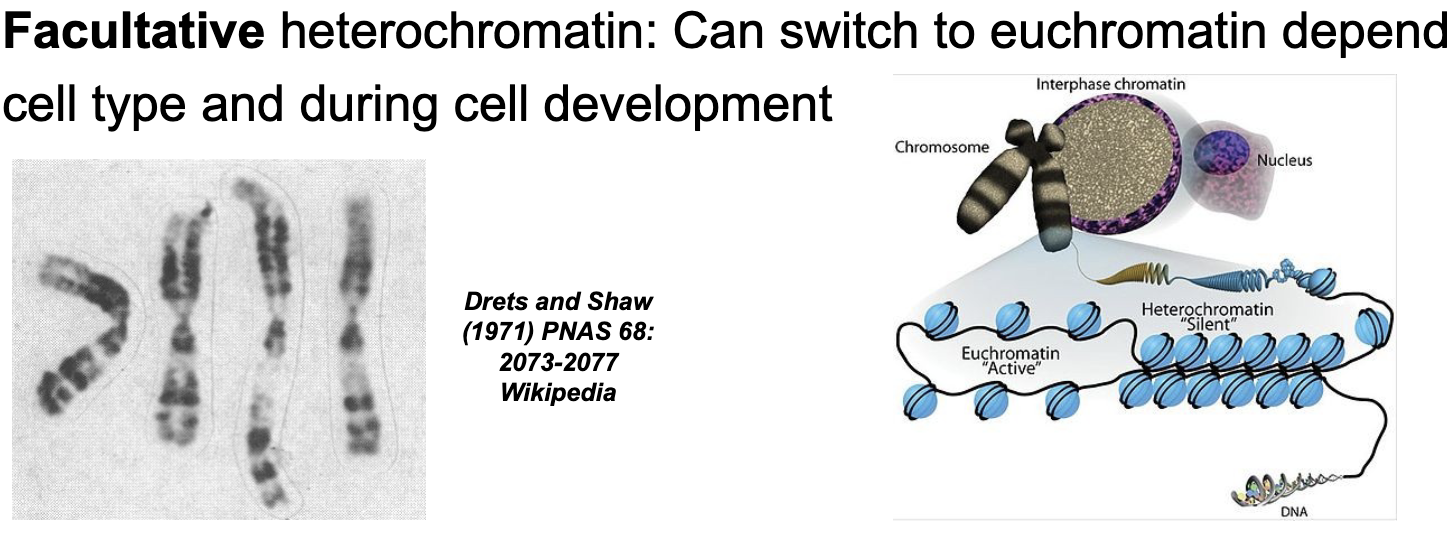
Heterochromatin are chromosomal regions of WHAT where gene expression WHAT
Heterochromatin are chromosomal regions of HIGH DNA COMPACTION where gene expression is CONSTITUTIVE or FACULTATIVE
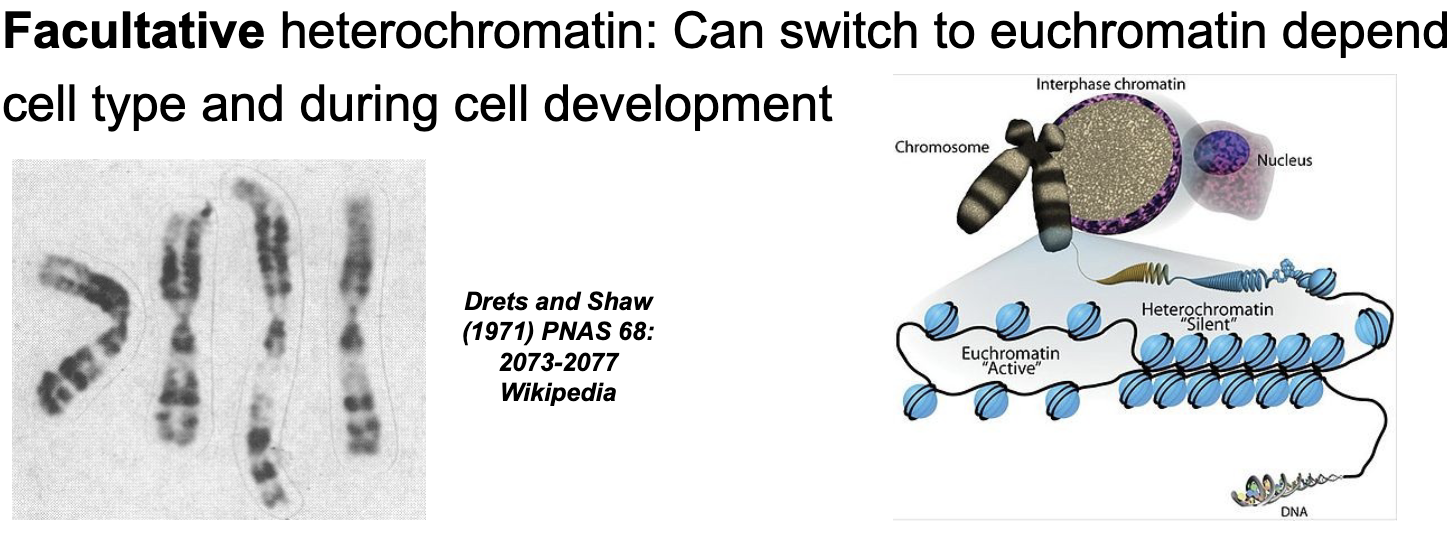
Constitutive (always) heterochromatin
DNA is ALWAYS highly compacted (centromeres and sub-telomeric regions)
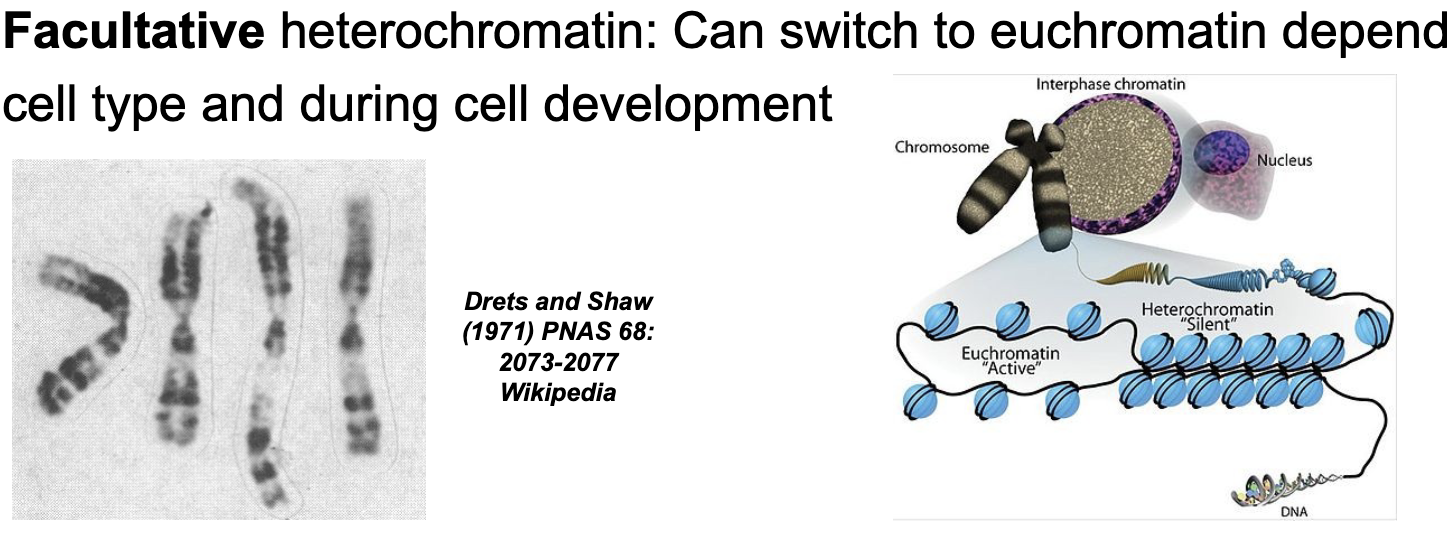
Facultative (sometimes) heterochromatin
Can switch to EUCHROMATIN depending on cell type and during cell development
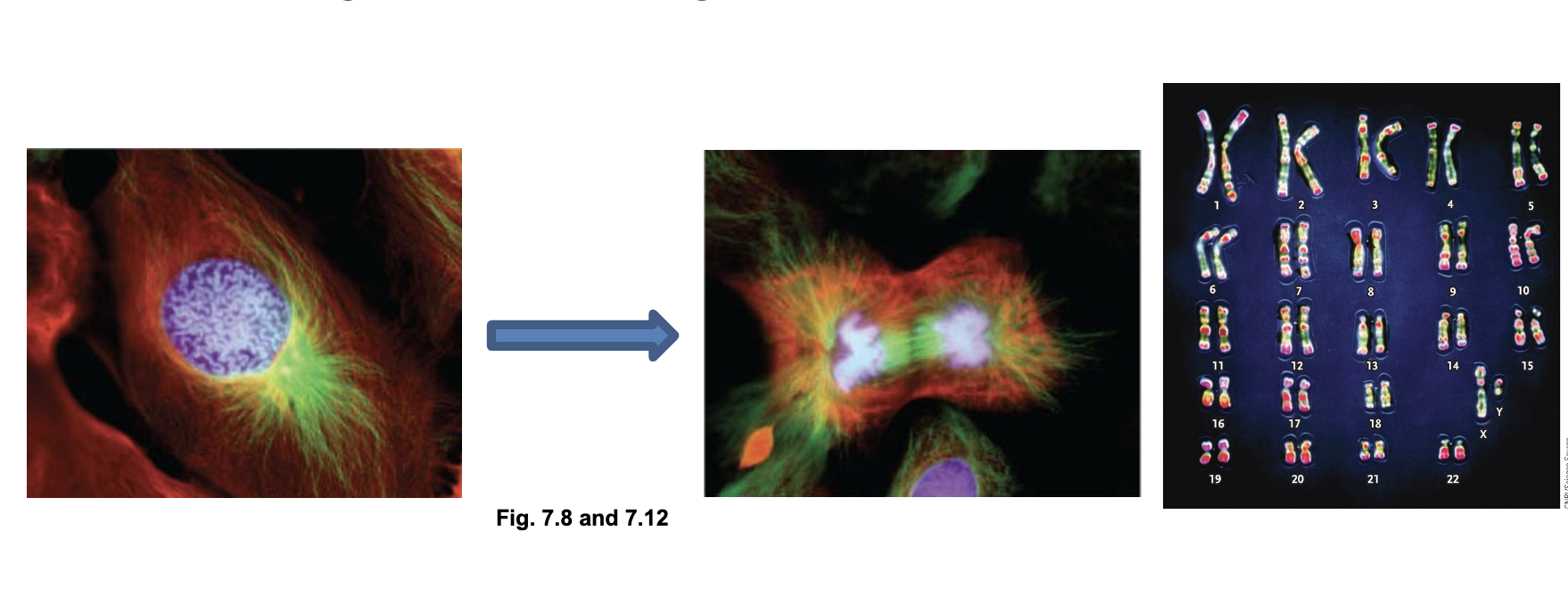
Chromosome organization:
Chromosomes are the WHAT from of DNA
Chromosomes structure WHAT DNA from WHAT
Chromosomes can be WHAT easily and transmitted to each WHAT during WHAT
Chromosome organization:
Chromosomes are the FULLY COMPACTED from of DNA
Chromosomes structure PROTECTS DNA from DAMAGE
Chromosomes can be SEPARATED easily and transmitted to each DAUGHTER CELL during CELL DIVISION
Chromosomes need to be fully WHAT (DNA WHAT) and properly transmitted to each WHAT during WHAT
Chromosomes need to be fully DUPLICATED (DNA REPLICATION) and properly transmitted to each DAUGHTER CELL during MITOSIS and MEIOSIS
Origins of replication
Multiple DNA sequences along chromosomes which INITIATE DNA REPLICATION
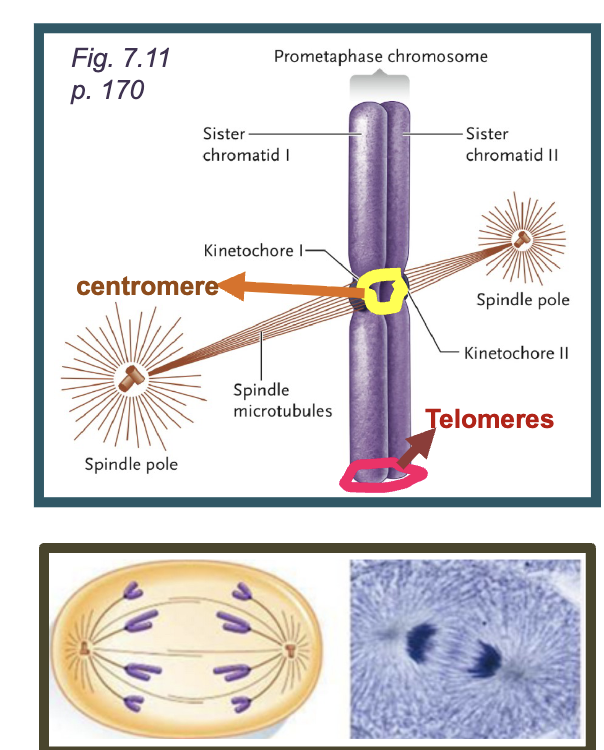
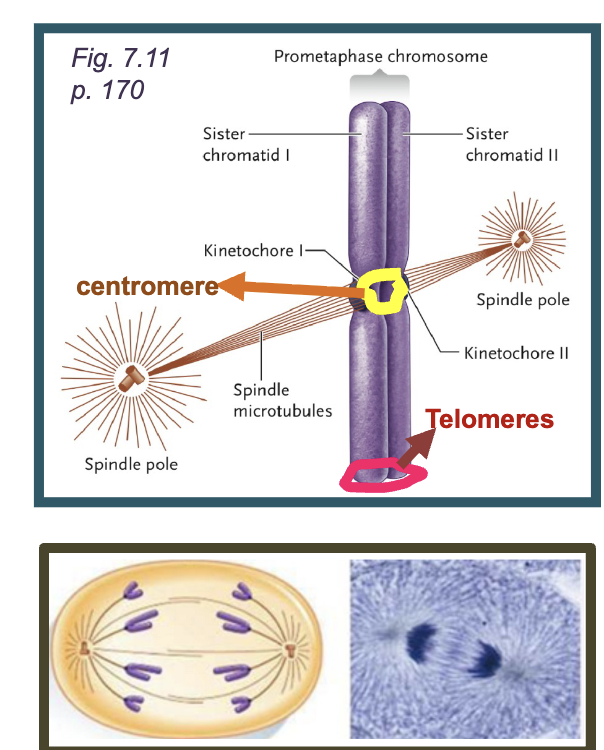
Centromere
DNA sequences required for correct segregation of chromosomes
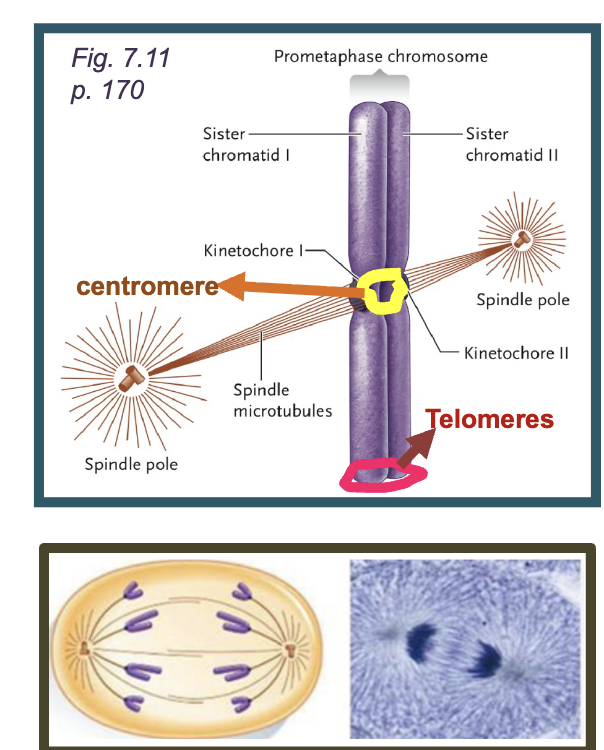
Telomeres
DNA sequences located at the ENDS OF THE CHROMOSOME that prevents degradation and allow proper replication
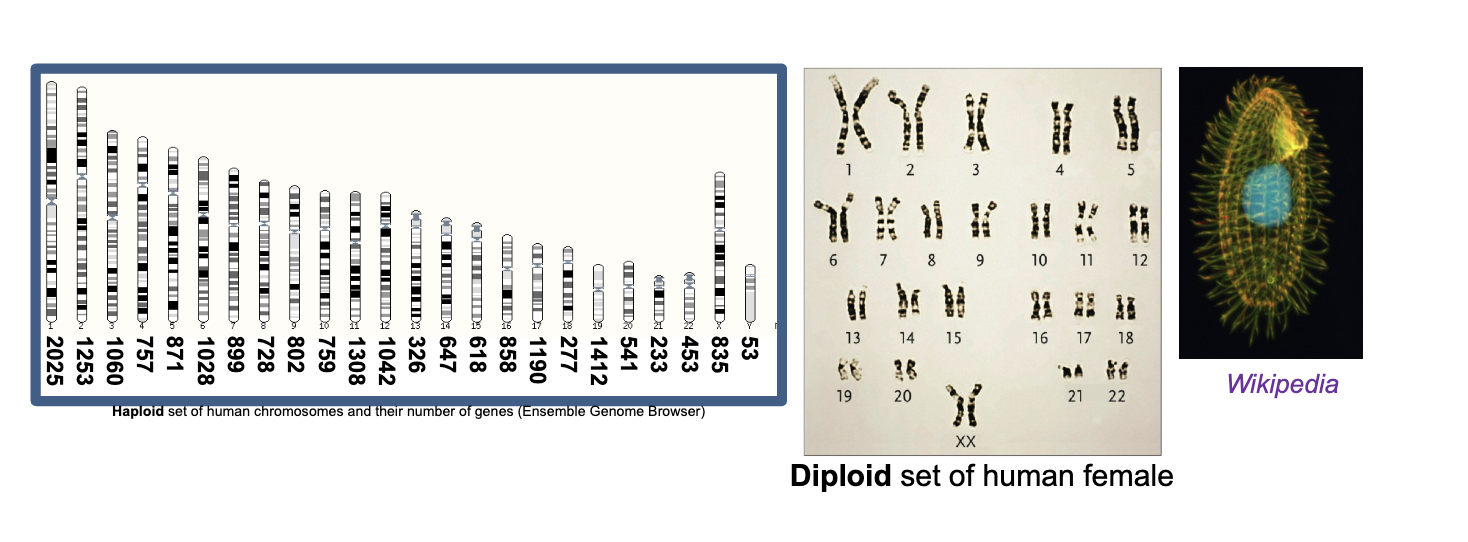
Majority of eukaryotic cells are WHAT (two copies of each WHAT chromosome)
Majority of eukaryotic cells are DIPLOID (two copies of each HOMOLOGOUS chromosome)
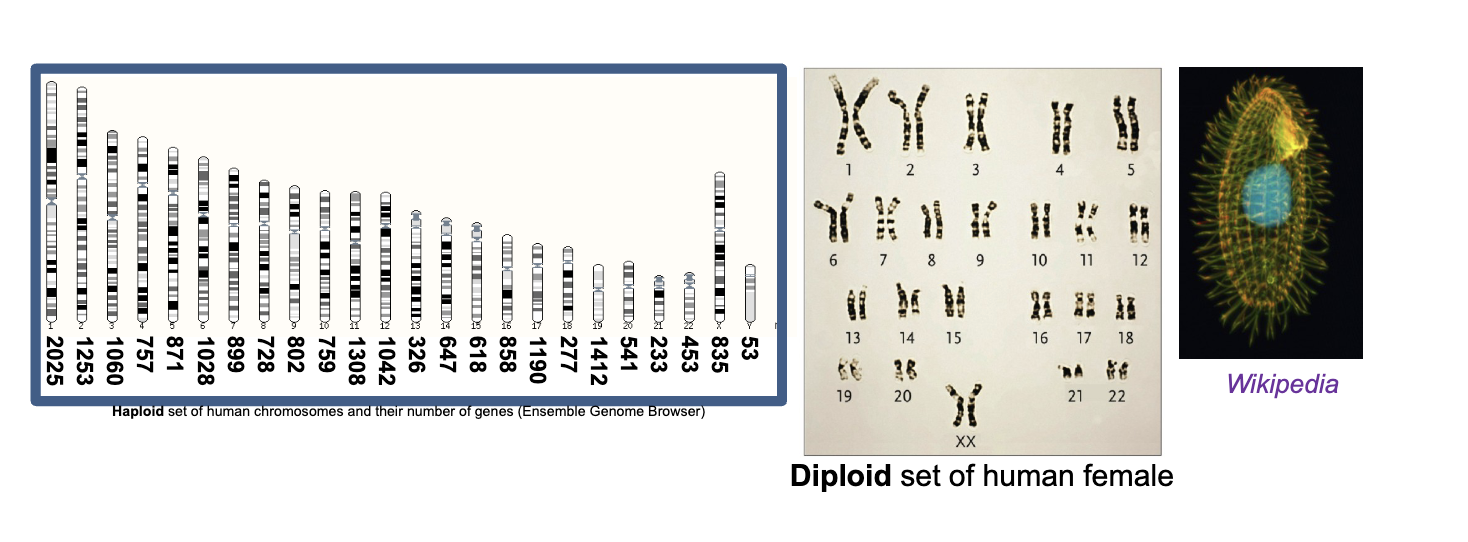
Only sexually-reproductive cells (sperm and ova) have a WHAT genome
Only sexually-reproductive cells (sperm and ova) have a HAPLOID genome
Some eukaryotes are WHAT (more than a pair of each chromosome) such as large WHAT and WHAT
Triploid = WHAT
Tetraploid = WHAT
Some eukaryotes are POLYPLOID (more than a pair of each chromosome) such as large PROTISTS and FLOWERING PLANTS
Triploid = 3 copies
Tetraploid = 4 copies
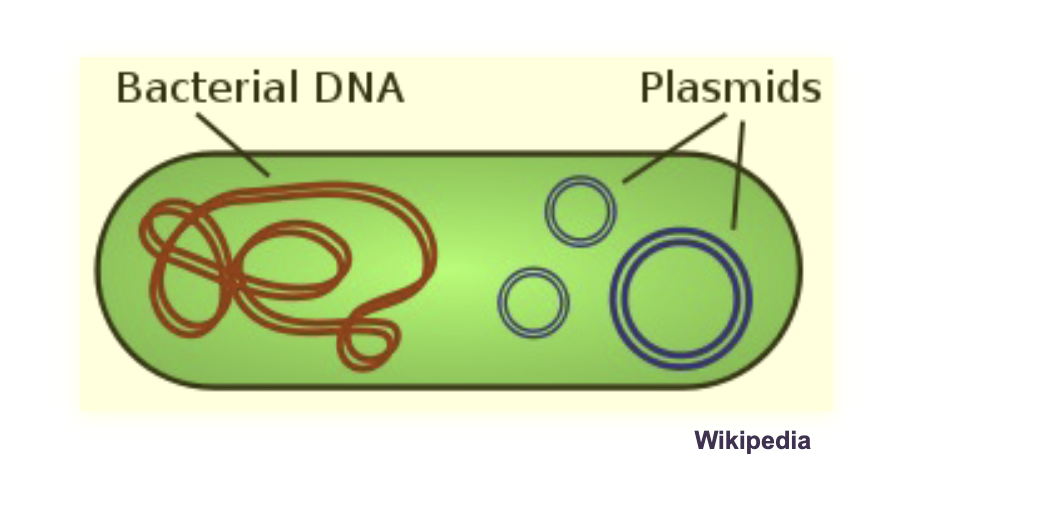
DNA organization is PROKARYOTES:
Prokaryotes typically have a WHAT DNA molecule found in the WHAT (no WHAT)
DNA organization is PROKARYOTES:
Prokaryotes typically have a SINGLE, DOUBLE-STRANDED CIRCULAR DNA molecule found in the CYTOSOL (no NUCLEUS)

DNA organization is PROKARYOTES:
Prokaryotic DNA does not need the same level of WHAT as eukaryotic DNA (because they are WHAT)
DNA organization is PROKARYOTES:
Prokaryotic DNA does not need the same level of COMPACTION as eukaryotic DNA (because they are CIRCULAR)
DNA organization is PROKARYOTES:
They use WHAT, also called WHAT (no WHAT)
DNA organization is PROKARYOTES:
They use HISTONE-LIKE PROTEINS (HLPs) , also called NUCLEOID-ASSOCIATED PROTEINS (NAPs) (no HISTONE)
DNA organization is PROKARYOTES:
Prokaryotes also have other small independent double-stranded circular DNA molecules called WHAT
DNA organization is PROKARYOTES:
Prokaryotes also have other small independent double-stranded circular DNA molecules called PLASMIDS
DNA organization is PROKARYOTES:
Each cell can have multiple different WHAT, as well as multiple WHAT of each plasmid
DNA organization is PROKARYOTES:
Each cell can have multiple different PLASMIDS (small DNA molecules), as well as multiple COPIES of each plasmid
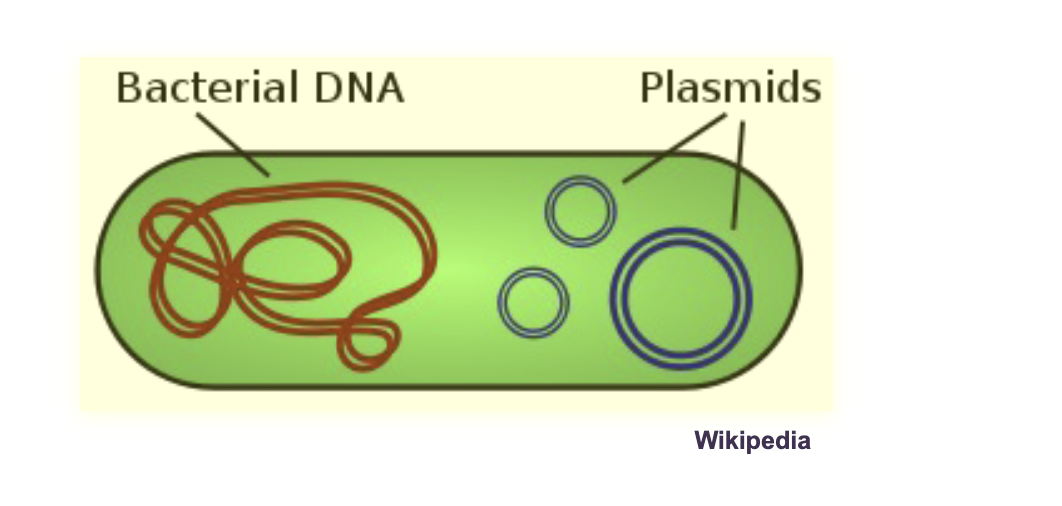
DNA organization is PROKARYOTES:
Plasmids can carry a few WHAT (1-10) - none WHAT
DNA organization is PROKARYOTES:
Plasmids can carry a few GENES (1-10) - none ESSENTIAL for life
DNA organization is PROKARYOTES:
Plasmids carry “WHAT” WHAT that give them an advantage in some environments
Such as
Metabolism of WHAT
WHAT (ability to cause disease)
WHAT
DNA organization is PROKARYOTES:
Plasmids carry “BONUS” GENE that give them an advantage in some environments
Such as
Metabolism of RARE CARBON SOURCE
VIRULENCE (ability to cause disease)
ANTIBIOTIC RESISTANCE
Uracil structure
Single (pyrimidines)
Only H
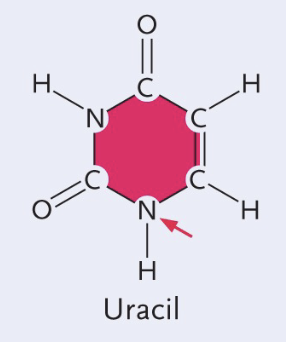
Thymine structure
Single (pyrimidines)
CH3 group
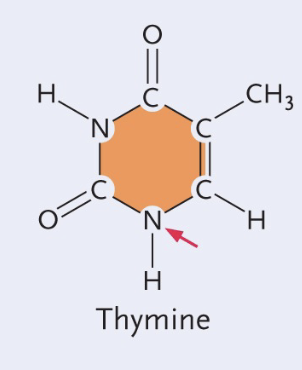
Cytosine structure
Single (pyrimidines)
NH2 group
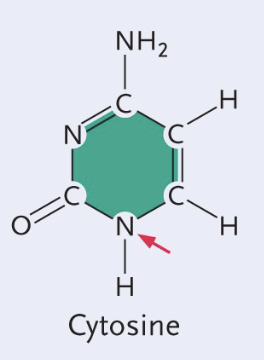
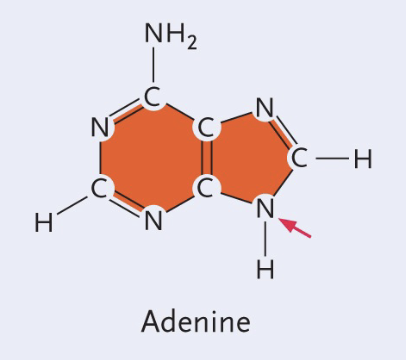
Adenine structure
Double (Purine)
No double bond O
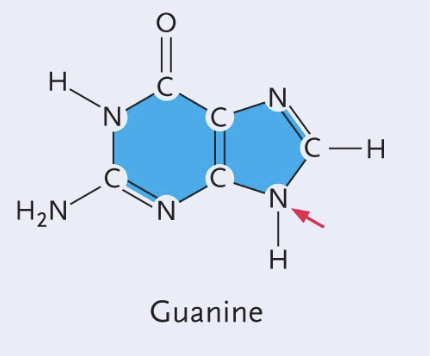
Guanine structure
Double (purine)
Double bond O