BIOL 3000 Transcription and Intron Processing
1/48
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
49 Terms
Central Dogma
Gene (genotype) → mRNA (intermediate) → “Protein”/polynucleotide (phenotype)
Transcription
DNA to mRNA
Where: cell nucleus (eukaryotes)
When: Either G1/S or G2/M (waves of transcription that coincides with different transition points during the cell cycle)
Doesn’t happen in mitosis
Can sometimes happen in S phase
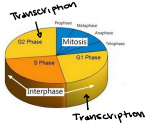
“Waves” of transcription
Times that make lots or little mRNA
Basic Rules of Transcription
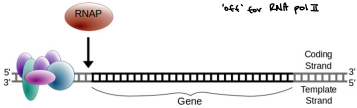
Only one of the two DNA strands serves as a template: template and coding strands
Selective process
Always in 5’→3’ direction; nucleotides added at the 3’ growing tip
Results in an RNA compliment (anti-parallel and unidirectional)
Template strand
The DNA strand that mRNA is built from
The strand that the enzymes are actually reading
Coding strand
“Exact” same sequence of nucleotides in the mRNA except T and ribose. Making a complimentary strand T-U
Selective process of transcription
Each gene has its own transcription protocol (has its own promoter)
Steps of Transcription
Pre-recognition: accessibility to DNA
Recognition: pre-initiation complex formation
Initiation: binding of RNA polymerase complex
Elongation: movement of RNA Pol II and formation of mRNA
Termination: cleavage of new transcript
Pre-recognition
DNA access
DNA is packaged in the nucleus as heterochromatin which we cannot make protein from this because we cannot get to the DNA
So it must be converted to euchromatin before transcription
Part of Gene Expression Regulation
Nucleosome (10nm fiber)
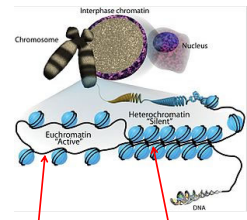
Nucleosome (10nm fiber)
Histone core is positively charged, and the DNA is negatively charged
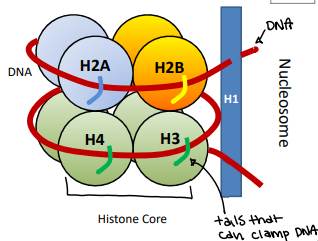
Lys (K) acetylation
Histone acetyl transferase (HAT)
Histone deacetylase (HDAC)
Allows the tail to open and close, releasing and loosing the DNA
Histone acetyl transferase (HAT)
Transfer acetyl group to the histone
This tightens the tail and holds onto the DNA
Increases the positive charge of the nucleosome
Histone deacetylase (HDAC)
Removes the acetyl group from the histone
This looses the tail and loosens the DNA and makes it able to move
Decreases the positive charge of the nucleosome
Recognition: pre-initiation complex (PIC) formation
Forms a large complex of ~100 proteins (PIC) required for RNA Polymerase II to bind to
TATA Binding Protein (TBP) binds the promotor region (TATA Box), once bound, it recruits the rest of the pre-initiation complex. Then the mediator complex arrives
General Transcription Factors (TF IID) recruited by TBP
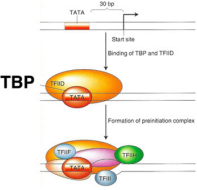
Initiation: binding of RNA polymerase complex
Other transcription factors (TF IIB, TF IIF, TF IIH) and RNA Pol II recruited to complex
Mediator complex: ~20 proteins
RNA Pol II binds to the Template Strand via its Active Site and is NOT on
Mediator complex
ATPase and helicase activity unwind DNA so that polymerase can read
One turn of DNA unwinds and forms a transcription bubble
RNA Pol II
Unphosphorylated at its carboxyl end (CTD)
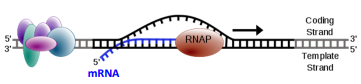
Elongation: Movement of RNA Pol II and formation of mRNA
RNA Pol II is phosphorylated at its carboxyl end (CTD) by Mediator Complex
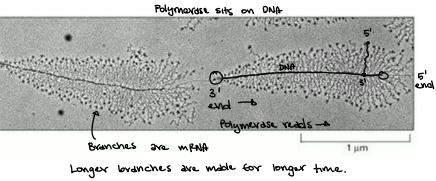
RNA Pol II traverses the template strand and creates an RNA copy
RNA Pol II traverse the template strand from 3’→5’
Exact copy of the coding strand (except that Thymines are replaced with Uracils, and the nucleotides are composed of ribose (5-carbon) sugar
READS
3’→5’
WRITES
5’→3’
Termination: cleavage of new transcript
Two new protein complexes carried by CTD recognize the poly-A signal (AAUAAA)
Once poly-A signal is made, the polymerase can stop
CPSF (cleavage and polyadenylation specificity factor)
CSTF (cleavage stimulation factor)
Other proteins recruited to carry out cleavage
Post transcriptional processing can now occur
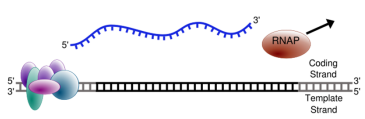
Image of Transcription
Basic Summary of Transcription
DNA to mRNA (specifically hnRNA)
Makes RNA 5’ - 3’ (ALWAYS ADDING TO THE 3’ END)
RNA polymerase
Immature mRNA consists of BOTH exons and introns
The two types of post-transcriptional regulation
5’ capping
3’ polyadenylation
5’ Capping
A guanine group is added to the 5’ end of the growing RNA chain at about 30 nucleotides long. This group caps the enzymes copying the DNA.
Happens co-transcriptionally
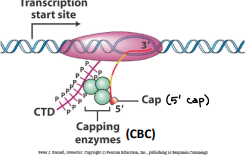
Functions of 5’ guanine cap
Regulates nuclear export
Promotes translation (helps the ribosome recognize the message)
Prevents the degradation of mRNA in the cytoplasm
Involves in intron splicing
3’ Polyadenylation
The RNA is cleaved by ribonuclease downstream of the AAUAAA site. A poly(A) polymerase adds adenine ribonucleotides to the 3’ end of the RNA molecule. The enzyme is not dependent on the template and is up to 200 bases.
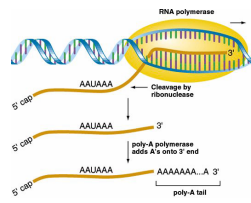
Functions of 3’ Polyadenylation
Enhances the stability of the RNA molecule
Regulates transport to the cytoplasm
RNA processing
Co-transcriptionally AND post-transcriptionally
Splicing
The mechanism by which introns are removed
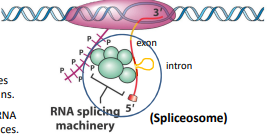
Introns
Intervening sequences of RNA not expressed in proteins
Exons
Retained in mature mRNA and are the expressing sequences
Splicesome
Protein/RNA complex that directs and ensures proper RNA splicing. Responsible for removal of introns from transcribed mRNA
Responsible for both cleavage of the intron from the RNA and ligation of the remaining exons
hnRNA
Pre-mRNA
Immature single stranded mRNA present in the nucleus (contains introns)
snRNA
“Snurps”
Small nuclear RNA molecules associated with specific ribonuclear proteins playing essential role in splicing
[U1,U2, U4, U5, U6]
mRNA
“Mature mRNA”
RNA that is translated to become a polypeptide chain
Four Distinct Types of Introns
Introns in protein coding genes, removed by splicesomes
Introns in tRNA genes, which are removed by proteins
(Group 1) Self-splicing introns, which catalyze their own removal from mRNA, tRNA, and rRNA precursors using guanosine-5’-triphosphate (GTP), or another nucleotide cofactor (ribozymes)
(Group 2) Self-splicing intros, which do not require GTP in order to remove themselves but do require assistance from proteins
Intron
Intervening sequences
Has GU, AG, polypyrimidine tract, and branch point sequence
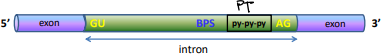
GU nucleotide sequence
At the 5’ splice site (donor site)
AG nucleotide sequence
At the 3’ splice site (acceptor site)
Polypyrimidine tract (PPT)
Just upstream of the 3’ splice site promotes the assembly of the spliceosome
It comes before the AG
Branch point sequence (UACUAAC)
The binding site for the snRNP-U2
Spliceosome Assembly (steps 1-5)
U1 binds to 5’ splice site
U2 binds to BPS
Trimer of U4, U5, and U6 recruited to 5’ splice site
U1 and U4 dissociate from hnRNA leaving U5 and U6 bound
U2 and U6 associate
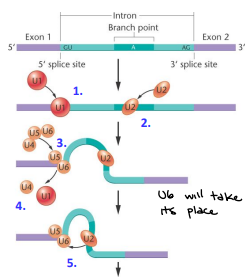
Spliceosome Assembly (steps 6-9)
“Lariat” of hnRNA is formed from the intron
5’ splice site is cleaved and U5 binds to 3’ splice site
3’ splice site is cleaved and U5 ligates exons together via ATP hydrolysis
snRNP’s are released along with spliced intron
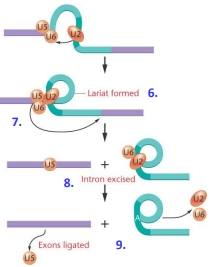
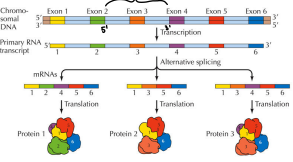
Alternative Splicing
A regulated process that results in a single gene coding for multiple proteins
It picks and chooses introns
It increases genetic diversity
The mechanism of removal is the same as the spliceosome
Under the control of specific factors
Trans-Acting Factors
Usually proteins that control gene expression and affects the DNA
Cis-Acting Factors
DNA sequences in the vicinity of a gene required for gene expression. Doing something to a nucleic acid
Primary splicing defects
Sequences in the pre-mRNA important for splicing are mutates (cis-activating factors)
Secondary splicing defects
Regulatory factors essential for splicing are mutated (trans-activating factors)
If the X protein acts on the RNA and covers the AG, then there will be more protein Z than protein Y