
Looks like no one added any tags here yet for you.
insertion; deletion; inversion
Site specific recombination can result in _______,__________or_______of DNA sequences. These are precise DNA rearrangements with defined limits and sites.
phage; plasmids; genetic expression
Site-specific recombination systems exist in many forms of cellular life and are also exploited by non-cellular replicators such as_______and________. They play roles in replication cycles of viral, plasmid and bacterial DNA and in the regulation of_________.
short DNA sequences; recombinase; homology
The minimum requirements for a site-specific recombination system are a pair of________ and a _________(usually structurally related to topoisomerase proteins). Some systems also include accessory proteins. These systems do not require large region of DNA sequence________, in contrast to homologous recombination.
inverted; non-palindromic; recombinase; recognition; homology; inversion; insertions; deletion; inversion; deletion
DNA Sequence Requirements:
The DNA sites recognized by site-specific recombinases usually include two_________repeat sequences of around 10-15 bp in length separated by a________core sequence. The sequence of the core can vary without affecting the binding of the________.
Recombination requires a pair of_________sites, and only pairs with identical cores can undergo recombination. This is the only sequence_________requirement.
The segment of DNA between the recognition sites can undergo_________, __________or __________depending on the orientation of the sites. If the sites are inverted relative to one another, the intervening DNA will undergo_______; if the sites are direct repeats, the DNA will undergo_________(or insertion if the sites are on different molecules)
inverted; cores
Each site comprises an_________repeat binding motif surrounding a core. Recombination occurs between a pair of sites with matching_______
recombinase; Tyr; Holliday; intermediate
Generic Pathway: Tyr Active Site
•The_______is a tetramer and each protomer recognizes a binding site (1).
•Pairs of active site_______attack the DNA and form covalent protein DNA intermediates (2).
•By exchanging partners, a_________structure is formed (3).
•Attack and resolution by the second pair of protomers resolves this_________(4 and 5).
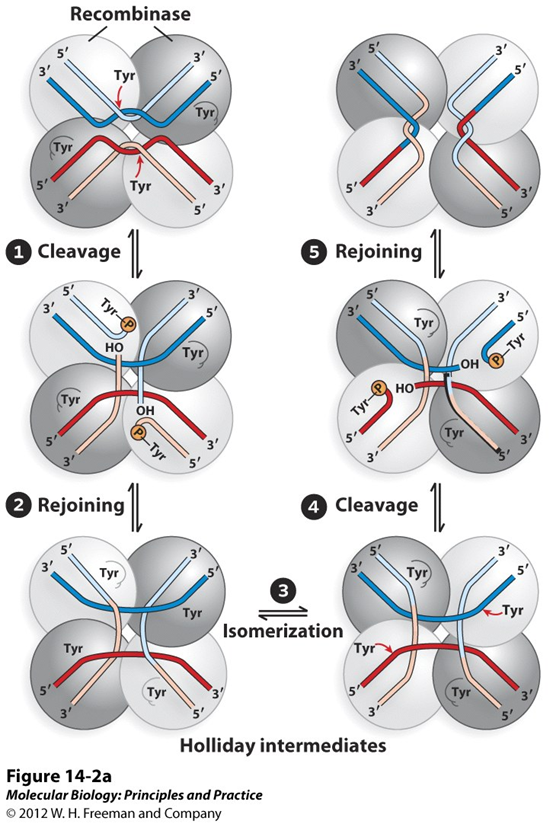
tyrosine; serine; phosphodiester; recombinases; homology; tyrosyl; Holliday
Protein Recombinases:
Site specific recombinases contain either a_______or a______in the active site. This residue will act as a nucleophile to attack the________backbone. The energy of the broken covalent bond is preserved in the phospho-tyrosyl (or phospho-seryl) bond generated between the enzyme and substrate. Thus the reaction is “energy neutral” and does not require a high-energy cofactor.
These_________acts as tetramers, which each protomer binding to a single half site (half of the inverted repeat flanking the core). Formation of the synaptic complex of the two sites relies on________between the two core sequences between the inverted repeats.
For________recombinases, the reaction takes place in two steps, with each protomer nicking the DNA at its bound half-site one time. The intermediate between the two steps resembles a________junction and requires limited branch migration in the region of core homology.
lysogenic; lytic
Two possible fates for a phage-infected host cell: ______pathway, _________pathway
dimeric; cross-over; site-specific recombination; resolved
Resolution of Replication Intermediate: E. coli XerCD/dif
The XerCD recombinase is responsible for resolving the________chromosomes that can result from__________ after recombinational repair in E. coli. XerCD binds to a chromosomal locus called dif whose sole purpose is to promote this__________reaction. XerCD is required for about 15% of chromosomal replication events, or about once every six cell divisions. This system is also linked to a segregation protein called FtsK, which assists in the segregation of the__________chromosomes.
lytic; lysogens; lambda; P1; circularizes; attP; attB; lambda integrase; IHF
Lambda and P1 Phage Lysogeny: Int/attPB and Cre/lox:
“Temperate” phage can carry out______growth—during which they amplify their genomes and kills the host cell—or can exist as________. A lysogenic phage is either integrated into the host chromosome (e.g. _________) or persists as a stable, autonomously replicating plasmid (e.g. _____). The equivalent of a lysogenic phage in eukaryotic viruses is a pro-virus, a very common stage in the retroviral lifecycle among others.
Lambda phage enters the host cell as a linear, monomeric genome that immediately___________via the complementary cos sites on both ends. Under appropriate conditions, the circular phage genome can then lysogenize using the________site on the phage and the_____site on the host chromosome, which is located between protein coding sequences and does not disrupt any cellular processes. Integration is carried out by the ___________ (Int) protein with the assistance of the_____protein. Excision requires additional accessory proteins.
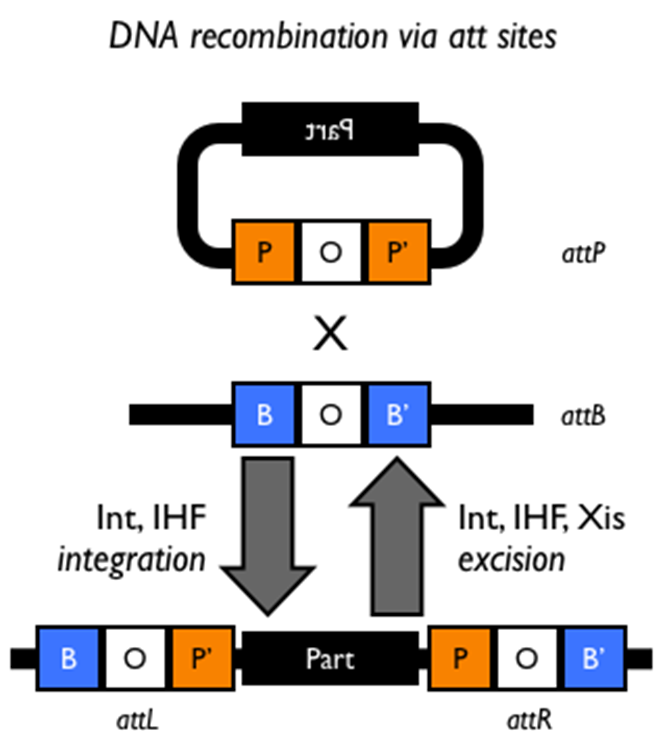
SSR recombinase; IHF; att
Lambda Lysogenization:
•Lysogenization and excision of l phage is based on a phage encoded ________ called l integrase as well as the host encoded_______.
•Note how the______sites on the phage and chromosome combine when the phage is integrated.
•The phage encoded Xis protein promotes excision of the phage genome under conditions that favor induction (eg UV irradiation).

rolling circle replication; loxP; Cre recombinase
The cre/lox system allows resolution of_______concatemers generated by____________into unit length circular phase genomes
P1 phage enter the cell as long linear concatemers, each flanked by a _____site, which exists as direct repeats relative to one another. Recombination between these loxP sites is carried out by the phage-encoded________, generating individual circular P1 plasmids. P1 can persist as a lysogen in plasmid form until the conditions are right for lytic growth at which point the plasmid is amplified by rolling circle replication.
Flp; FRT; copy number; replication; inverted-repeat; rolling circle replication; Flp; FRT
Amplification of Plasmid Copy Number: 2µ (two micron) Plasmid Flp/FRT system:
The 2µ plasmid of yeast encodes the_________ site-specific recombination system that allows it to modulate its_______. The two FRT sites on the plasmid are located asymmetrically relative to its________origin. Thus after bidirectional replication has initiated at the origin, one site is replicated prior to the second. Recombination between the two_________FRT sites inverts about half of the plasmid sequence and changes the orientation of one of the replication forks. The two forks then chase one another around the plasmid in a manner similar to_________, amplifying the plasmid sequence. ______then recognizes pairs of directly-repeated _______sequences and resolves the replication product into individual 2µ plasmids.
Hin/hix; host specific; FljB; FljC; inverted; transcription; Hin recombinase; FljC; topological; recombination
Regulation of Gene Expression: S. typhimurium phase variation:
Salmonella is a pathogen of the human intestine that uses the ______site-specific recombination system to evade the _________ immune system. The bacterium can switch expression between one of two flagellin proteins (___and______) in a process called phase variation. Flagellin expression is based on the orientation of a promoter flanked by_______repeats of the hix sites. In one orientation, the promoter drives expression of FljB as well as FljA, a repressor protein that binds to the FljC promoter and prevents it’s______. When the promoter region is inverted through the action of the ______, FljB and the repressor FljA are no longer expressed. In the absence of FljA, _____can be transcribed, ultimately leading to a change in the proteins that make up the bacterial flagellum.
The Hin/hix system undergoes a “________filter” which constrains the system to sites that are present on the same, supercoiled molecule. This prevents_______from occurring between hix sites on two different copies of the chromosome, potentially causing deletions and rearrangements outside of the Flj ABC operon. The topological filter is mediated by additional auxiliary proteins.
flagellin; promoter; reversed
Phase variation in Salmonella: Hin recombinase and hix:
•Phase variation relies on alternating expression of fljB and fliC genes that encode for________proteins.
•Altered expression occurs when the orientation of the_____for the flj operon is__________.
knock-outs; knock-ins; transgenes; targetable vectors; homologous recombination; site-specific double-strand; CRISPR; chimeric; transgene
Homologous Recombination Applications:
Genetic _______ (inactivations), ________(replacements) and______are introduced by the process of homologous recombination, relying more or less exclusively on functions already present in the target cell/organism.
__________consist of a positive selectable marker (e.g. antibiotic resistance gene) flanked by long regions of homology. They may also include a negative selection marker (e.g. a lethal proteins) outside of this region.
The vector is introduced into murine embryonic stem cells and, hopefully, incorporated into the desired locus via________. The cells are then grown out under positive selection, and surviving clones are analyzed to confirm that the vector has been correctly targeted. __________breaks in the target locus can be introduced using_______technology, significantly increasing the likelihood of accurate targeting.
Confirmed ES clones are injected into mouse blastocysts (from a different strain of mouse) which are transferred to female mice to produce_______offspring. These are interbred to identify mice in which the ________-has entered the germline. Such mice can be further interbred to analyze the phenotype.
CRISPR; Cas9
CRISPR/Cas 9
•Anti-phage system discovered in S. pyogenes. Bacterial adaptive immune system.
•_________: Clustered regularly interspersed short palindromic repeats
•_______: CRISPR associated (gene/protein) 9
•Integration of segments of previously encountered phage leads to phage destruction upon reinfection.
Cre recombinase; direct repeats; Cre recombinase; conditional knock-outs
Site-Specific Recombination Applications:
Simple site-specific recombination systems, such as the Cre-lox system, have been exploited for manipulation of the (murine) genome as well as numerous other applications. Many mouse strains have been developed that express the_________under the control of a tissue-specific promoter (e.g. it is only expressed in developing neurons). These mice can then be bred to strains in which the gene of interest has been modified such that it is flanked by lox________. Thus the gene will only be deleted in tissues where the___________is expressed. Such “________” are particularly useful for analysis of proteins for which the regular knock-out is embryonic lethal (e.g. proteins that are required for early embryonic development but may or may not be required in certain adult tissues).
knock-out; double-nicking; Cas9
CRISPR/Cas9 Applications:
A)Introduction of a dsDNA break can lead to gene_______or replacement.
B)More complex gene replacement can be achieved with______strategy using a Cas9 mutant.
C)_______fusion proteins can be specifically targeted and used for a variety of purposes.
homing endonucleases; Zn Finger Nucleases; TALENs
Other Gene Editing Systems:
_________: Large sequence-specific endonucleases. Limited Flexibility.
________: Customizable DNA triplet sequence specific-proteins fused to the FokI cleavage domain. Moderate flexibility.
______: Customizable single nucleotide sequence-specific proteins fused to FokI cleavage domain. Good flexibility.
double stranded breaks; target; know
Gene Editing Systems:
•They introduce__________ (or sometimes single stranded nicks) at specific sequences.
•They have off________effects. This must be controlled for in any experimental use of the technology, and can be prohibitive for therapeutic uses.
•You have to_______what to edit.
A; X; homology; Y; dsDNA
Knock-Ins and Knock-Outs:
•The gene of interest (____) is disrupted by a positive selectable marker (___) flanked by long regions of ______. The negative selectable marker (____) determines the boundary for HR and guards against random insertion.
•Accurate targeting can be increased by introduction of a ____ break at the target site using gene editing technology.
mutant; blastocyst; chimeric
Engineering a KO mouse:
Introduce DNA containing_______gene
Inject altered ES cells into_________stage of embryo from brown mouse
Implant embryo into surrogate mother mouse
Surrogate mother gives birth to_________pup; this pup has cells from the black strain and cells from the brown strain
embryonic; recombinase
SSR: Conditional KOs
•Conditional knock-outs are particularly useful for gene that are________lethal. They allow the researcher to bypass development and look in a specific tissue.
•__________expression can be under tissue-specific control or can be drug- or light-inducible.
attsite recombinase; monoclonal
SSR: Protein Therapeutic Production:
•________technology allows for integration of genes of interest into a specific site in the CHO cell line genome.
•This leads to high levels of protein expression, sufficient for industrial scale production of therapeutic proteins such as monoclonal antibodies.
site-specific recombination
Each neuron undergoes stochastic__________at two cassettes (one on each chromosome). This leads to a rainbow of fluorescent neurons. The long axons can be easily distinguished and traced.