BCH210
1/427
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
428 Terms
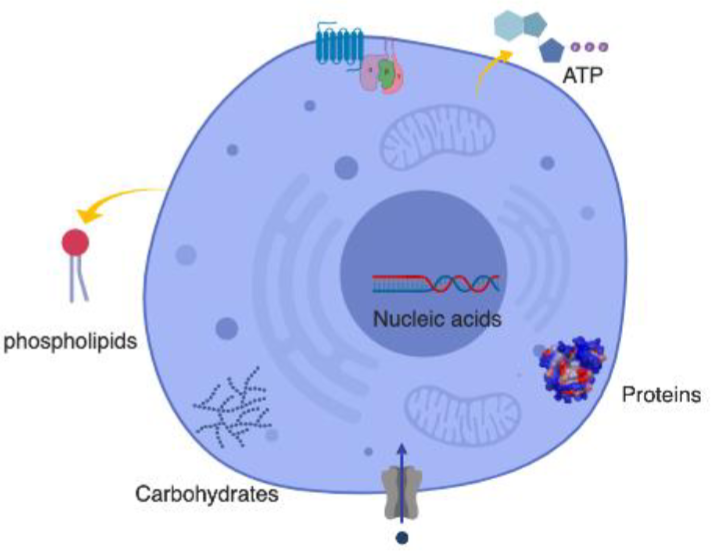
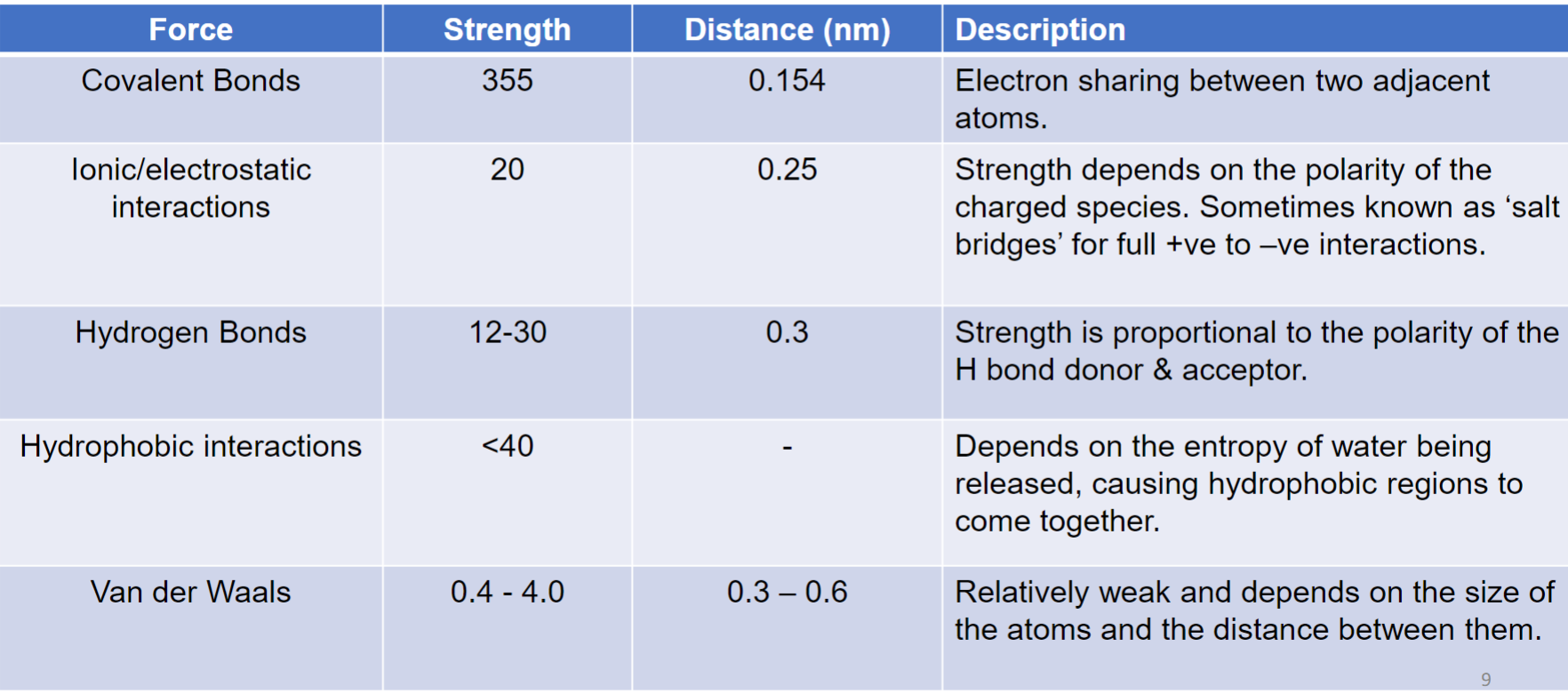
* Lipids (fats, cholesterol, hormones)
* Sugars (nrg, cell struct)
* Nucleic Acids (DNA, RNA)
* Small molecules (vitamins, metabolites)
* Ions (K+, Na+, Ca2+, Cl-)
* Water (H2O)
* structure of protein/sugar/lipid is important for it's function
* cellular diseases arise when a genetic mutation affects the structure and function of a protein
* proteins play a variety of roles including catalysis, structure, and recognition
* can be recognized through structure or cofactors
* non cov allow chains to fold into final struct
* cofactors may be bound cov/noncov
* transporters may also bind molecules non-cov
* chem func groups are responsible for mediating this binding
* 2 types: (can also be loosely or tightly bound to protein)
1. essential ions (inorganic)
2. coenzymes (organic)
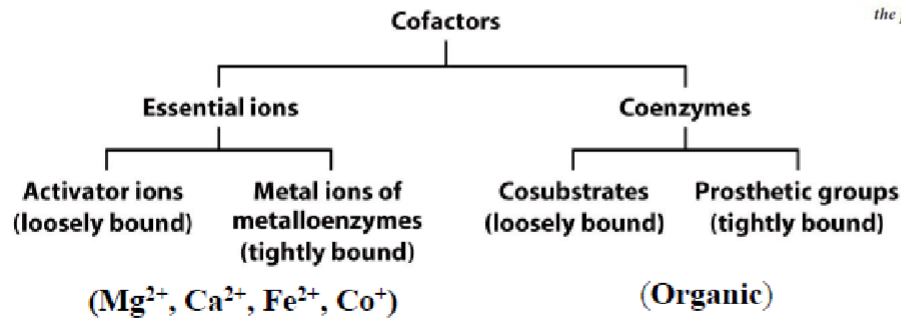
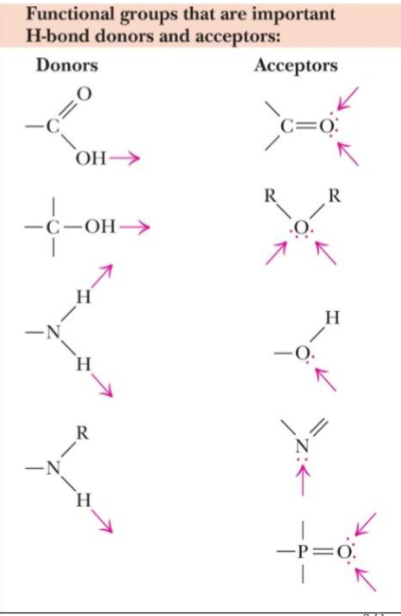
* acceptors: EN atoms (has lone pairs)
* donors: H atoms covalently bound to EN elements (O, N, F, Cl, Br)
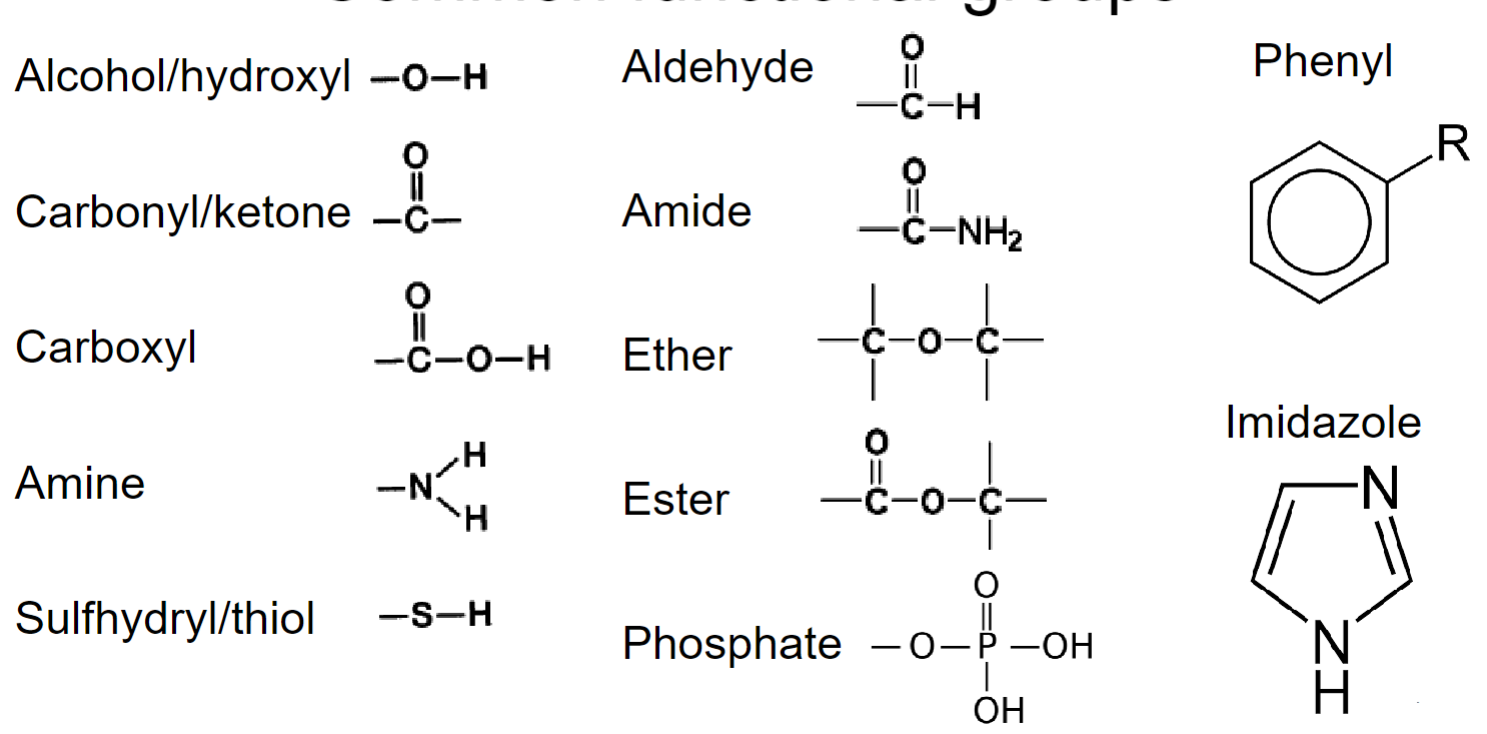
* __**amides:**__ important for protein struct
* __**Phosphate:**__ important for regulation (very polar)
* __**Phenyl:**__ important for R group (amino acid side chains), impedes ability to form H-bonds (resonance)
* important for molecule solubilization and formation of complex struct
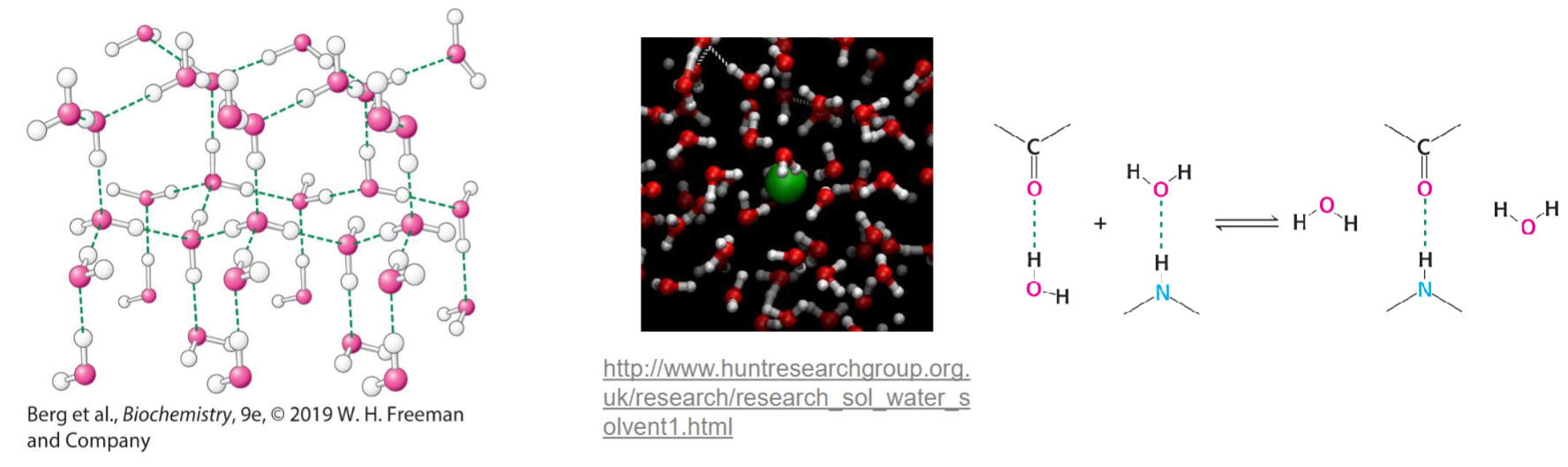
* water can solubilize molecules that have hydrophilic func groups (polar)
* amphiphiles: both hydrophobic/philic
* main driving force behind formation of macromolecular struct (helps w/ protein folding
* seems to contradict 2nd law of thermodynamics (entropy (disorderness) always increases in spontaneous rxn)
* but decrease of entropy of struct causes increase in water’s entropy (leading to overall increase in entropy
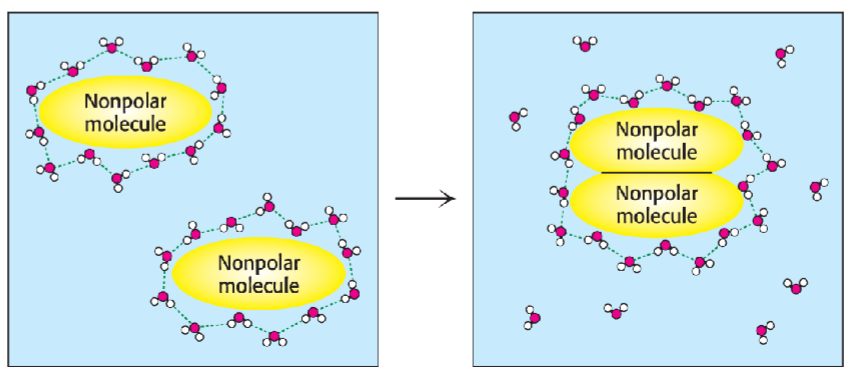
* depending on how macromolecules fold, rxns could be prevented from occurring but maximizing # of interactions of hydrophilic func groups w/ H2O for solubility
* binding interactions + intermolecular forces are important considerations
* presence/absence of func groups can assist w/ solubility (water/lipids) and binding (H-bonds, ionic, hydrophobic)) to specific targets
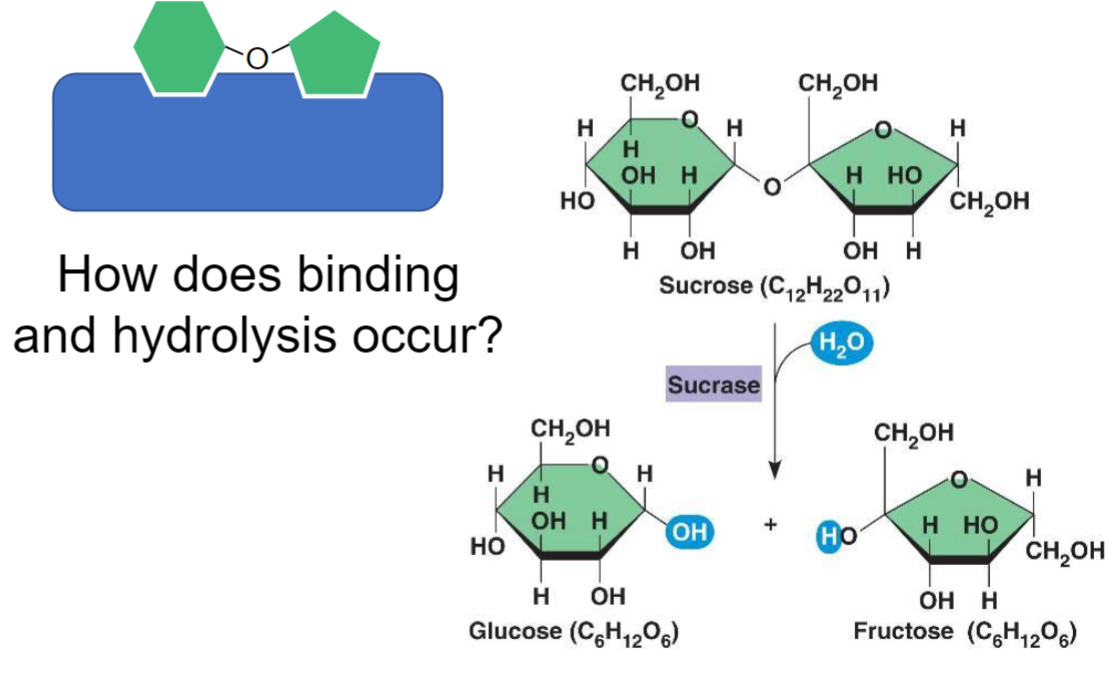
Major categories:
1. proteins
2. carbohydrates (most complex, variety of subunits + branched struct)
3. nucleic acids
1. basic units: (a.a., sugars, nucleotides)
2. connection bond: (peptide, glycosidic, phosphodiester)
3. structure: linear vs. branched
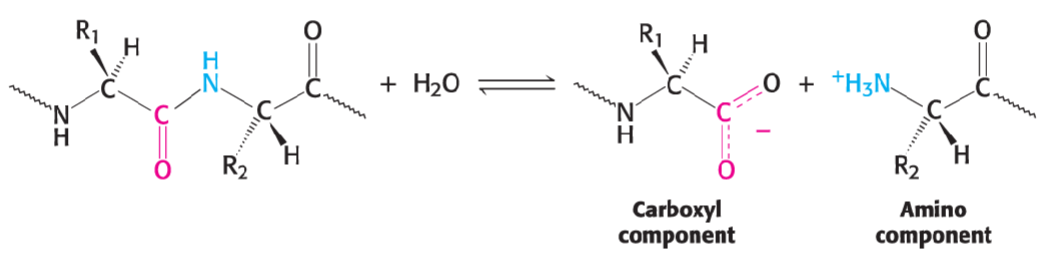
* amino group (NH3) (+)
* carboxy group (COO) (-)
* R side chain
*peptide bond made in condensation rxn btwn amino group and carboxyl group*
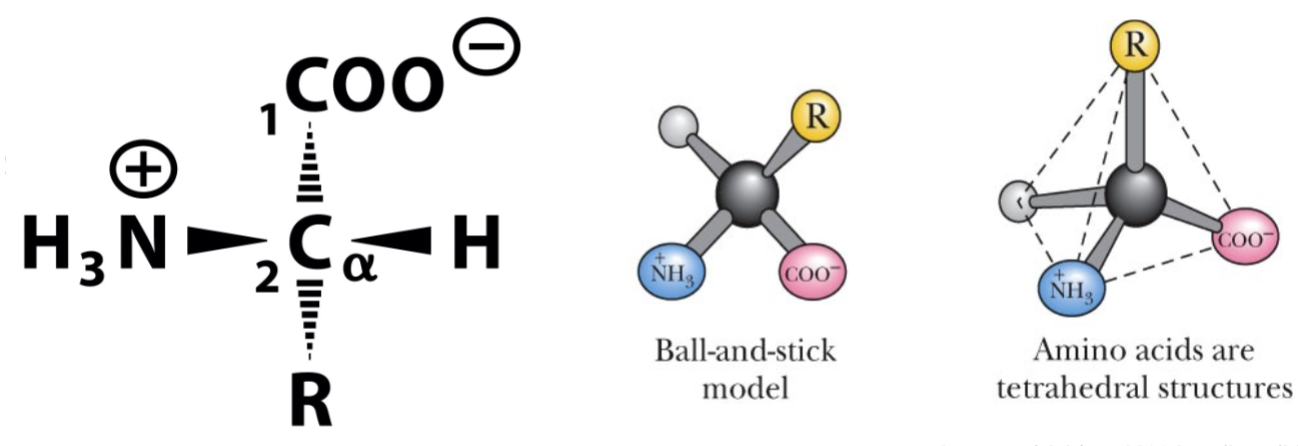
* chiral molecules: non-superimposable, mirror images (enantiomers)
* isomers exhibit optical activity
* __**L amino acids are physiologically relevant**__ in plant and animal proteins
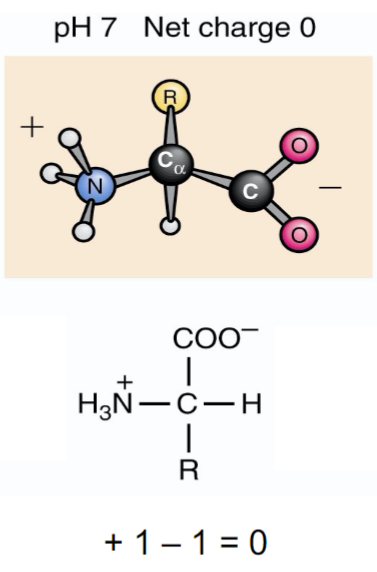
* R side chain needs to be uncharged for amino acid to be zwitterons
* hormones
* neurotransmitters
* nitrogenous bases (DNA/RNA)
* Energy-producing intermediates
* non-cov/cov interactions btwn func groups r improtant for holding protein together + its ability to interact w/ other molecules
* interaction of hydrophillic amino acids w/ H2O help solubilize some proteins
* H-bonds can also form btwn amino acid side chains w/in protein’s struct (every 4 residues) (folds into more compact struct)
* hydrophobic interactions can occur btwn aliphatic + hydrophobic side chains
* __**ionic interactions**__ are important for __**ligand, cofactor and/or substrate binding in enzymes**__
* salt bridges can form btwn positively + negatively charged amino acids
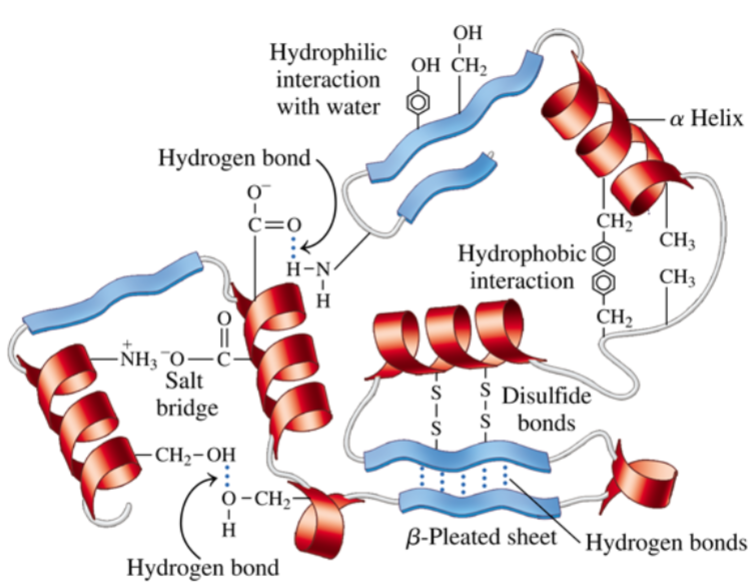
* disulfide bridges can be intrachain/btwn diff polypeptide chains
* these linkages can __**stabilize struct**__
* Protein Disulfide Isomerase (PDI) enzymes help catalyze this oxidation rxn
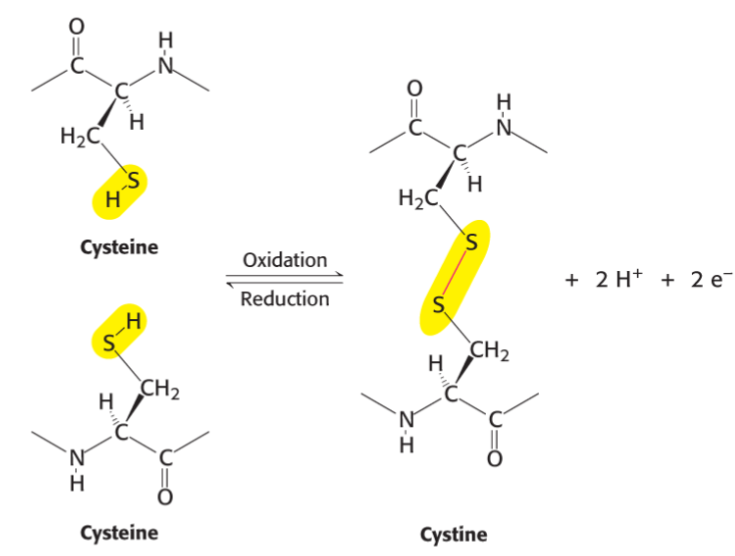
* cytosolic proteins usually contain cysteines due to reducing nature of cytosol
* disulfide bonds can be broken by reducing agents in cytosol or lab
* disulfide bond formation is type of post-translational modification
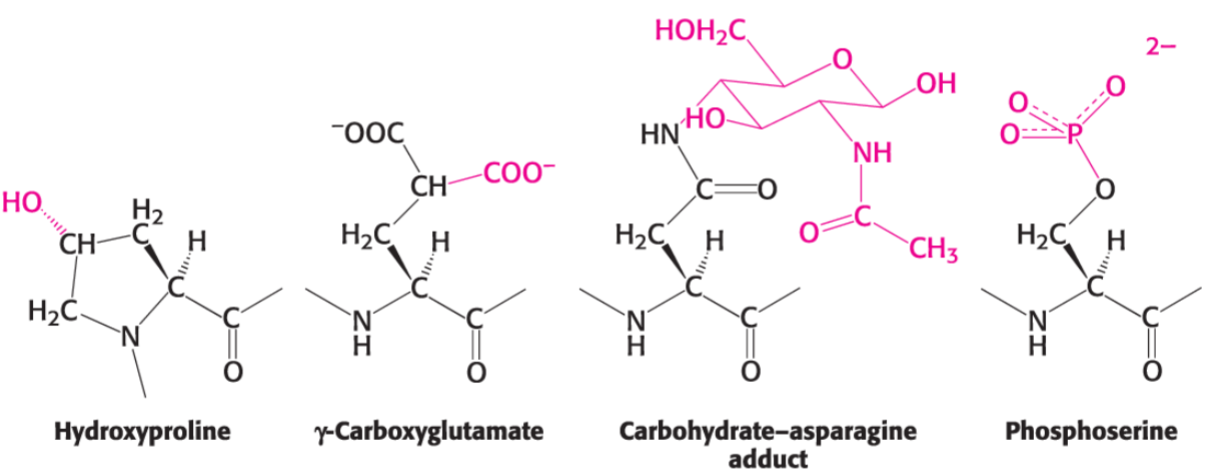
* important covalent modifications:
* phosphorylation
* ubiquitination (proteins tagged w/ ubiquitin for degradation)
* glycosylation
* acetly, methyl, hydroxyl, carboxyl
* cofactor/ligand binding also important for struct/func
* amino acid changes can influence absorbance spectra
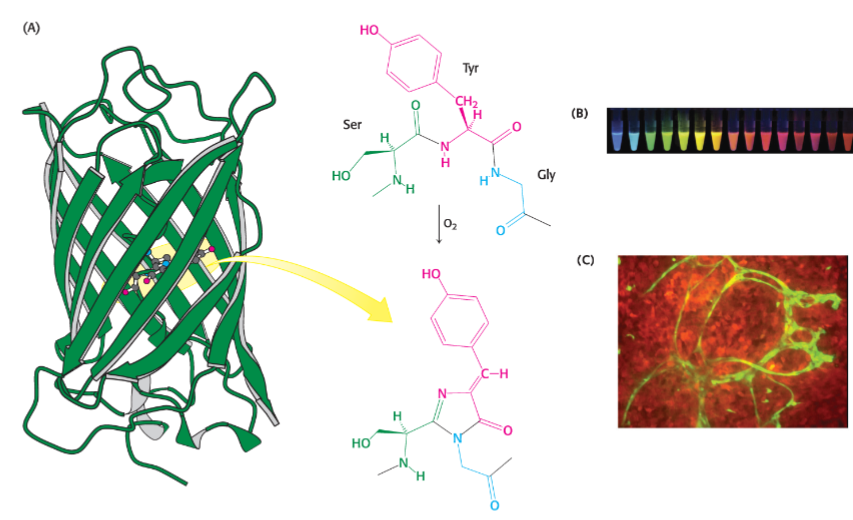
* can be used to elucidate proteins func using homology searches
* mutations in primary sequence can lead to change in func or disease
* are conserved for struct and/or func
* pH = -log \[H+\]
* at lower pH values, more H+ are present to protonate diff func groups
* pH of solution important since func groups can also act as weak acids/bases, losing + accepting H+ at diff pHs
* proton removal of weak acid HA is described by:
* Ka = (\[H+\]\[A-\])/\[HA\], pKa = -log Ka
* strong acids have high Ka (high dissociation) and low pKa
* pKa value tells you the pH at which func group loses/gains its H+
![* used to measure strength of acid (HA)
* proton removal of weak acid HA is described by:
* Ka = (\[H+\]\[A-\])/\[HA\], pKa = -log Ka
* strong acids have high Ka (high dissociation) and low pKa
* pKa value tells you the pH at which func group loses/gains its H+](https://knowt-user-attachments.s3.amazonaws.com/23a1a3b9d176484bb6240bbbf2bc9899.jpeg)
* derived from acid dissociation constant eqn (Ka)
* pH = pKa + log (\[A-\]/\[HA\])
* can be used to determine pH of mixture of weak acid and conjugate base
* proteins contain func groups that must be protonated/ deprotonated to allow for non-cov interactions to take place
* buffers r solutions of weak acids + conjugate bases that can resist changes in pH, maintaining protein’s struct and/or func
* buffers can neutralize small additions/loss of H+, keeping pH of solution stable
* buffers maintain pH +/- 1 pH unit around pKa
* consider chem stability (are there any chem rxns that may occur producing molecules that will affect your protein, cell or experiment)
* does buffer interfere w/ any of your experiments
* cost/availability?
* Blood pH: 7.35-7.45
* Stomach: 1.5-3.5
* Intracellular pH:
* cytoplasm: 7.2-7.8
* lysosomes: < 5.5
* golgi: 6.0-7.9
* mitochondria: 7.8
* some cells may also survive in acidic/alkaline environments
* 3 buffering regions exist around 3 pKa values
* isoelectric point (pl) is pH when charge of molecule is zero
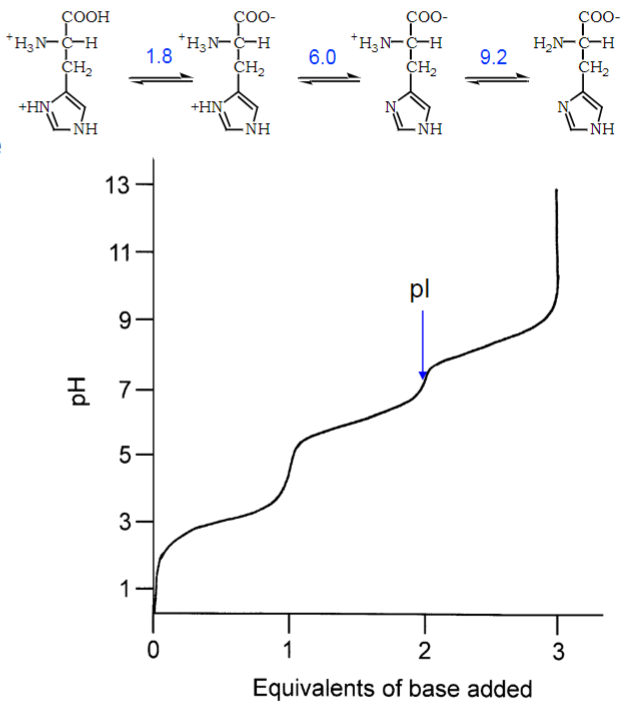
* pH also affects H-bonding that is important for enzymatic func and ability to interact w/ H2O and other binding partners
* chem environment of func group can influence pKa value
* buffers are important for maintaining stable pH environment in order to study protein/biological process
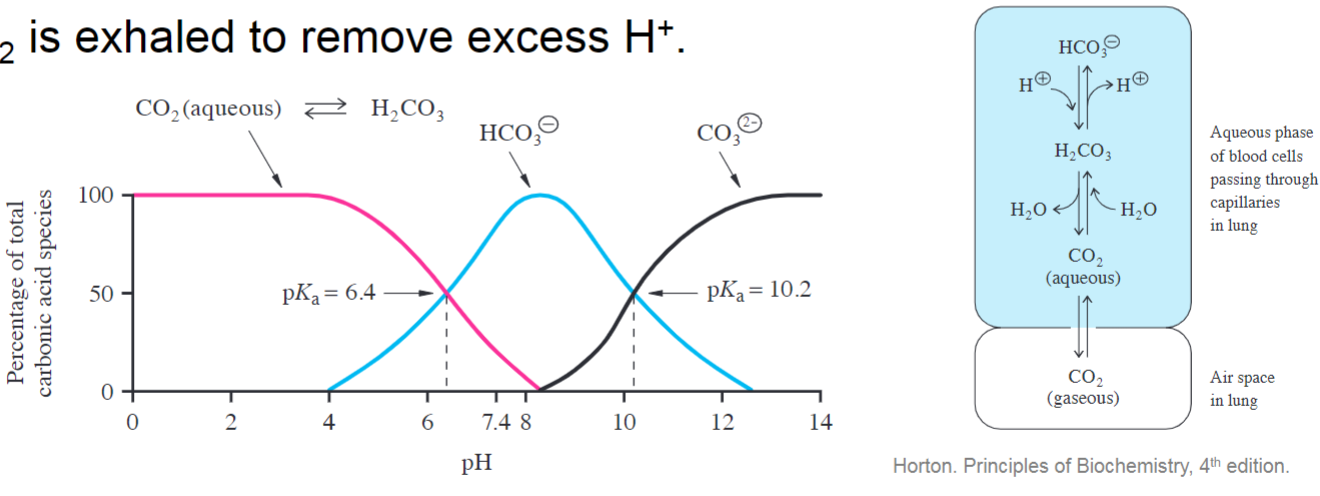
* bicarbonate is main buffering species, accepting and donating H+ to prevent changes in pH
* CO2 is exhaled to remove excess H+
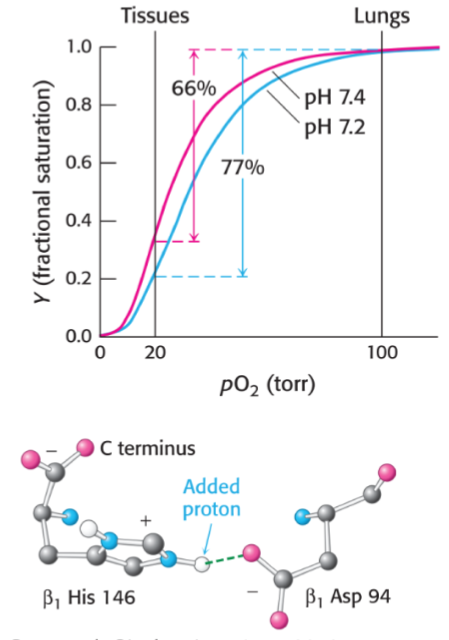
* when CO2 is made in tissues and combines w/ H2O to make bicarbonate and H+, this helps O2 release
* at lower pH, His146 is protonated and creates a salt bridge to Asp94
* favours the deoxygenated struct of hemoglobin
Essential vs nonessential Amino acid (tut 2)
Essential:
amino acids can’t be produced by body and need many chemical reactions to be made
must come from food sources
Nonessential:
can be produced by body, easily made from intermediate metabolites
Protonation state (tut 2)
pH < pKa = protonated
pH = pKa = 50% mix of both species
pH > pKa = deprotonated
Levels of protein structure (lec 6)
Primary (deg. 1): linear sequence of amino acids encoded by DNA
Secondary (deg. 2): periodic regular structures (alpha helix, beta strands/turns) (H-bond interactions form these structs)
Tertiary (deg. 3): folding of secondary structures into define protein motifs and domains
Quaternary (deg. 4): assembly of distinct chains into multi-subunits structures
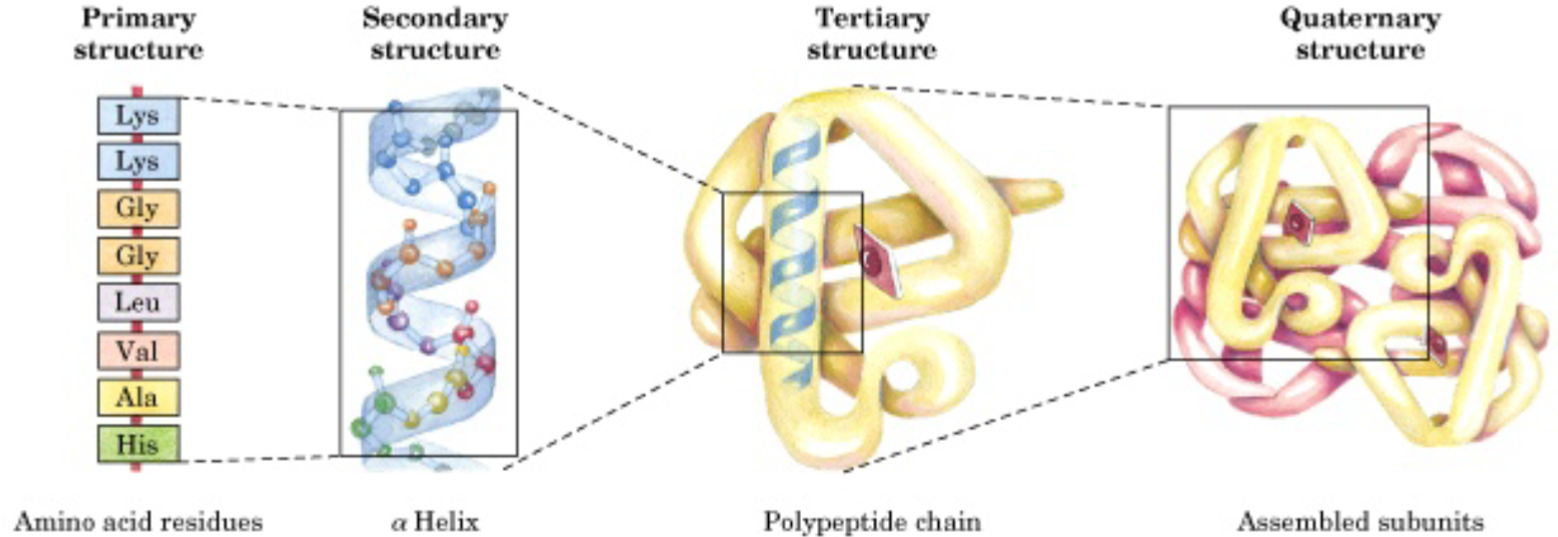
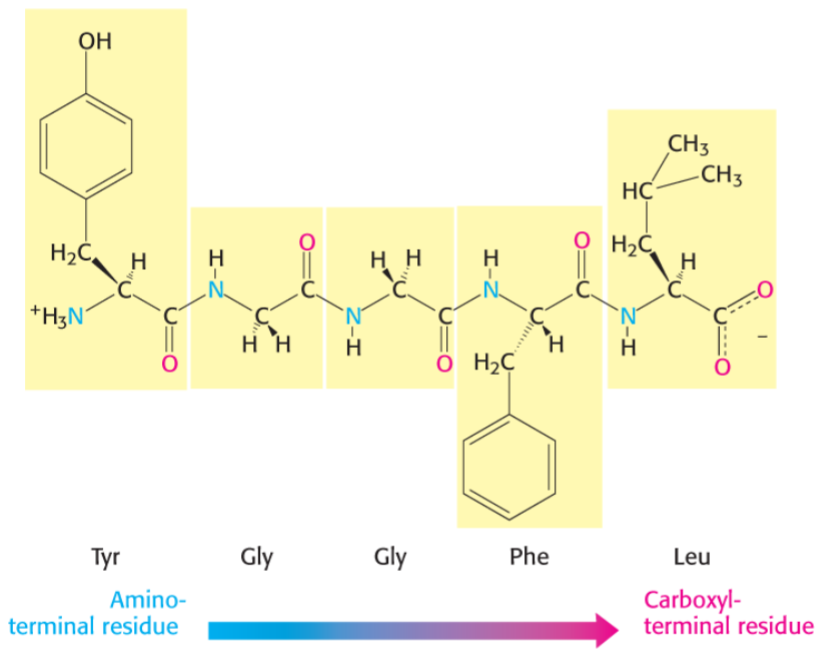
Primary structure directionality (lec 6)
amino acids are joined enzymatically in condensation rxn
polypeptide chain has directionality, amino terminal is start of chain
backbone consists of peptide bonds and alpha carbons, while variable part are distinct R side chains
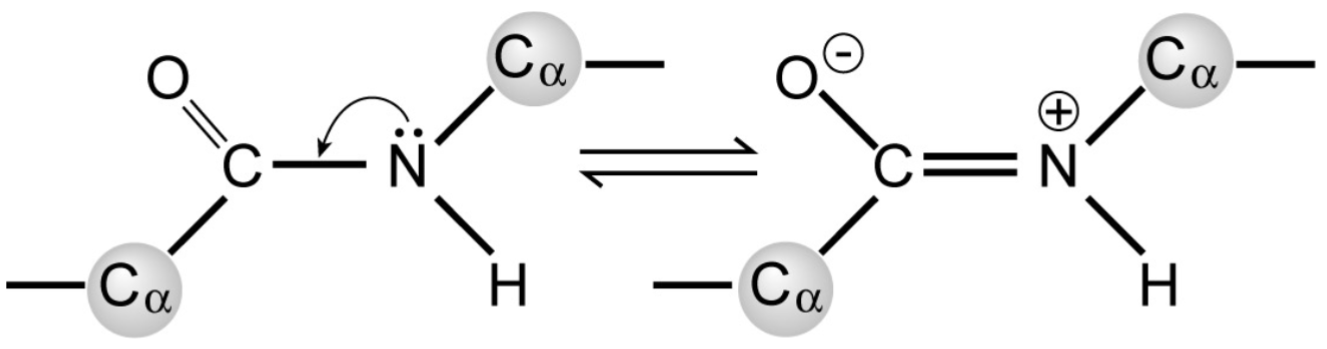
Peptide bonds (lec 6)
are planar
polar but uncharged
have partial double-bond character due to resonance, preventing rotation of peptide bond
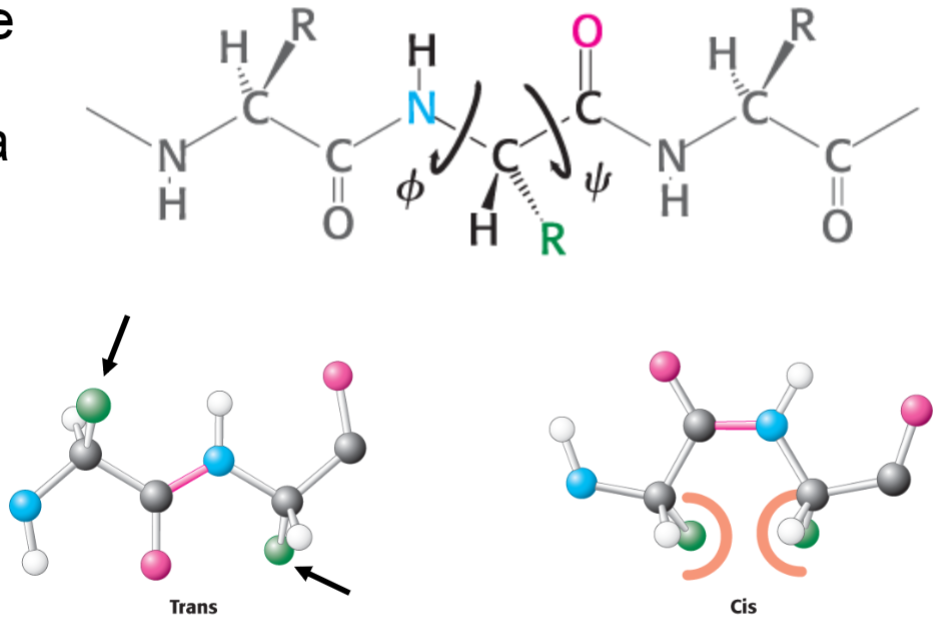
Polypeptide chains (lec 6)
peptide bonds are planar but rotation can occur around alpha carbon (amide to carbon, carbonyl to carbon)
angle range from -180 deg to 180 deg, but not angles are permitted
steric clashes are minimized when side chains are trans to one another
Secondary structures (lec 6)
arise from non-cov interactions btwn func groups
alpha helix
beta strands and sheets
beta turn
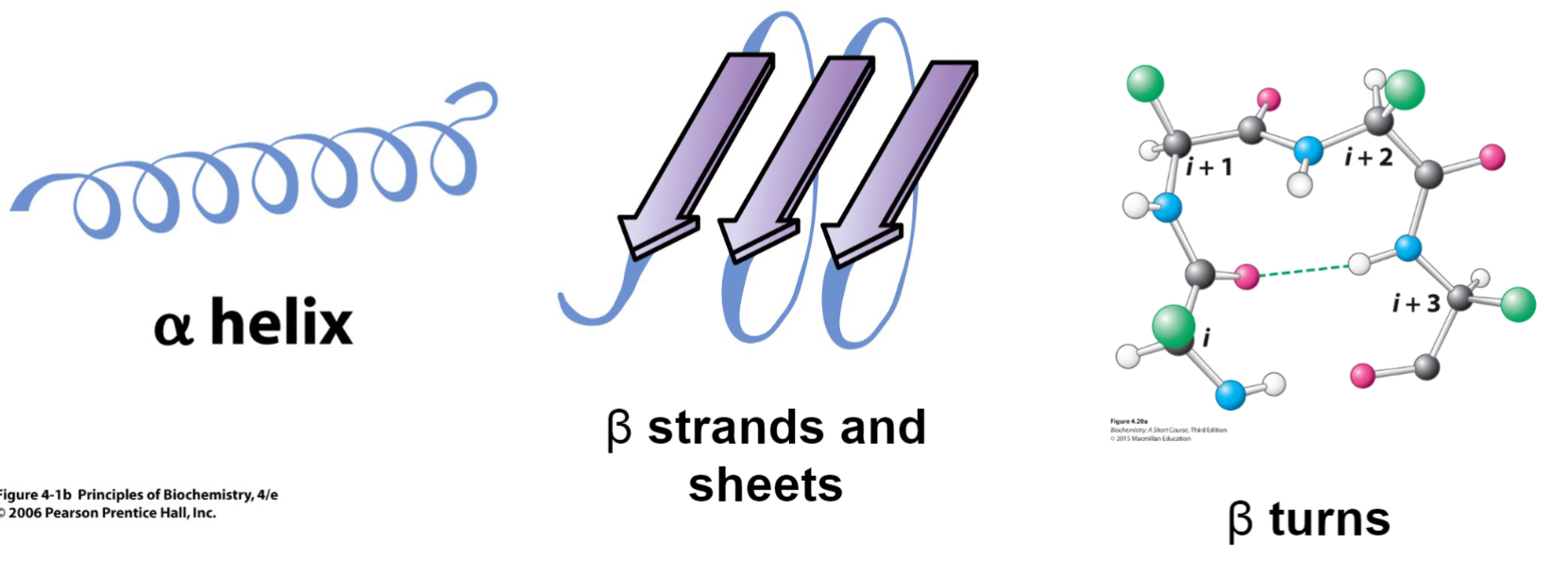
alpha helix (lec 6)
right handed helix w/ side chains pointing out
intra-strand hydrogen bonds form btwn backbone C=O and N-H groups (i and i+4), buried down centre of helix
there 3.6 residues per 360 deg turn and each residue is 1.5 A high
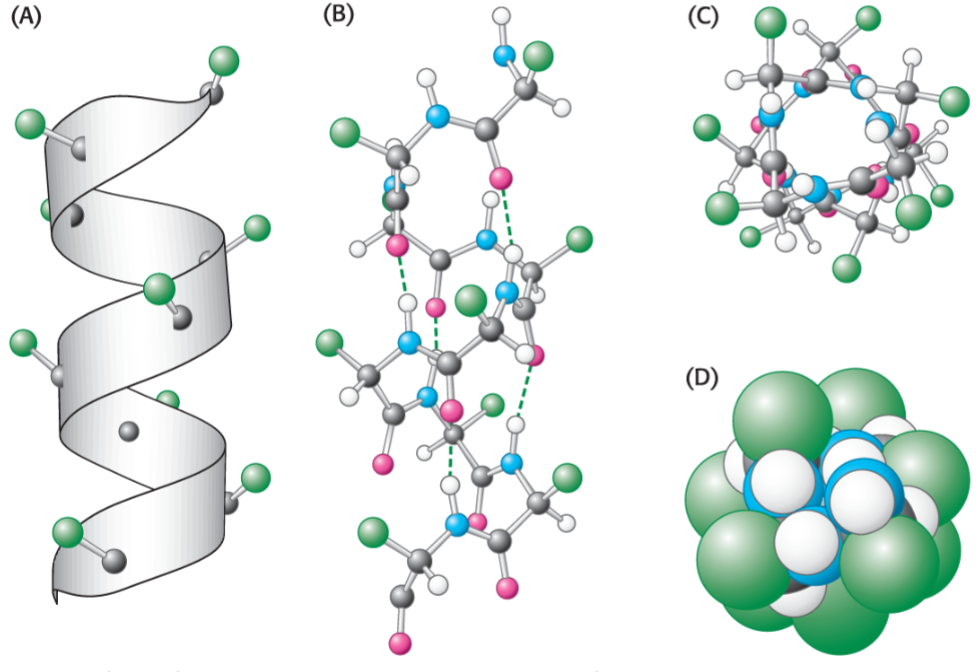
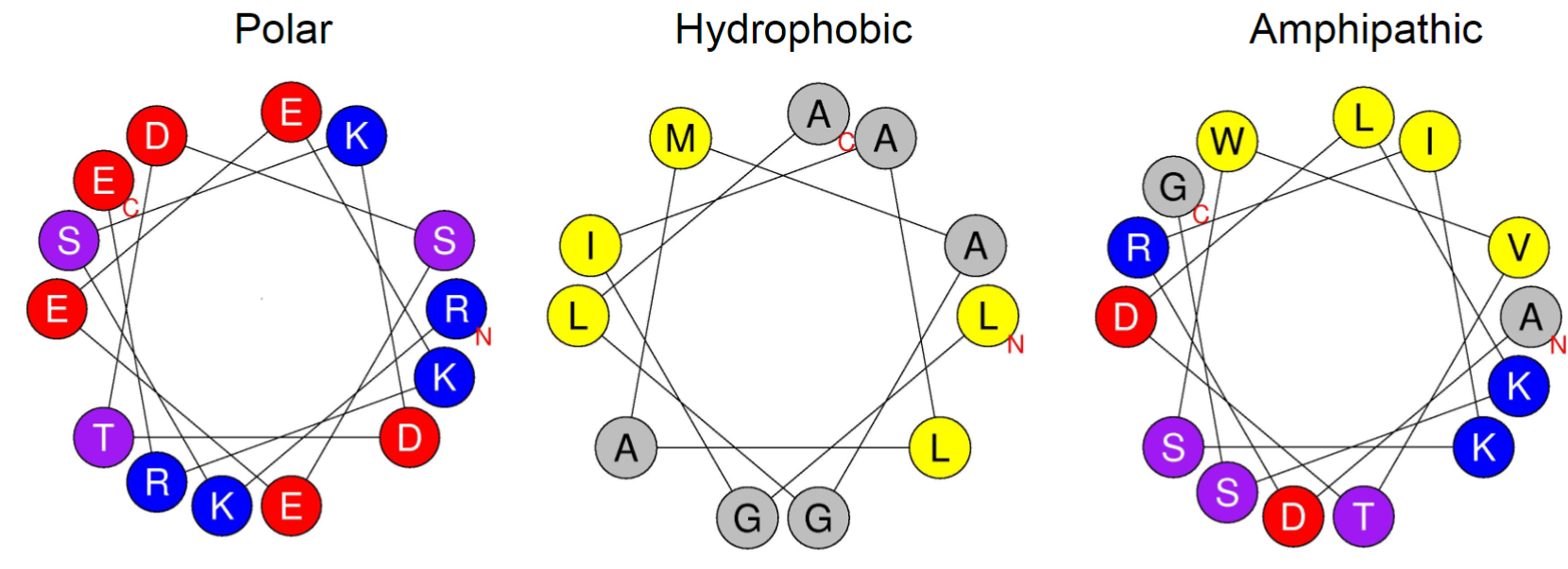
Properties of alpha helix (lec 6)
properties depend on side chains
amphipathic most common
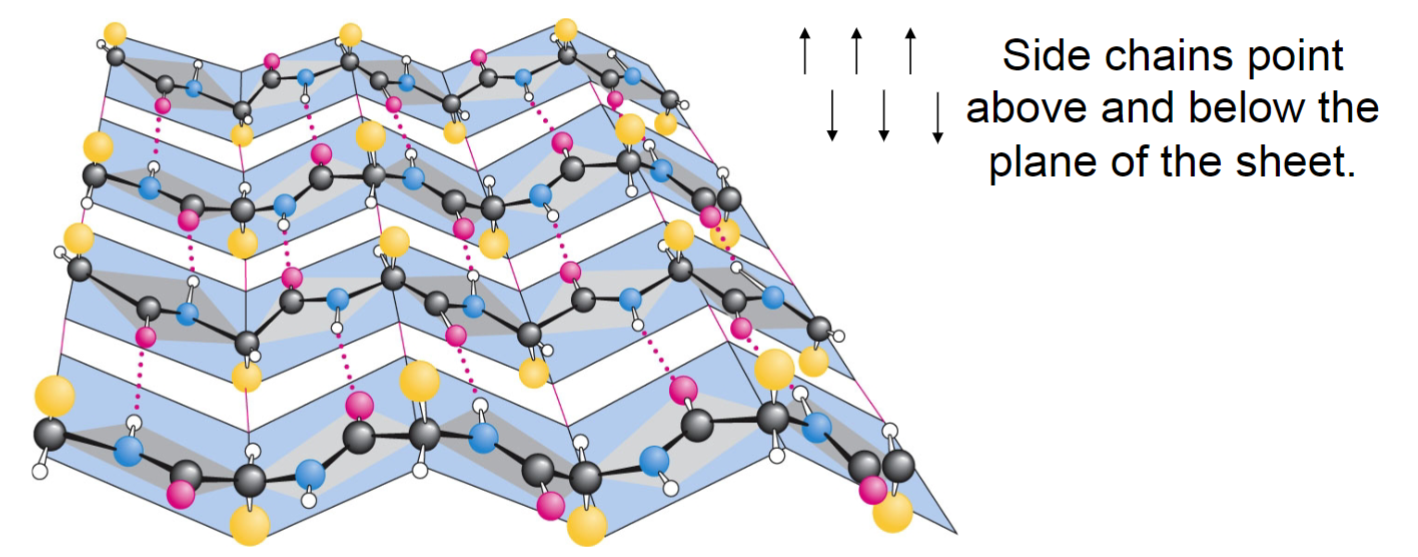
Beta strands and sheets (lec 6)
more extended structure where intermolecular H-bond link 2/+ beta strands to form beta strands
H-bonds occur btwn carbonyl and amines
strands may be parallel, antiparallel(shown in picture) or mixed
can bring distant parts of protein together
dimensions are more extended, allowing for diff interactions to occur
adjacent R groups alternately point up and down causing each side of the sheet to have diff properties
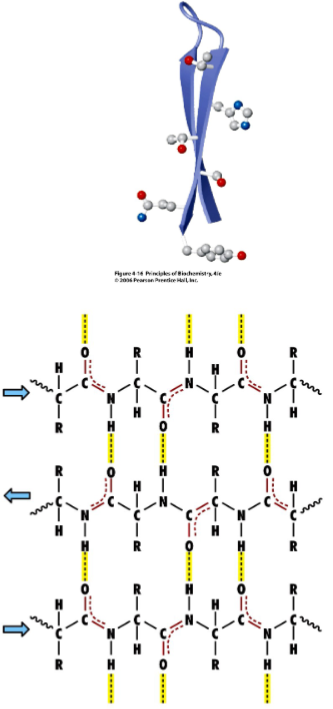
Beta pleated sheet (lec 6)
Beta turns (aka reversed turns) (lec 6)
join 2 beta strands to form sheet
4 residue segment that allows peptide chain to turn 180 deg
can be found on surface of globular proteins, connect secondary structures
H-bonds form btwn carbonyl O and amine H
Pro (P) is common at position 2
Gly (G), Asn (N), Ser (S) also seen frequently in turns (N and S have good modifications which make them more common)
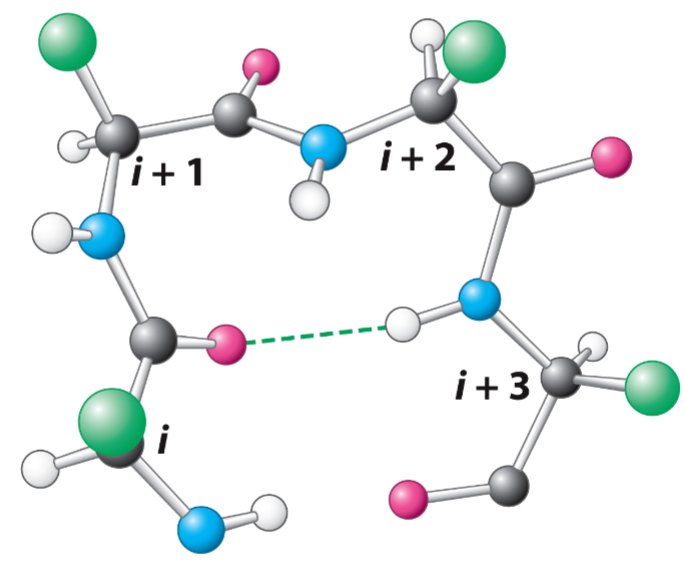
Higher levels of organization (lec 6)
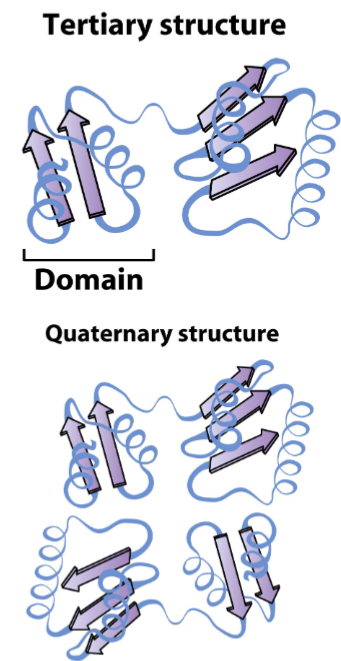
secondary structures come together to form stable, 3D tertiary structures called motifs/larger domains linked by flexible linker segments
domains may function independently of the rest of the protein
ligands may assist by bringing distinct regions together
disulfide bonds can stabilize both tertiary and quaternary structures
quaternary structure involves arrangement of multiple subunits of distinct polypeptide chains
tertiary structure is final lvl if only 1 polypeptide chain
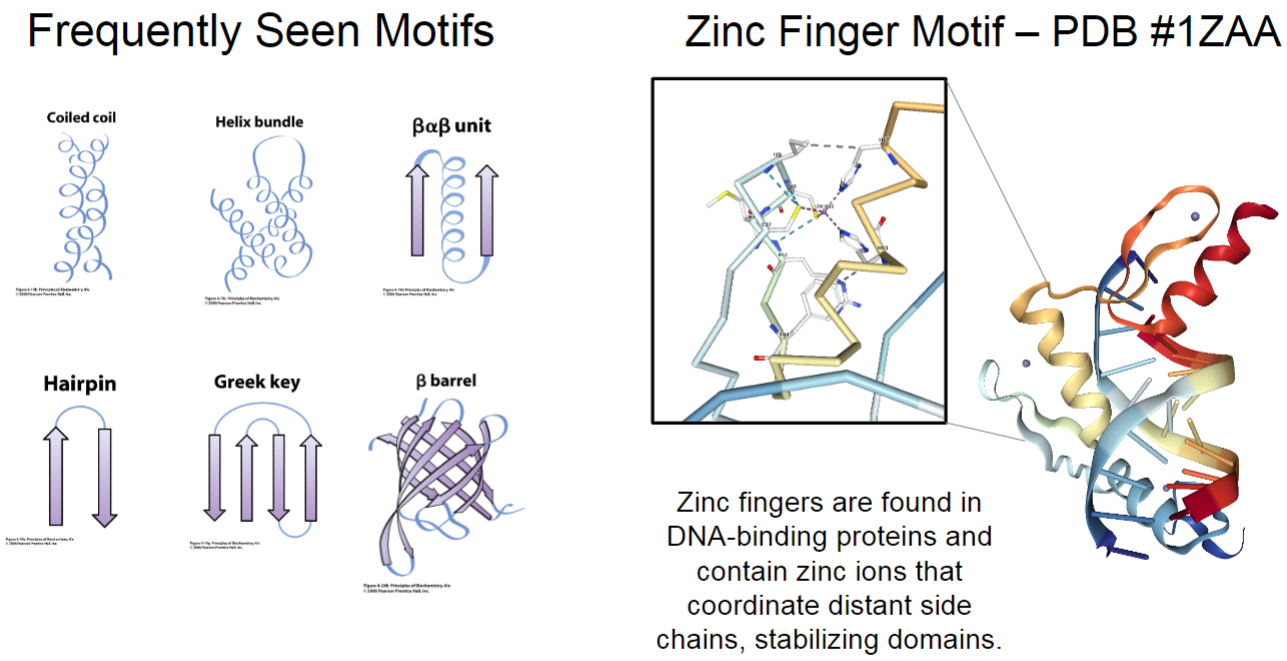
Frequently seen motifs (lec 6)
coiled coil (2 helices wrapped around each other)
helix bundle
beta alpha beta unit
hairpin
Greek key
beta barrel (sheet wrapped so pore forms)
zinc finger motif
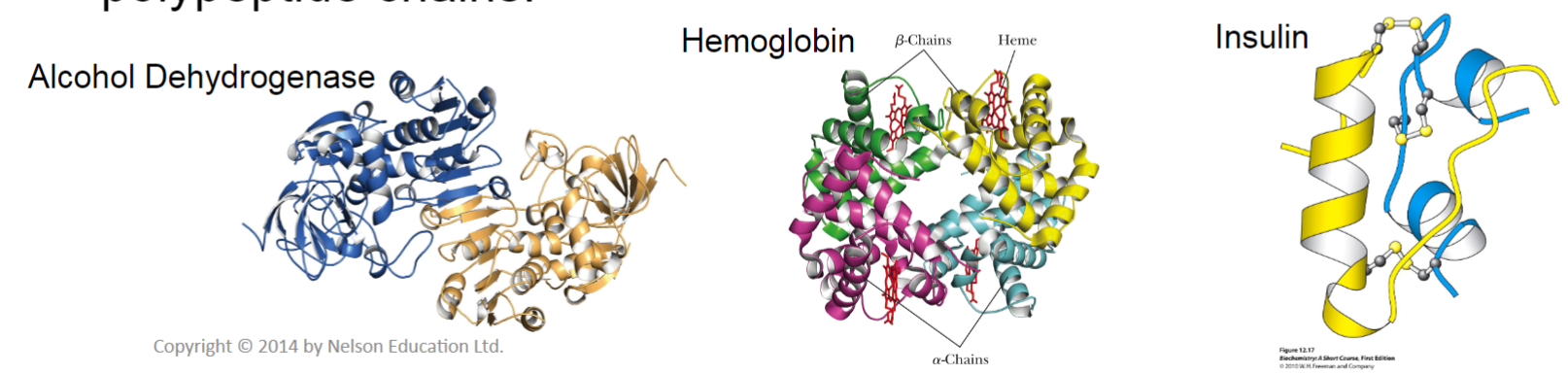
Quaternary structure (lec 6)
multi-subunit proteins may consist of identical/non-identical polypeptides held together cov/non-cov
diff subunits may arise from multiple genes/due to post-translational cleavage of precursors
larger macromolecules can also form due to interactions btwn polypeptide chains
very complex
How do proteins fold? (lec 6)
secondary structures form due to favourable H-bonding
non-cov interactions (H-bond, ionic, VdW) and disufide bonds play key role in tertiary and quaternary struct
hydrophobic effect is primary driving force in protein folding (forming of more stable structures)
random coils not necessarily random, they may be stable structures
chaperones prevent aggregation of newly synthesized and unfolded proteins by binding to exposed hydrophobic regions (transient interactions)
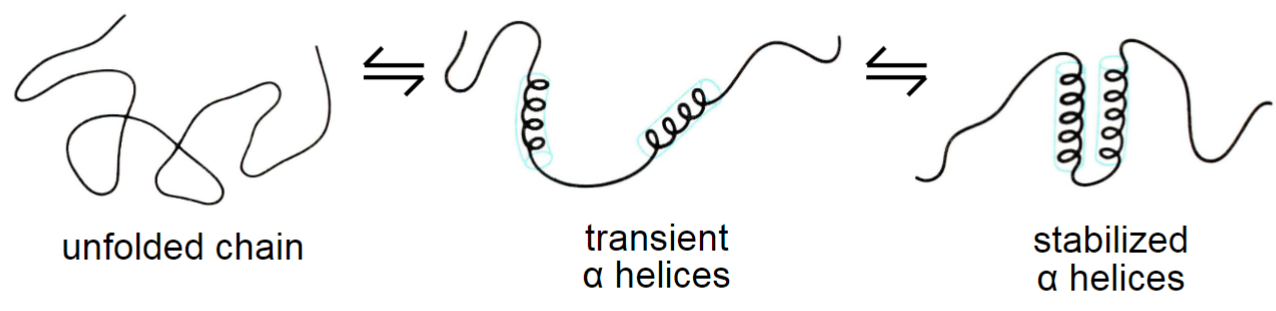
Protein structure and drug design (lec 6)
knowing a protein struct at all levels can help design molecules that interact and inhibit protein's func
inhibitors can be designed to mimic an enzyme’s substrate but prevent rxn from occuring
drugs can also block and prevent binding interactions btwn proteins necessary for signaling or a downstream event, or even modify amino acid side chains and change a protein’s struct
molecules can also block necessary non-cov interactions required for structure (ex: chelators of metal binding)
Protein purification (lec 7)
cell is a crowded environment w/ ~ 10^10 proteins per mammalian cell
in order to study a protein’s struct and/or func, we need to be able to purify it from other cellular components
other molecules/ions/proteins may interfere/modify your protein, creating a heterogenous population
ability to purify protein is first step in understanding its struct and/or func
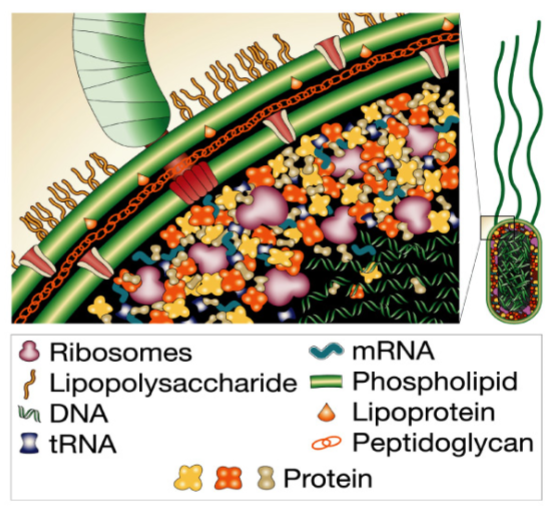
preparation of crude extract (lec 7)
choosing correct cell/tissue source is important for obtaining adequate quantity and quality of sample
purifying intracellular proteins require lysing cell to generate crude extract consisting of mixture of proteins and cellular contents
centrifugation can be used to produce a supernatant of soluble materials and pellet other large organelles/insoluble precipitants
molecular bio can also be used to genetically engineer your protein of interest
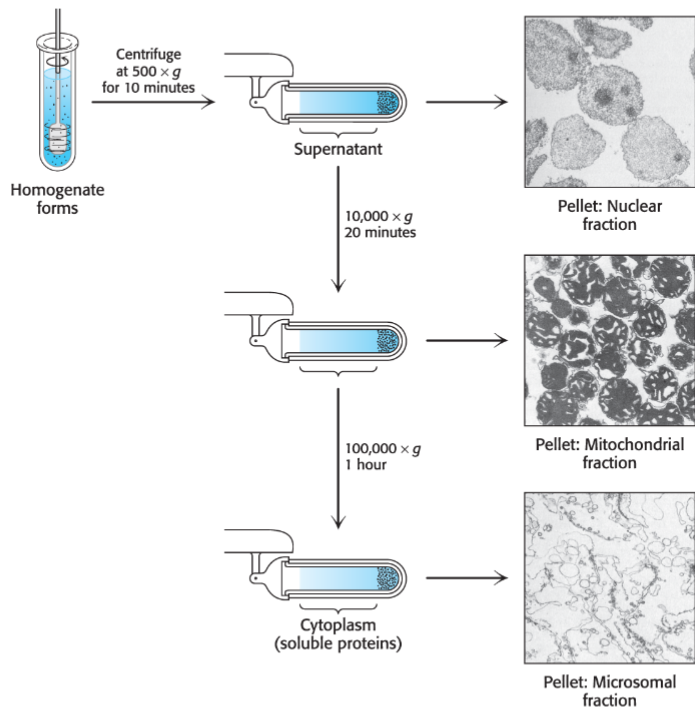
Cell lysis (lec 7)
mechanical/physsical methods:
grinding
sonification
vortexing w/ glass beads
osmotic pressure (water flows into cell and breaks it apart)
chemical bases (ex: detergents)
Extra considerations:
lysing cells may release proteases, enzymes that may cleave your protein of interest
conditions (pH/temp) may alter your protein’s structure and lead to denaturation
Types of chromatography (lec 7)
Chromatography: diff partitioning of molecule btwn mobile (buffer) and stationary (column) phase
proteins can be purified based on diff in chemical properties
Size/shape: Size-exclusion/gel filtration chromatography
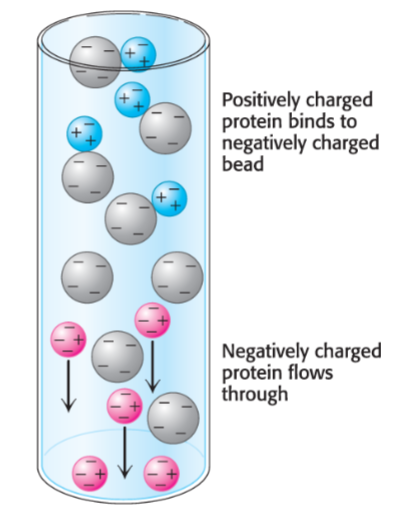
Charge: ion exchange chromatography
binding interactions: affinity chromatography
hydrophobicity: RP-HPLC (Reverse Phase High Pressure Liquid Chromatography)
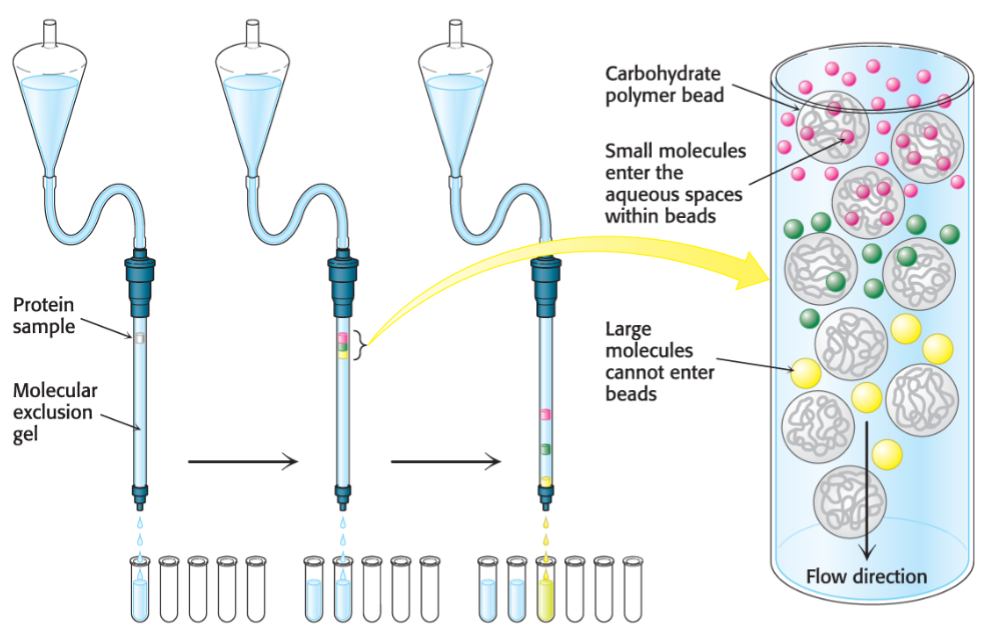
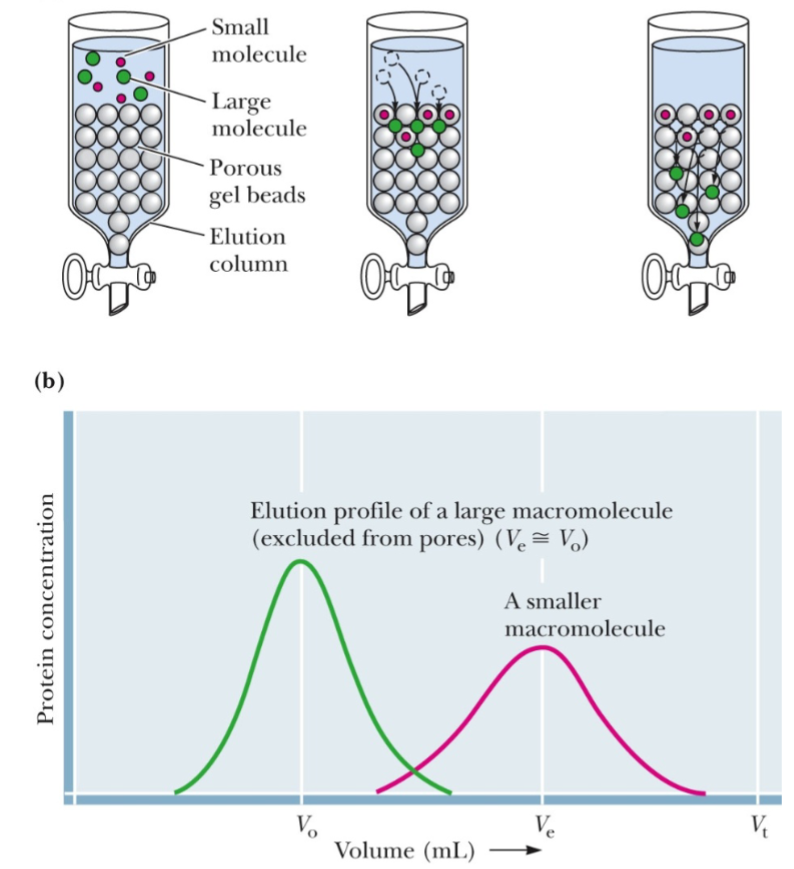
Size exclusion chromatography (lec 7)
proteins are separated on basis of size
column containing a resin of porous beads allow smaller proteins to enter the beads, while larger proteins are excluded and exit first
elongated proteins may appear larger as their tumble through buffer and also be excluded, eluting faster
calibration w/ proteins known of MW is required
Protein absorbance (lec 7)
biomolecules absorb light at characteristic wavelengths ad this can be measured using spectrophotometer
most proteins are colourless and don’t absorb visible light (380-750 nm)
concentration of protein in solution can be measured based on absorbance at 280 nm, arising from aromatic amino acids
stains like Coomassie Blue can also be used to visualize proteins/quantify protein concentration in Bradford assay
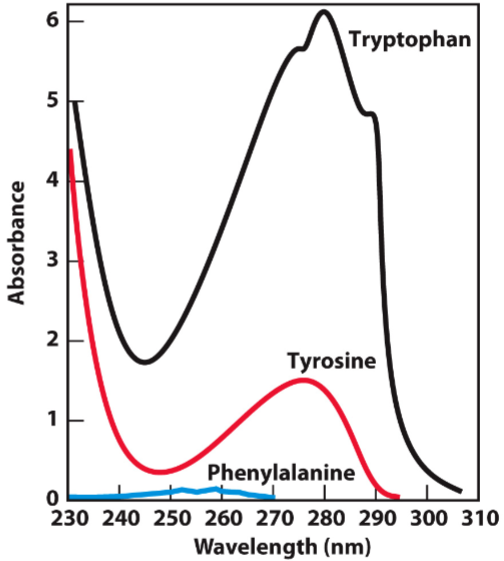
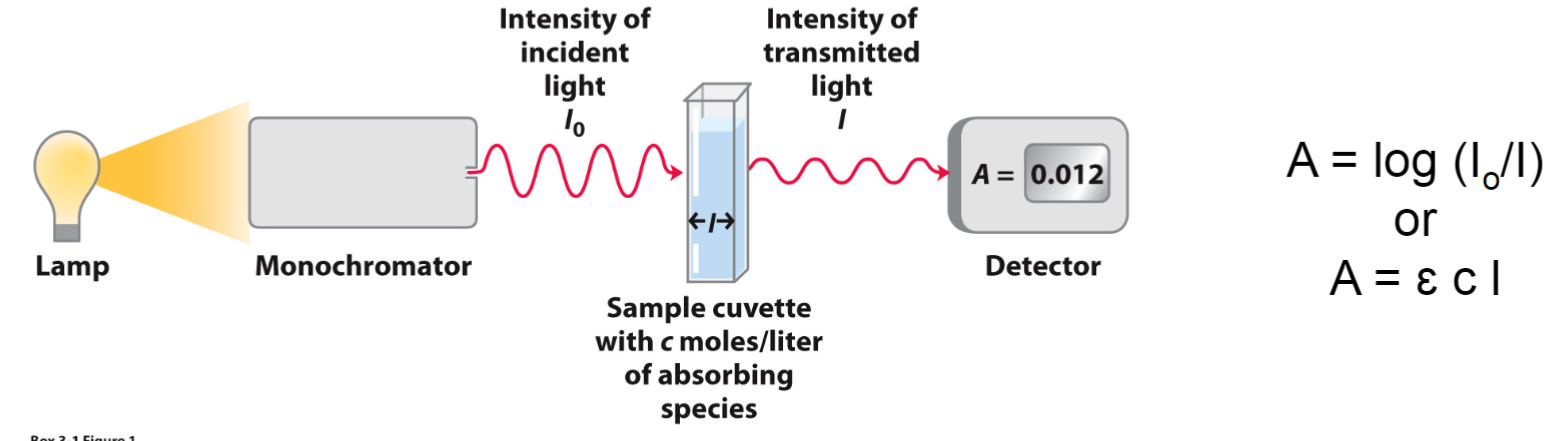
Beer-Lambert Law (lec 7)
E: molar extinction coefficient (1/Mcm) depends on # of Trp + Tyr (Phe-minimal)
c: concentration (M)
I: path length of cuvette (cm)
A = log (Io/I)
Size-exclusion chromatography example (lec 7)
Vo: void volume, anything larger than column’s fractional range goes straight through
Ve: elution volume of molecule
Vt: total volume of column
Ion-exchange chromatography (lec 7)
separates amino acids/peptides based on net charge
pl = pH when polypeptide is neutral
cation exchange resins binds (+) charged peptides while anion exchange resins attracts and binds (-) charged polypeptides
proteins can be eluted by increasing salt concentration/changing pH
Affinity Chromatography (lec 7)
proteins bind column based on their affinity for specific molecules/chemical groups
resin contains covalently bound molecules/ligands that recognizes certain proteins in mixture and interacts via non-cov interactions
bound protein is released by passing solution containing free molecules to compete for binding
useful for concentrating proteins
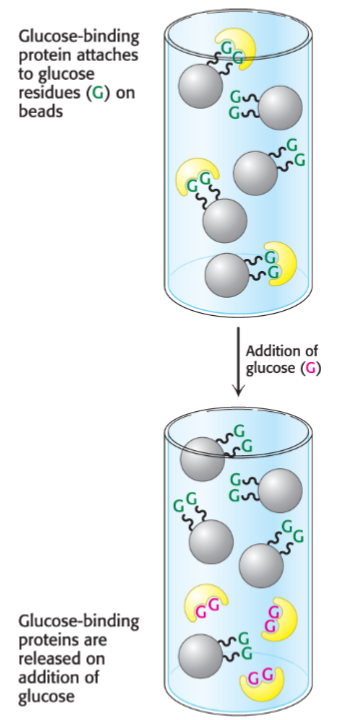
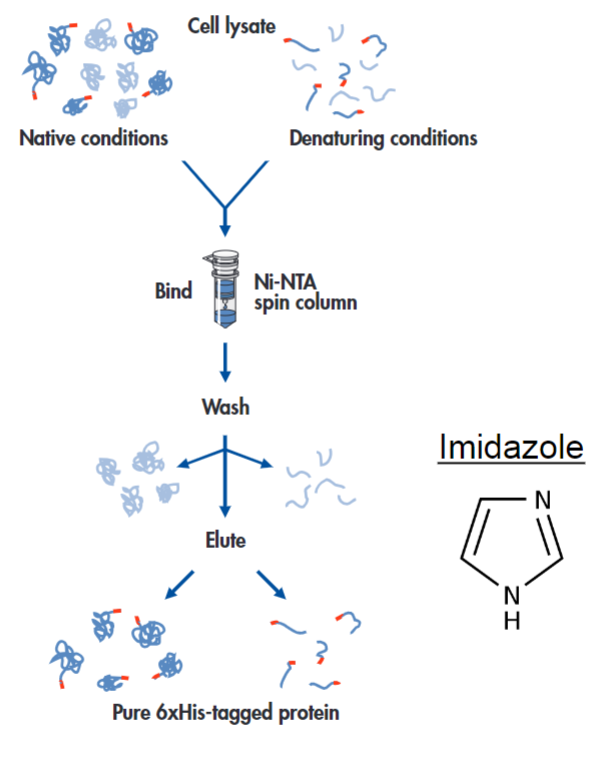
Example of affinity chromatography: His tags and nickel-NTA resin (lec 7)
histidine side chains can bind to Ni2+ when bound to NTA (nitrilotriacetic acid) resin
His tages: 6-10 His residues can be added to proteins to help w/ purification
free imidazole can be used to elute protein of interest
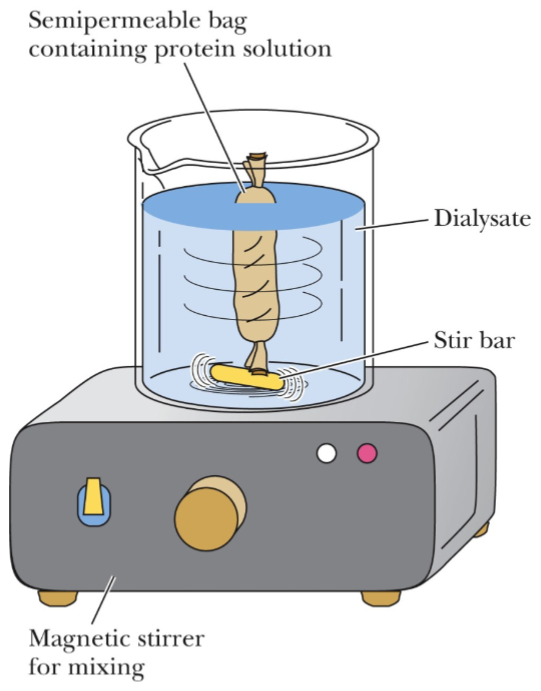
Dialysis for protein purification (lec 7)
can be used to remove small moleules
high salt may interfere w/ other experiments/assays
pH of buffer may also (de)protonate side chains involved in chem rxns/interactions
buffer exchange can be done to change pH of buffer as well
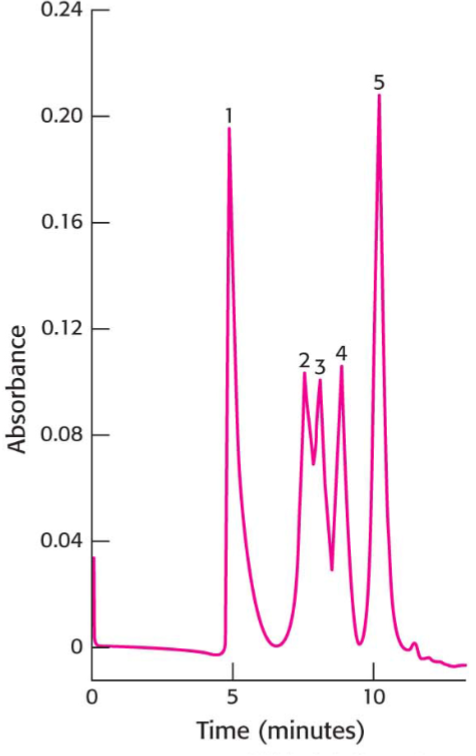
High Pressure Liquid Chromatography (HPLC) (lec 7)
uses very fine beads and high-pressure pumps to move sample through column achieving higher resolution of peaks
resin choice determines separation basis, usually silica covered in HCs
aka Reverse Phase PHLC when separating based on hydrophobicity
in RP-HPLC, hydrophobic compounds interact stronger w/ column and have longer retention time
Determining purity of sample (lec 7)
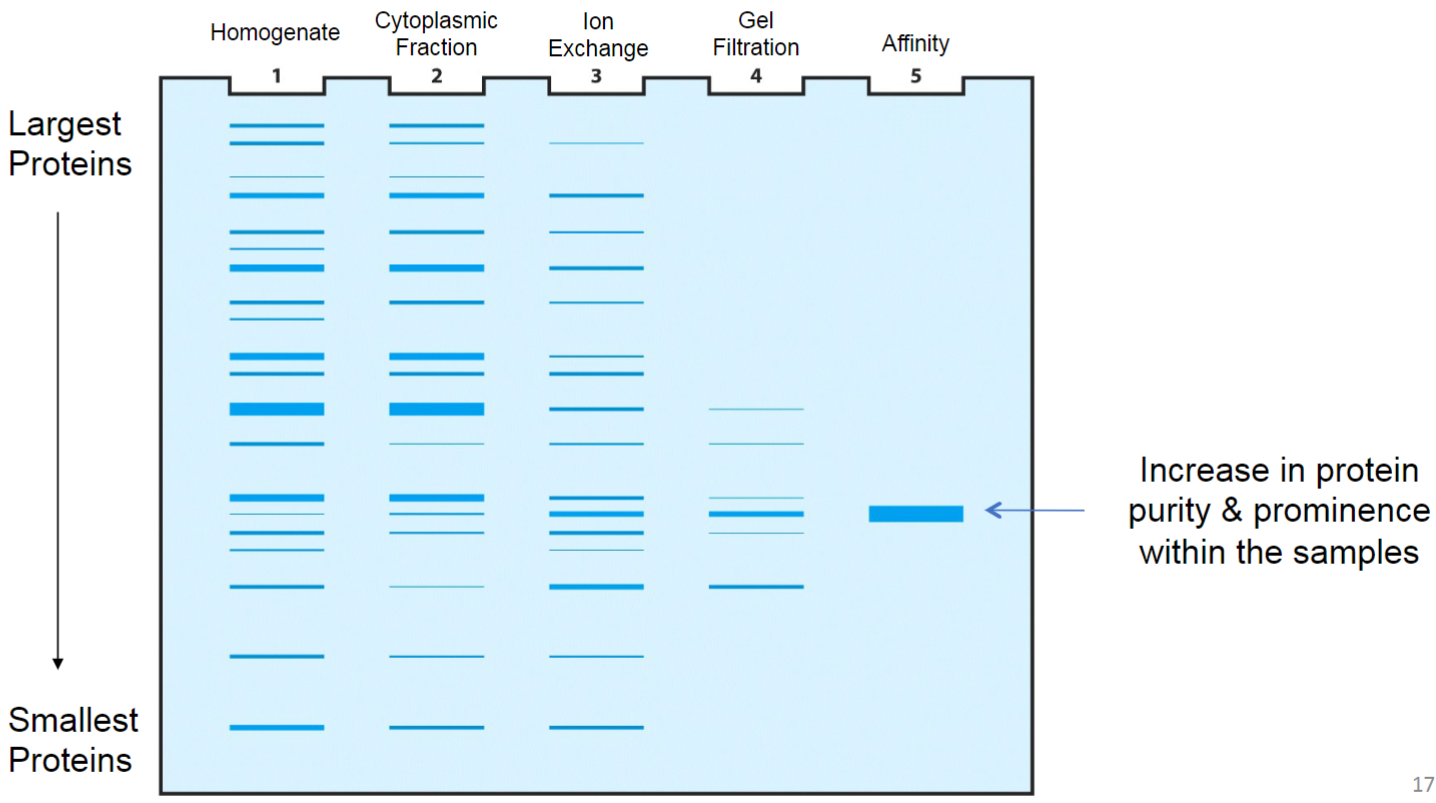
SDS-PAGE (lec 7)
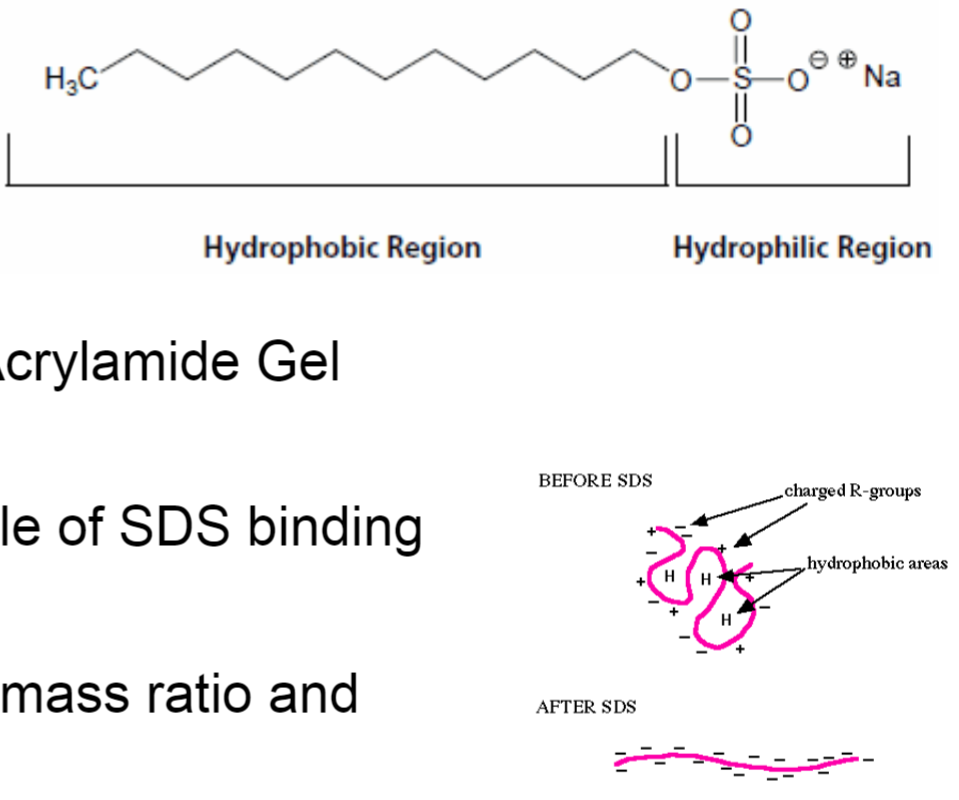
Sodium Dodecyl Sulfate (SDS) - PolyAcrylamide Gel Electrophoresis (PAGE)
SDS denatures proteins w/ 1 molecule of SDS binding every 2 amino acids (amphipathic)
proteins will have same charge: mass ratio and migrate in gel towards anode
polyacrylamide gel creates mesh/sieve of cross-linked molecules that separate subunits based on size
size can be deduced by comparing MW markers
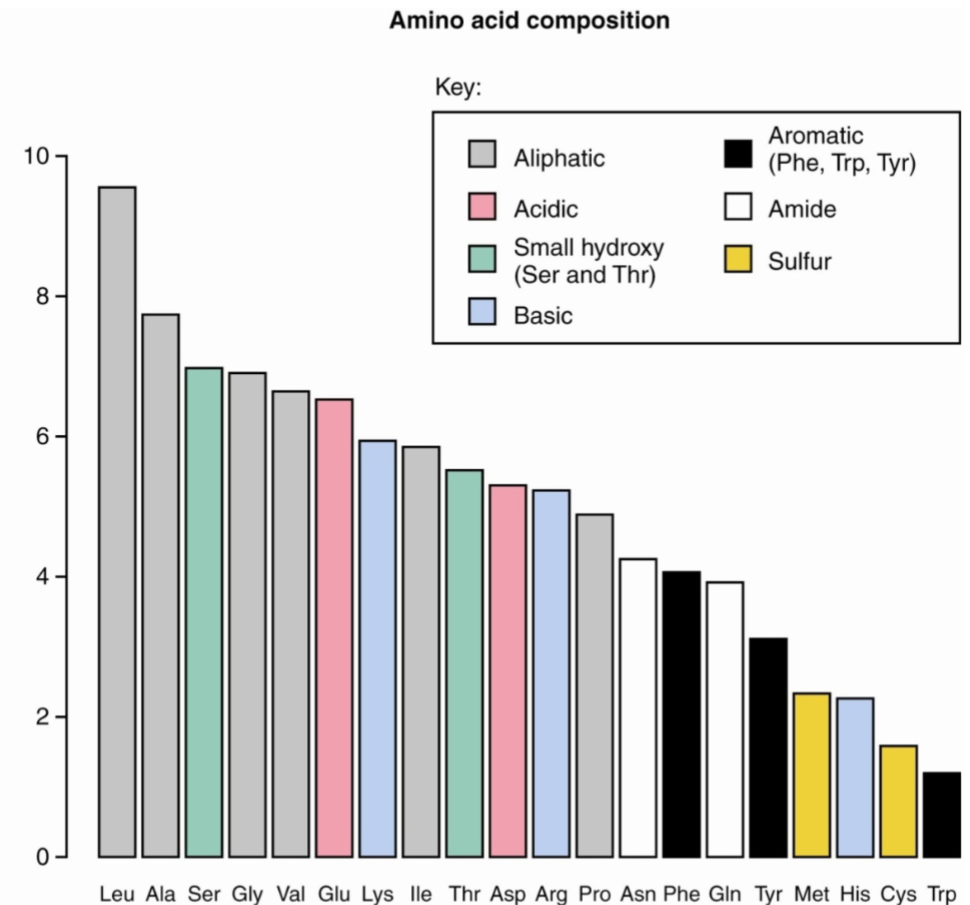
Amino Acid composition vs. sequence (lec 8)
composition of protein could be obtained by hydrolyzing peptide bonds and quantifying amino acids
however, may not tell much abt the protein unless it has unique composition (ex: gelatin)
structure of protein may not necessarily tell what protein is present as many share common domains
one way proteins can be identified is through amino acid sequencing
Protein sequencing (lec 8)
sequence could be obtained indirectly via DNA sequencing, however, post-translational modifications may occur
samples must first be purified before sequencing can occur
Edman degradation can be used to sequence amino acids from N-terminus and carboxypeptidase for C-terminus
Edman degradation (lec 8)
series of chem rxns
N-terminal labeled w/ PLTC
low pH to remove peptide bond
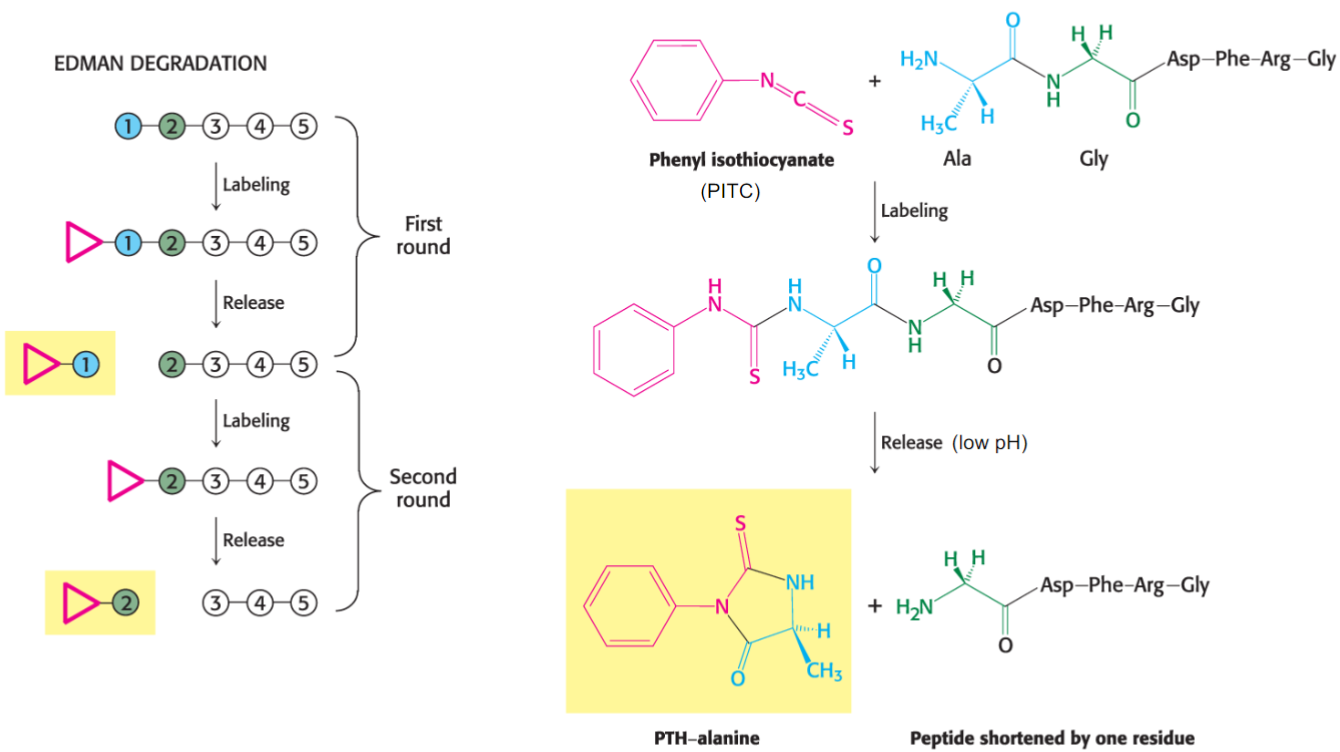
Limitations of Edman Degradation (lec 8)
limited to ~100 amino acids
post-translational modifications may block N-terminus and other complementary techs may be required
proteins can contain thousands of amino acids and smaller fragments may need to be generated, purified and then sequenced
Other chemicals and proteases can be used to generate smaller fragments for Edman degradation
Enzymatic and chemical cleavage (lec 8)
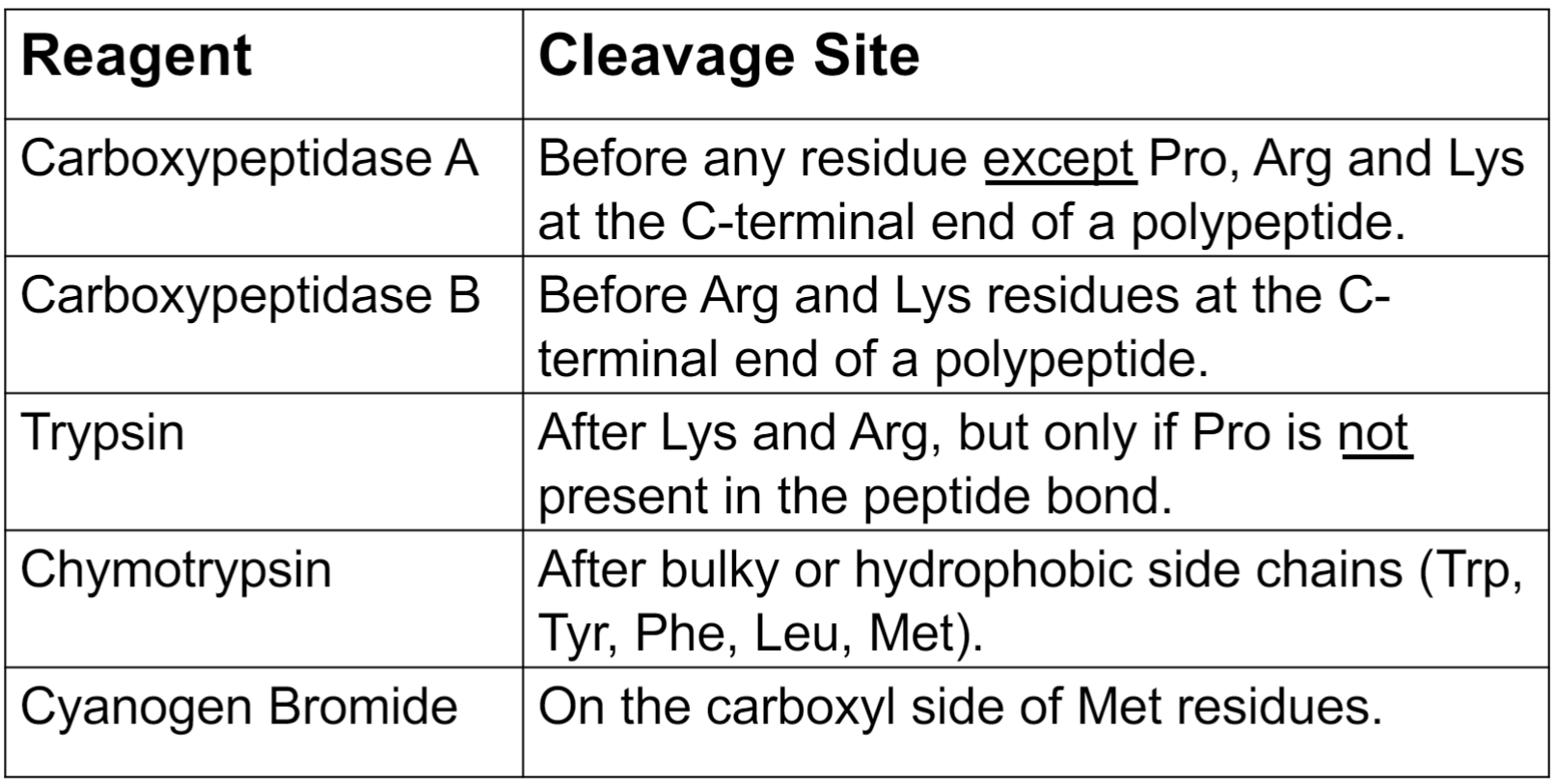
Protein cleavage example (lec 8)
15 amino acid polypeptide is treated w/ trypsin to generate the following 3 peptides, sequenced using Edman Degradation:
EH QSVVWK and AVFNDYR
cleavage w/ chymotrypsin generates following 4 peptides:
KEH NDY RQSVVW and AVF
what is sequence of og polypeptide:
AVFNDYRQSVVWKEH
Identifying peptides (lec 8)
smaller fragments can be sequenced using Edman Degradation
SDS-PAGE and immunoblotting (Western Blotting) can be used to identify larger peptides containing specific sequence using antigen-specific antibodies
Polypeptides can be ionized, separated, sequenced and identified based on their mass: charge ratio using (Tandem) Mass Spectrometry
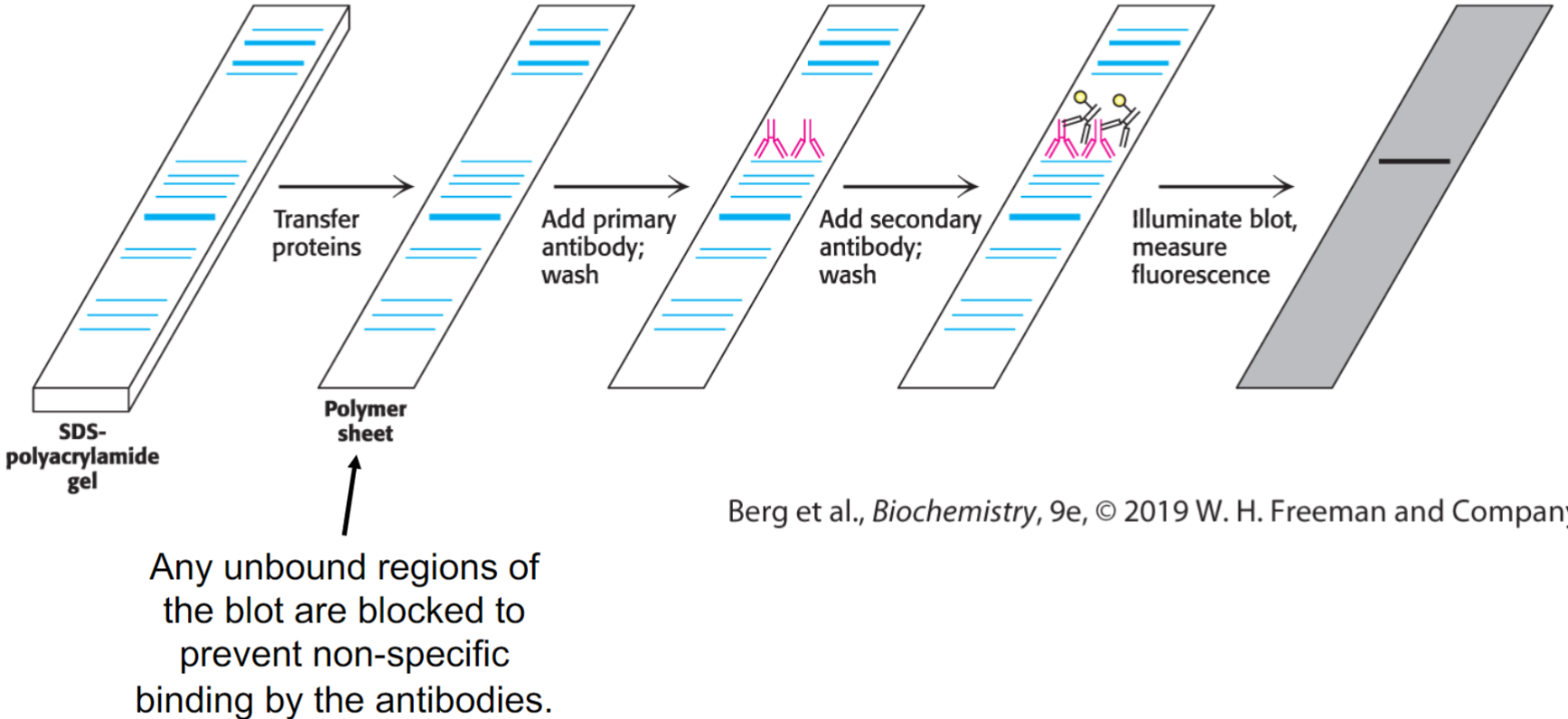
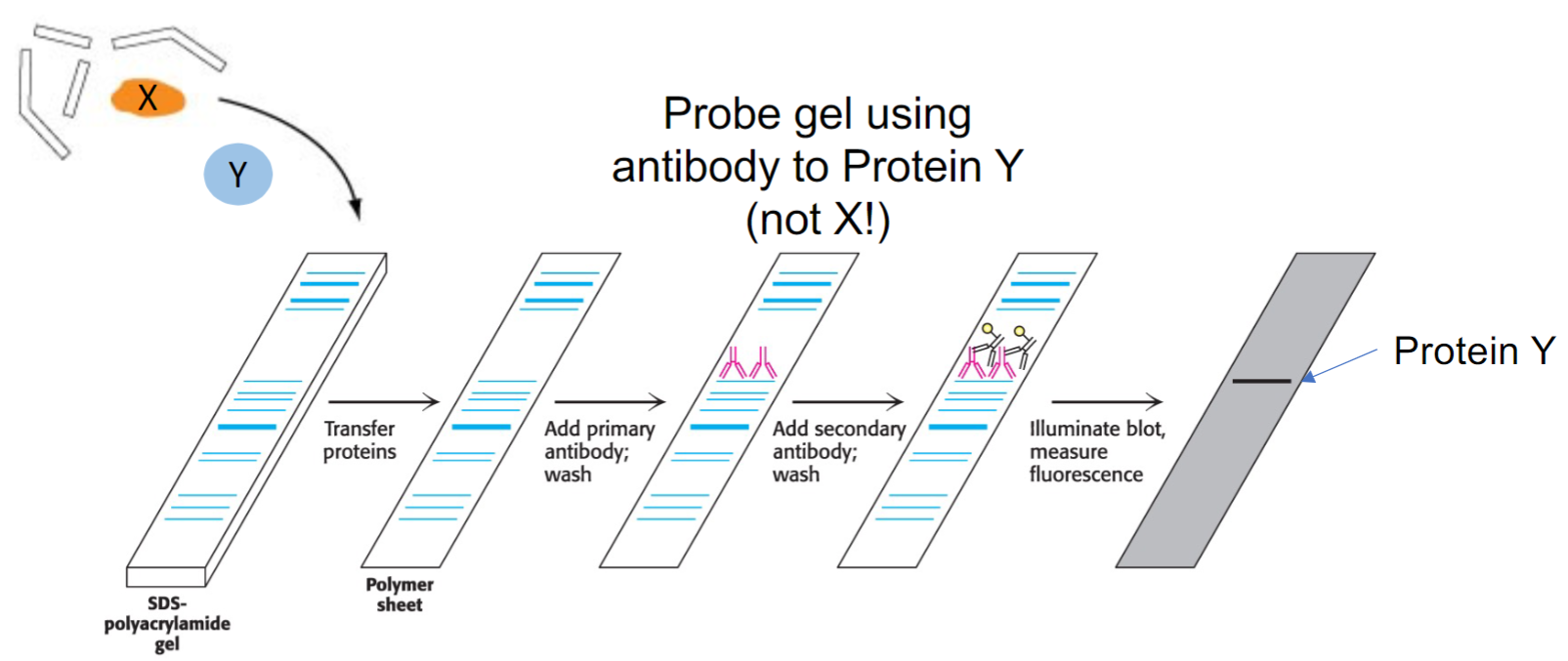
Immunoblotting (western blotting) (lec 8)
proteins are separated by SDS-PAGE and transferred to solid-support membrane (blotting)
primary antibody, specific for protein of interest, is added to recognize either linear sequences of amino acids
secondary antibodies are specific for Fc domain of primary antibody and are attached to fluorescently labeled tag/enzyme that generates chemi-luminescent product
will reveal whether or not protein of interest is present in sample and it’s potential size based on where it migrated on gel
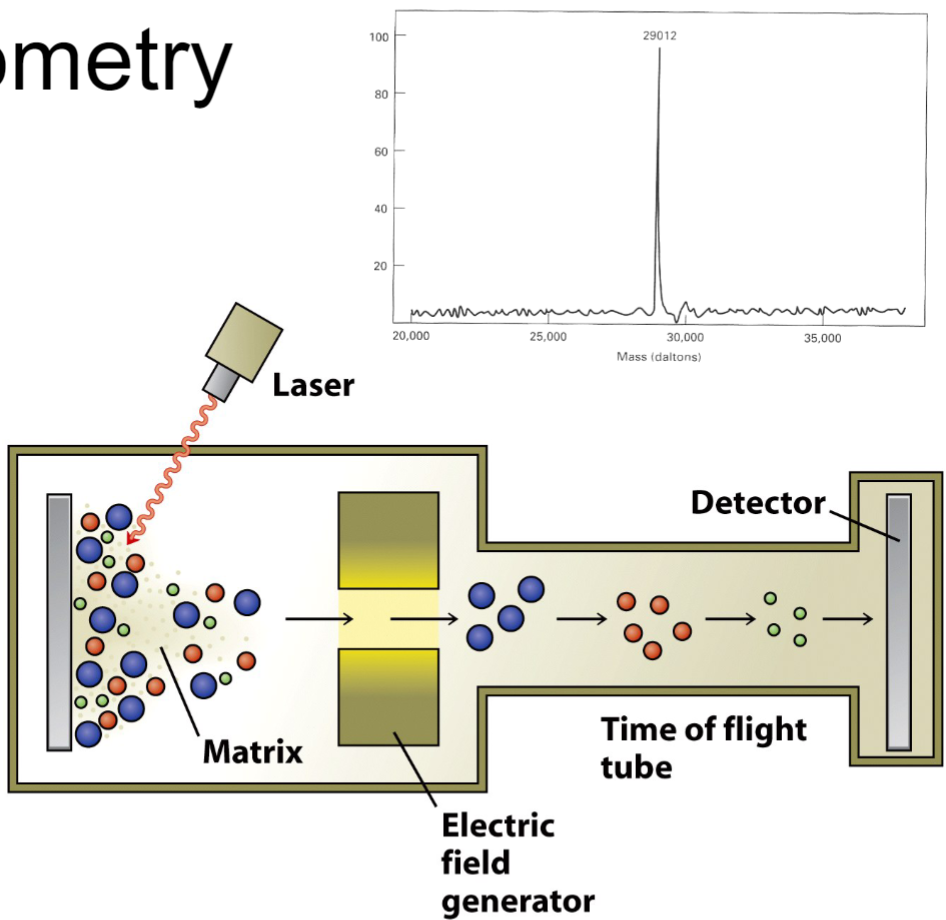
Mass spectrometry (lec 8)
peptides are bombarded by laser/high energy electron beam to create ionized fragments
products are attracted to charged plate detector and analyzed in mass analyzer by their time of flight
their time of flight depends on charge and mass of molecule
comparison to known peptides can elucidate mass and sequence of polypeptide
Sequencing using Tandem Mass Spectrometry (MS/MS) (lec 8)
used to determine sequence of amino acid
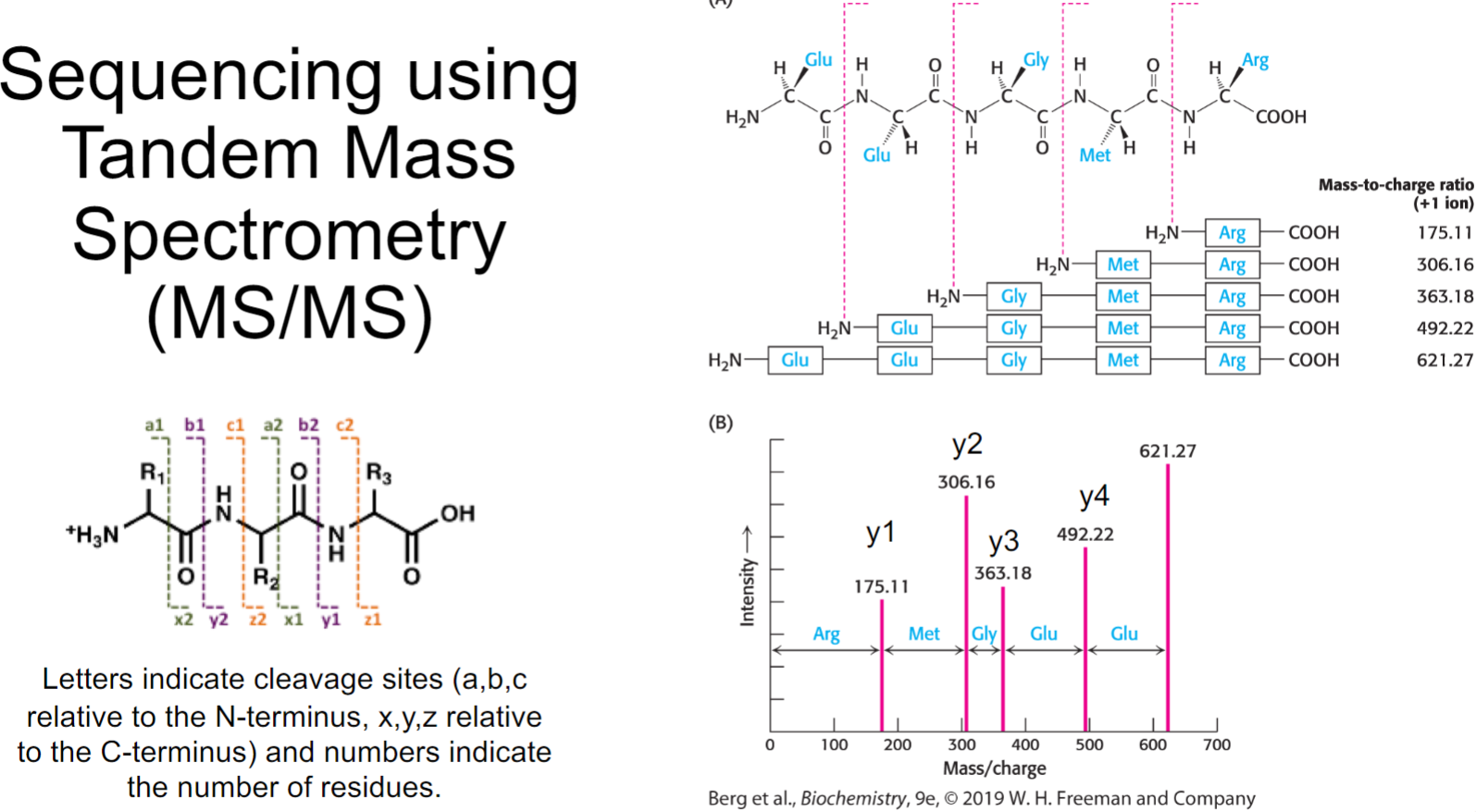
Sequencing using Mass spec (lec 8)
Studying Purified Proteins (lec 8)
function:
enzymes assays
inhibition of function
Interactions:
binding interactions
transport assays
structure
expression
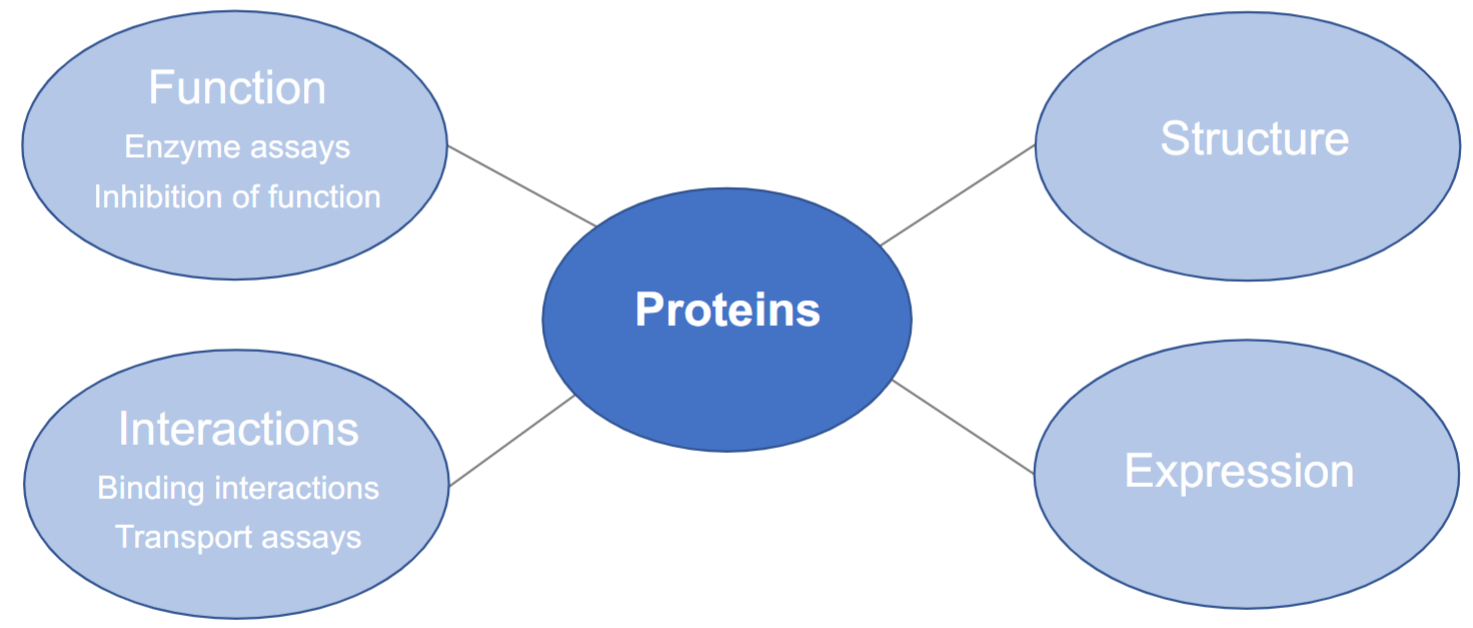
Tryptophan Fluorescence (lec 9)
presence of indole ring also allows Trp to fluoresce when excited w/ UV light (270-295 nm)
emission of Trp occurs btwn 310-355 nm and is sensitive to polarity of its local environment in protein
in a polar environment, fluorescence is ‘red-shifted’ to longer wavelengths and be less intense (opposite for ‘blue-shifted)
Protein Chromophores (lec 9)
chromophore groups usually contain conjugated double bonds
chemical groups absorb ultraviolet (UV) and/or visible light at characteristic wavelengths (can give proteins colour)
aromatic rings and amide carbonyls are important chromophores found in proteins
protein’s structure influences accessibility of these groups to light and can be used to characterize protein’s struct
Protein structure techniques (lec 9)
Localized primary and secondary structure:
Fluorescence spectroscopy
Infra-red (IR) spectroscopy
Circular Dichroism (CD) spectroscopy
overall 3D structure:
X-ray crystallography
Nuclear magnetic resonance (NMR) spectroscopy
Electron microscopy (EM)
Infrared (IR) spectroscopy (lec 9)
proteins contain vibrating, strecthing, and bending groups
these motions lead to an absorption of infrared radiation
most prominent bonds, N-H and C=O groups in peptide bond, contribute most to absorption seen
IR spectrum an be used to understand secondary structures that are present and influence H-bonds
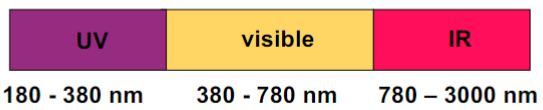
H-bonding and IR Spectra (lec 9)
band position of amide I band (C=O stretch, NH bend) can distinguish alpha helix and beta sheet structures
stronger H-bonds, weaker C=O bond, lowering position of IR spec.
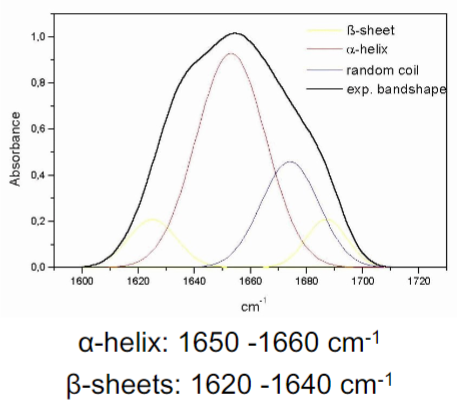
Circular Dichroism (CD) (lec 9)
asymmetry of proteins results in diff (molar ellipticity) in absorption of left and right circularly polarized UV light
chiral alpha carbons and secondary structures preferentially absorb 1 direction of light over the other
protein folding and secondary structure content can be assessed based on observed spectra:
alpha helix: 222nm and 208nm (negative), 195nm (positive)
beta sheet: 217nm (negative)
Random coil: 198nm (negative)
Studies proteins in solution to stimulate cell environment
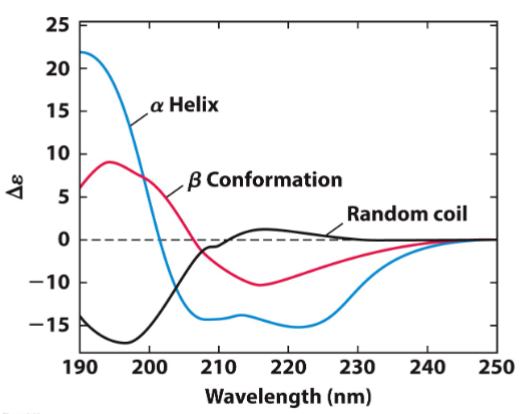
CD experiment - Antibiotic peptide (lec 9)
CD spectra was determined in presence of water (aq) or sds, which mimics the hydrophobic environment of membrane
transition from random coil to alpha helix can be seen
what interactions are mediating this change?
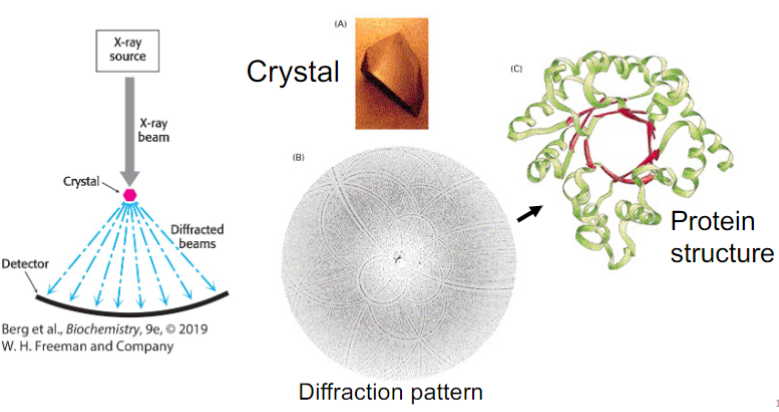
X-ray crystallography (lec 9)
some proteins can form ordered crystals under varying conditions (pH, high salt, etc)
crystals placed in X-ray diffractometer produce diffraction patterns that be interpreted in terms of atomic positions (x,y,z)
the (𝜙, Ѱ) angles of each residue defines protein fold
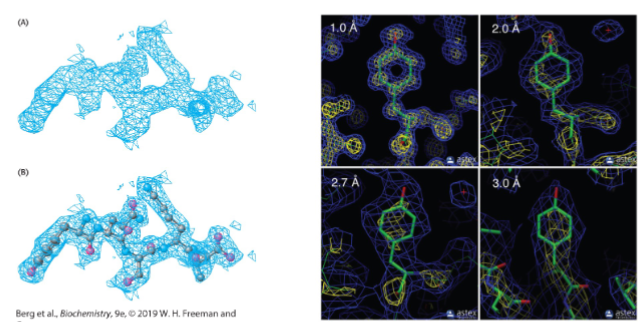
3D structures of proteins can be reconstituted at high resolution (2A), including backbone and side chains
structures are deposited in Protein Data Bank (PDB)
Electron density maps and resolution (lec 9)
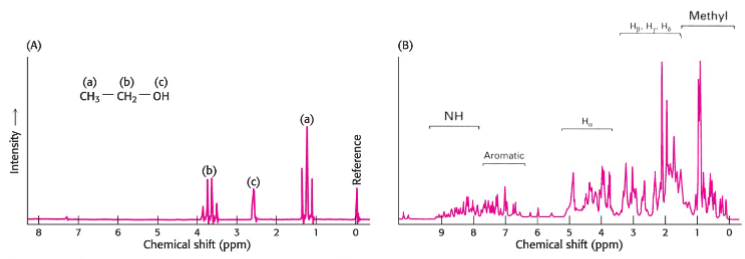
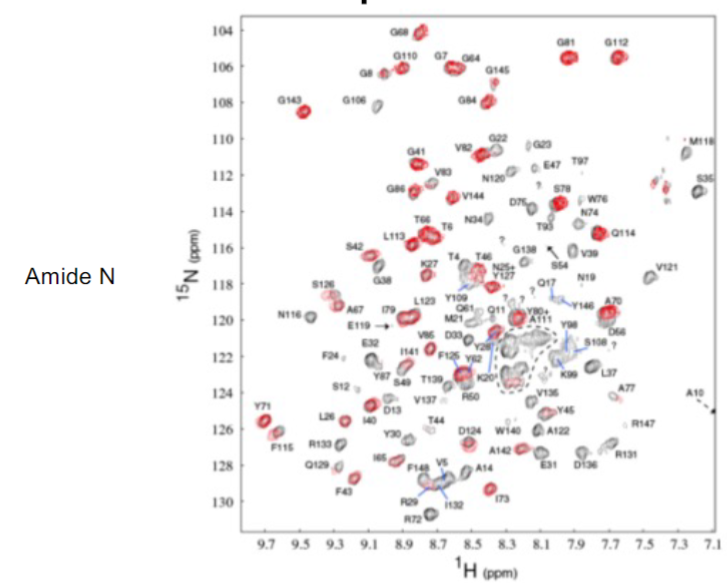
Nuclear Magnetic Resonance (lec 9)
carried out on proteins in solution
used to be limited to smaller proteins (<25 kDa), but now can be done on larger proteins
can monitor conformational changes, proteins folding and interactions w/ other molecules
NMR is based on nuclear spin of certain nuclei (1H, 13C, 15N) that can be measured in a strong, static magnetic field
absorption of electromagnetic radiation can be used to deduce environment of nucleus and determine protein’s structure
1D NMR of ethanol vs larger protein (lec 9)
2 NMR Spectra of protein (lec 9)
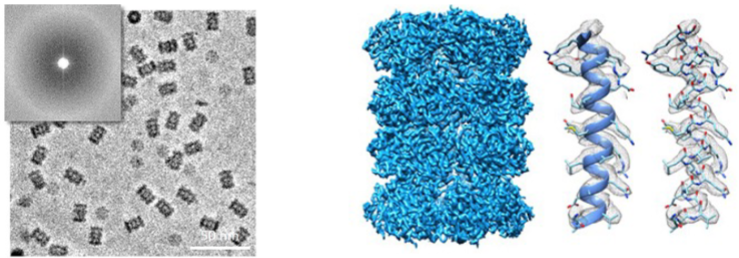
Cryo-electron microscopy (lec 9)
larger complexes can be visualized (>100 kDa)
thin layer of protein solution is prepared on fine grid and frozen (cryo) very quickly to trap molecules in ensemble of orientations
high powered microscopes measure beam of electrons that pass through protein sample. Diffraction in beam can be used to elucidate structure
structure of single particle can be obtained and multiple structures are averaged out to build 3D representation of protein
Studying proteins experimentally (lec 10)
understanding protein’s physiological environment is important for designing experiment that mimics cellular conditions (temp., pH, detergents, cofactors/substrates, etc.)
experiments can be done in test tube (in vitro) w/ purified proteins, or in live cells cells (in vivo) to determine its intracellular localization/binding partners
assays (experiment) can take advantage of non-cov interactions, chemical catalysis and other biochem properties to elucidate protein’s function
control experiments are just as important when analyzing data
Important biomolecular interactions (lec 10)
interaction of protein w/ protein/molecule is crucial for its function and numerous biochem processes:
protein folding/unfolding
cellular localization
post-translational modifications
signalling pathways and regulation
metabolism
complication: non-cov interactions may be transient and short lived
Studying biomolecular interactions (lec 10)
in addition to structural studies, common techs used in biochem:
Protein-Protein interactions: Co-immunoprecipitation and pull-down assays, cross-linking reagents, 2-hybrid screening, FRET
Protein-molecule interactions: transport assays, enzymatic reactions
visual output is most common way of detecting occurence of biomolecular interaction
enzymes and fluorescent proteins can also be exploited to generate secondary visual response when used as a reporter (ex: luciferase, beta-galactosidase-based assays, bioID)
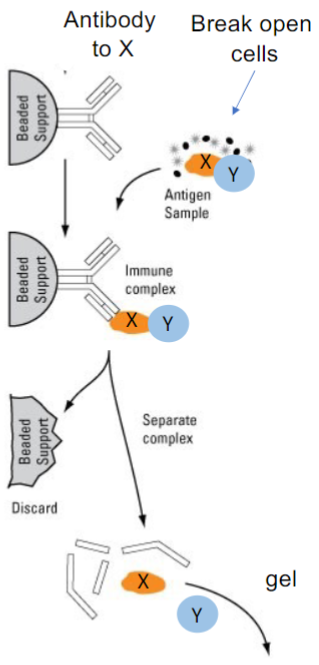
Pull-down Assays (Co-immunoprecipitation) (lec 10)
Question: does protein X interact w/ protein Y in cells?
Approach: pull down X w/ and see if Y comes along
Tools required: Antibody specific to X + antibody specific to Y
Experimental steps:
immobilize antibody to X to a solid support (ex: beads)
break open cells and mix w/ antibody
isolate beads, wash and detect bound proteins by Western blot
Detecting binding partners by immunoblot (lec 10)
often to be sure, you would do the experiment in reverse: pull down Y and probe for X on the blot
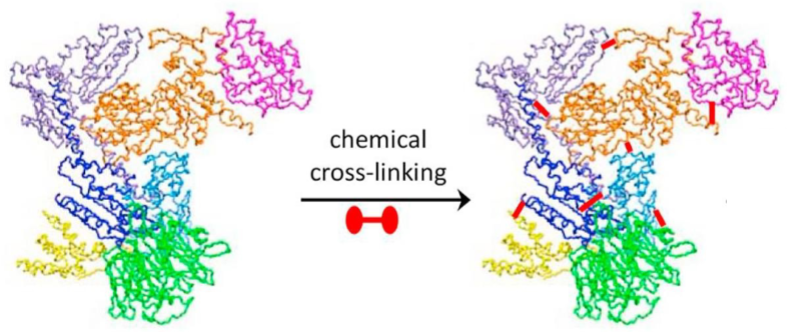
Capturing Complexes w/ chemical cross-linking (lec 10)
chemical cross-linking can form cov bonds btwn molecules
cross-linking studies can be used to reveal the inter and intra- (molecule organization of amino acids based on their locations)
Formaldehyde is an example of non-specific chemical crosslinker
cross-linking can also be nonselective using photo-reactive groups
reactive side chains of amino acids can be targeted specifically for cov bond formation:
primary amines
carboxyls
carbonyls
sulfhydryls