MCAT Biochemistry Flashcards
1/160
Earn XP
Description and Tags
This time, I'm just going to manually do everything. I'll only include flashcards from things I did not know before.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
161 Terms
hnRNA
pre-mRNA (unprocessed form of eukaryotic mRNA)
snRNA
Small nuclear RNA, RNA that aids in splicing during post-transcriptional modifications inside of the nucleus
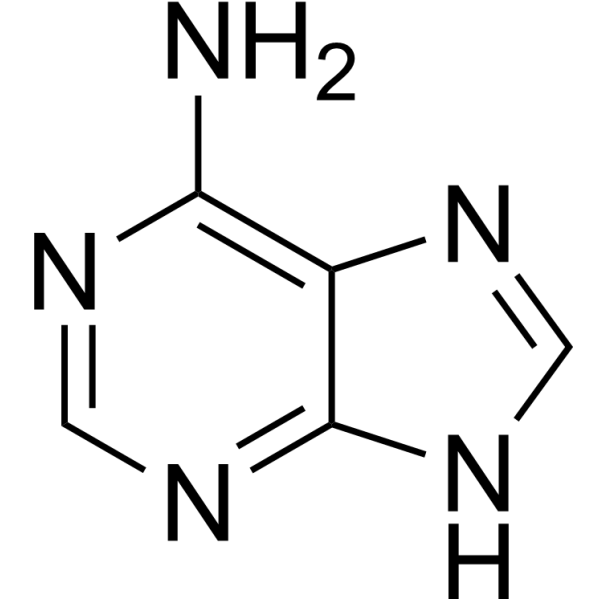
Adenine
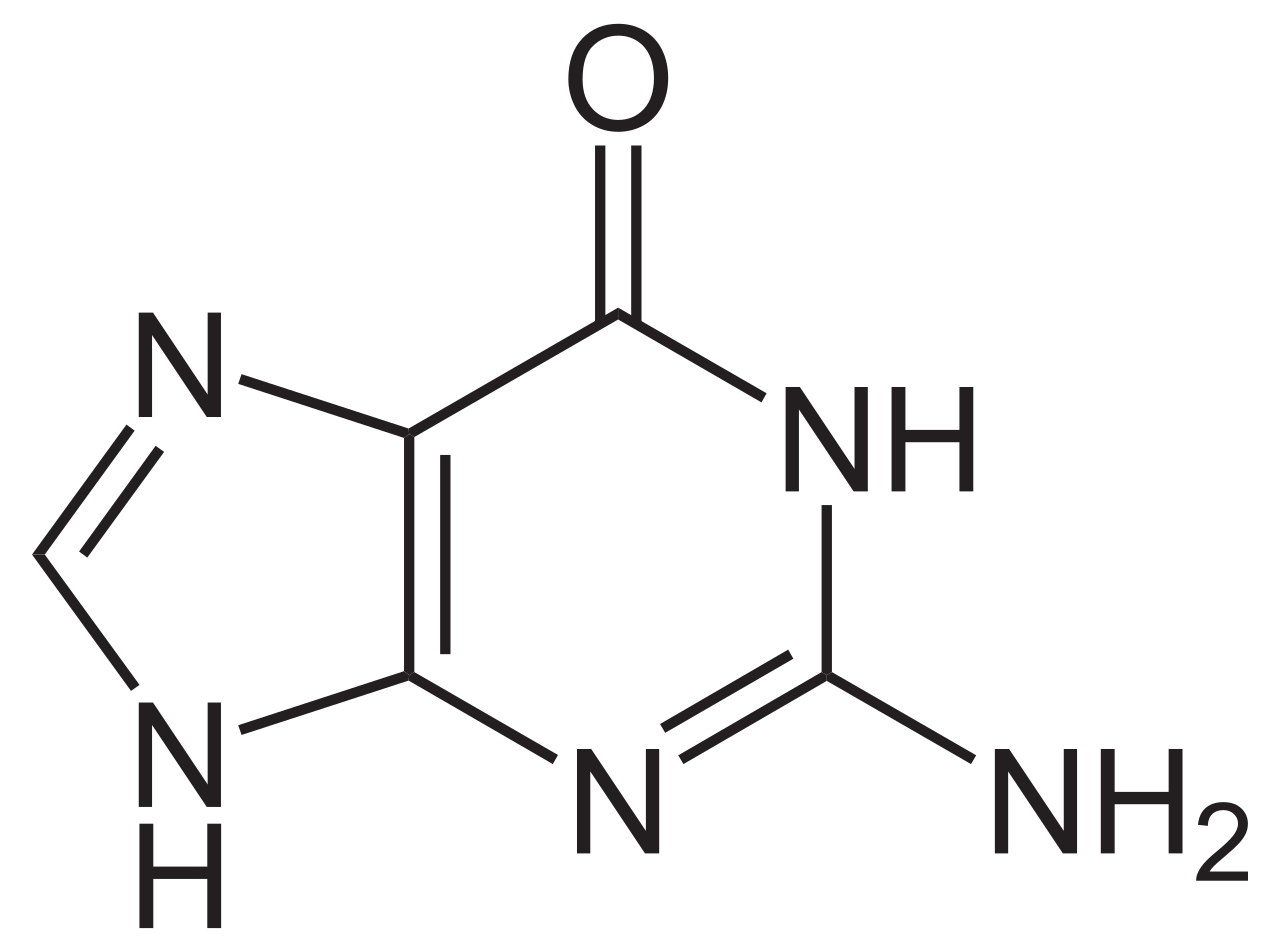
Guanine
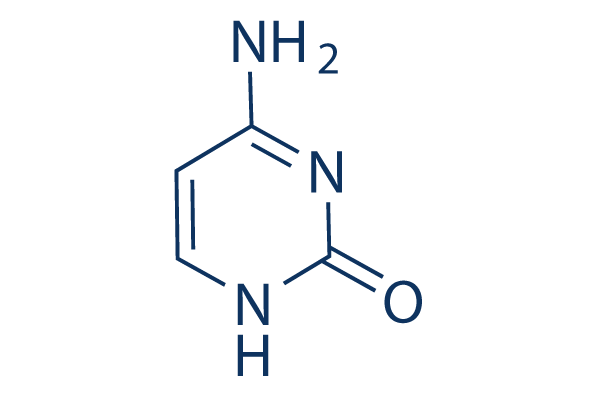
Cytosine
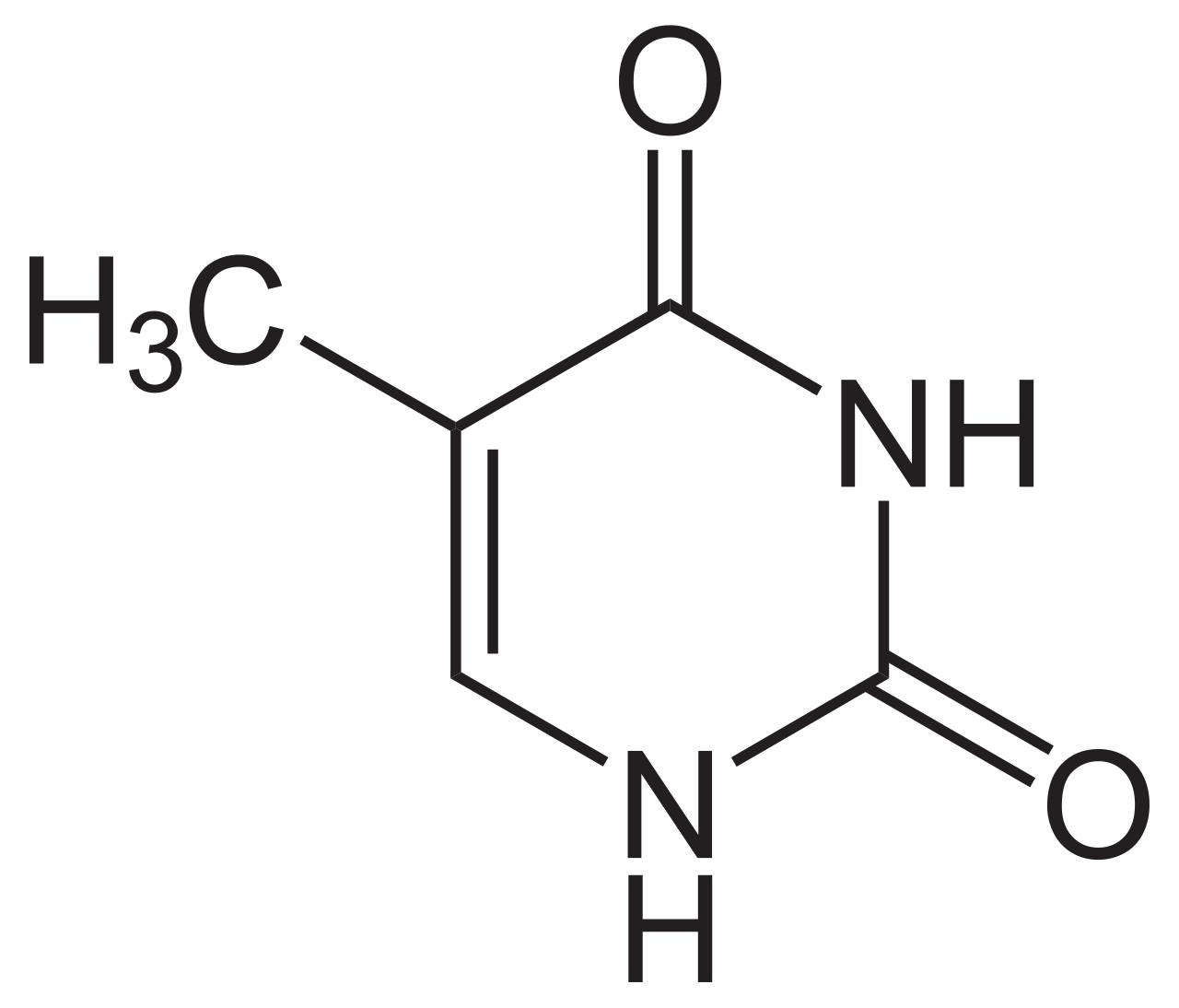
Thymine
Which of the nitrogenous bases are purines and pyrimidines?
Purines: A and G
Pyrimidines: C and T
Memorize “Pure As Gold” and “Purines have two rings.”
Helicase, topoisomerase, DNA Pol, Primase, and Ligase
Helicase - unzips DNA
Topoisomerase - Relives torsional strain
DNA Pol - adds dNTPs in 5’ to 3’ direction (specifically DNA Pol III)
Primase - Adds RNA primers to signal DNA Pol where to start
Ligase - Seals nicks
DNA Repair Mechanisms
Mismatch Repair (DNA Pol can detect wrong nucleotides after replication has completed)
Endonucleases (cut DNA to refill the correct base)
Methylation (in prokaryotes, methylation indicates what is the parent strand)
What are the post-transcriptional modifications in eukaryotes for mRNA? What about prokaryotes?
5’ m7G (methylguanosine cap)
Splicing (remove introns, join exons together)
Poly A tail
Prokaryotes do not do any post-transcriptional modifications
Ribosomes are made of two subunits, a large and a small. One set is 60S and 40S to give you an 80S ribosome, while the other is 50S and 30S to give you a 70S ribosome (math just doesn’t math here). Which one corresponds to prokaryotes and eukaryotes?
Prokaryotes - 70S
Eukaryotes - 80S (eukaryotes even in the tens place; 4, 6, 8)
Start and stop codons
Start - 5’ AUG 3’ (Met)
Stop - UAA, UGA, UAG
In post-translational modifications, what is lipidation, ubiquitination, glycosylation, and phosphorylation?
Lipidation - addition of a lipid to the protein
Ubiquitination - Addition of ubiquitin, a regulatory protein that often marks proteins for degradation/recyling
Glycosylation - addition of a sugar to the protein
Phosphorylation - addition of a phosphate to the protein
Proteolysis activation
When you cut the protein in order to activate it
Chaperones
Proteins that aid in the proper 3D folding of a protein by helping facilitate the noncovalent interactions in secondary through quaternary structures
In translation, the addition of each amino acid requires the input of _____ ATP across the various steps of translation.
4
Polycistronic vs monocistronic
Polycistronic - prokaryotes are capable of making multiple proteins from a single strand of mRNA at one given time
Monocistronic - eukaryotes are only capable of making one protein from a single strand of mRNA at a given time
Alternative splicing (in transcription)
We can customize a protein by only splicing together certain exons (so while the gene leads to only one protein, we can make it a little different by splicing together different exons). We call these isoforms
Euchromatin vs heterochromatin
Euchromatin are more loosely wound around histones, making them more easily accessible to transcriptional proteins and enzymes. Heterochromatin are more tightly wound around histones, so they are less accessible.
I remember this by “Eu” = “Easy for proteins”
P53
Tumor suppressor gene that sends the cell into arrest if there is damage to DNA at the growth checkpoints (if this is mutated, the cell cycle isn’t stopped if there is damage, leading to tumor growth)
Proto-oncogenes vs oncogenes
Proto-oncogenes are normal versions of genes that promote regular growth patterns, while oncogenes are cancerous versions of proto-oncogenes that promote uncontrolled and enhanced cell growth
Inducible vs Repressible Operons
Inducible Operons are those that are normally off with a repressor bound to the operator site, preventing transcription. A repressible operon is normally on, but can be shut off by the binding of a repressor
Aliphatic Amino Acids
Hydrocarbon R groups that aren’t aromatic (G, A, V, L, I, M, and P)
Doesn’t include F and W (those are aromatic)
Essential Amino Acids
The amino acids we can’t synthesize on our own (F, V, T, W, I, M, H, K, L)
Pi-Stacking Amino Acids
Have pi bonds in aromatic rings (F, Y, and W)
Commonly Phosphorylated Amino Acids
S, Y, and T (anything with OH)
Ketogenic vs Glucogenic amino acids
Ketogenic are amino acids that can be broken down into Acetyl CoA to be sent to the Krebs Cycle (L, and K)
Glucogenic are amino acids that, upon being catabolized, are turned into intermediates in glucogenesis (D and R)
Cysteine vs Cystine
Cysteine is the amino acid, while cystine is the oxidized form (contains S-S bond)
pI (isoelectric point)
the pH at which the zwitterion is reached (the overall charge of the amino acid is neutral)
Alpha helices are most commonly ____-handed. H-bonds are between every _____ amino acid, and a turn of an alpha helix is completed roughly every _____ residues.
right; 4th; 3.6
Parallel vs Antiparallel Beta Sheets (name kind of gives it away, but how can you recognize it from a structure)
In antiparallel beta sheets, the NH and C=O are right in line with each other, while in parallel beta sheets, they are staggered
Tertiary structures can have different types of turns that are classified based on how far away the hydrogen bonding amino acids are (i vs i + N). Match the following with it’s name (greek letter)
i → i + 2
i → i + 3
i → i + 4
i → i + 5
gamma
beta
alpha
pi
(Get Back And Practice)
How would you disrupt each level of protein structure?
1 - use enzymes or hydrolysis reactions
2 - denaturation (pH, increasing T, salinity, using SDS or sodium dodecyl sulfate, or using urea)
3 - Denaturing agent or condition (if you want to disrupt disulfide bonds, you need reducing conditions)
4 - Denaturing agent or condition (same as 3)
Localization sequences vs signal sequences (in proteins)
Localization sequences are a certain strain of amino acids within any part of the peptide sequence that destines the protein for movement towards a certain organelle or region of the cell
Signal sequences are similar to localization sequences, but specifically are regions of a peptide sequence typically near the N terminus that will imbed themsleves into the plasma membrane
Cofactor
Nonprotein components that are added to proteins for proper functioning of the protein (like iron to hemes). They need these cofactors or else they can’t function
Apoprotein vs holoprotein (use the term prosthetic group in your response)
Apoprotein is the inactive form, and when a prosthetic group is added, it becomes active, which is the holoprotein
Ordered vs Random ordered vs Ping-Pong Mechanism
Ordered - the substrates will bind to the enzyme in a specific, fixed order, with the substrates bound to the enzyme at the same time
Random - The order in which the substrates bind to the enzyme doesn’t matter
Ping-pong - A specific substrate will bind first in order to slightly modify the enzyme, then being released. The now modified enzymes can then bind the second substrate to create the product (substrates bind at different times)
Michaelis Menten Equation
v = (v_max [S]) / (K_M + [S])
Enzyme Kinetics Assumptions
[E] is constant (steady-state assumption)
[S] >> [E]
The reaction is irreversible
When studying the rate at a given [S], we only measure the initial rate
Efficiency of catalyst
k_cat / K_M
Turnover number (enzyme kinetics)
k_cat = v_max / [E]
Positive cooperativity. What curve does it yield?
The binding of one active site makes the binding of other active sites easier. It yields a sigmoidal curve (S-shaped)
Hill’s Coefficient
The number used to measure cooperativity (> 1 is positively cooperative, < 1 is negatively cooperative, = 1 is non-cooperative)
Describe how K_M and v_max change in competitive, uncompetitive, and non competitive inhibitors.
increase; no change
decrease; decrease
no change; decrease
Zymogens
Enzymes that are formed and secreted in their active form, later activated by a cleavage reaction from a protease or environmental conditions
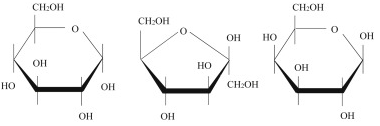
What the structures from left to right?
Glucose; fructose; galactose
What makes up maltose?
Two glucose monomers linked via an alpha-1,4 glycosidic linkage
What makes up sucrose?
One glucose and one fructose monomer linked in an alpha-1,2 linkage
What makes up lactose?
One glucose and one galactose monomer linked in a beta-1,4 glycosidic linkage
What makes up trehalose?
Two glucose monomers linked via an alpha-1,1-alpha linkage
What makes up glycogen?
Contains tons of D-glucose monomers linked in alpha-1,4 linkages, as well as alpha-1,6 linkages for branching points
In the anomeric carbon, alpha means the OH group is pointing _____, while in beta, it’s pointing _____.
down; up
What makes up starch?
Less often branching (alpha-1,6) bonds than glycogen, but it’s overall very similar to glycogen
What makes up cellulose?
Glucose monomers with beta-1,4 linkages
In the Fischer projection, substituents to the left will be pointing _____, while substituents to the right will be pointing _____ when converted to the ring form.
up; down
Ring flip
Flip the chair, and all substituents shift clockwise, but change from axial to equatorial and vice versa
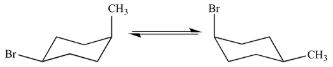
Mutarotation (in sugars)
The conversion of one anomer to the other (alpha to beta or beta to alpha)
D and L enantiomers. D is the same thing as an _____ configuration, just specifically applying to the penultimate carbon. L, on the other hand, is the same thing as an _____ configuration.
R; S
Mammals have _____ carbohydrates and _____ amino acids.
D; L
Do unsaturated fatty acids or saturated fatty acids have a higher boiling point?
Saturated (unsaturated fatty acids like oil are liquid at room temperature, while butter, a saturated fatty acid, is a solid at room temperature)
Amphipathic vs Amphoteric
Amphipathic - having both polar and nonpolar portions of a molecule (fatty acids are mostly nonpolar, but the carboxyl group is polar)
Amphoteric - a substance that can act as an acid or base
Triglyceride Synthesis
React glycerol with a carboxylic acid (fatty acid). The OH on glycerol attacks the carboxyl group to give you the ester
Adipocytes
Cells specifically designed for fat storage
Chylomicrons
A type of lipoprotein (fat-carrying protein) that aids in the transport of fats, cholesterols, or fat soluble vitamins (KADE) through the blood stream
Wax
A super long fatty acid esterified to long alcohols, super hydrophobic (you form this by a normal esterification reaction where a carboxylic acid and an alcohol are added together to form an ester)
Terpene
Precursors to steroids, made form isoprene units
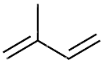
Isoprene
Steroid
Hydrophobic molecules important in hormones or other signaling pathways, based on the structure of cholesterol
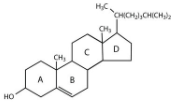
Cholesterol
Vitamin
Organic compounds used as cofactors or other important reactants in many biological processes, can be lipid soluble (hydrophobic) or water soluble (hydrophilic)
What are the fat soluble vitamins? Indicate how many rings or other notable structures in them.
K - important blood clotting factor, 2 rings with quinone
A - carotene, helps with vision in the eyes, 1 ring
D - becomes calcitriol, regulates calcium and phosphorus (vitamin D deficiency causes Rickets), 3 rings
E - Tocopherol, important aspect of antioxidants (prevent reactive oxidative species from causing damage), 2 rings, non quinone
Remember Fat KADE
List what each of the B vitamins are the precursor to
B1 - TPP
B2 - FAD (FADH2)
B3 - NAD (NADH)
B5 - CoA (acetyl CoA)
B6 - PLP
Hmm in order, TFNCP
The Fox knows (N) C/P on the MCAT??? LOL
Agarose Gel Electrophoresis
Uses a gel made of agarose instead of polyacrylamide to study the size of DNA molecules
Polyacrylamide Gel Electrophoresis (PAGE)
Uses polyacrylamide (PA) in the gel to study proteins
Polyacrylamide = polypeptide (protein)
Isoelectric Focusing (IE)
Type of gel electrophoresis that will separate proteins on the basis of their charge (pI)
Acidic amino acids will have a pI below 6, while basic amino acids will have a pI above 6
Native PAGE. Indicate what proteins are on top and what are on bottom
When nothing is added to the proteins before being added to the gel
The heaviest proteins stay near the top, and the lightest proteins migrate the furthest toward the bottom (in general, heavy = top, light = bottom)
Nonreducing SDS PAGE vs Reducing SDS PAGE
Nonreducing SDS PAGE is a specific type of PAGE where SDS (sodium dodecyl sulfate) is added to the proteins before they are run, causing them to have a uniformly negative charge, so they are only separated by the size or shape. It also disrupts non-covalent interactions that may be holding together the quaternary structure of proteins together.
Reducing SDS PAGE is the same thing, but you add a reducing agent that reduces and breaks covalent disulfide bonds (remember, cysteine has an SH group that when reduced, is bonded to another cysteine sulfur atom)
Western Blotting
A blotting technique that uses reducing SDS PAGE more-so used to identify whether or not a specific protein of interest (of a known weight) is present in a solution of proteins
It shows you relative changes in concentration, but can’t tell you the exact concentration of a protein
Ion-Exchange Chromatography, Anion-Exchange Chromatography, and Cation-Exchange Chromatography
Used to separate proteins based on their charge, either anionic or cationic, by passing a solution of proteins over beads coated either with negative or positive charges.
In anion-exchange chromatography, the beads are positive, so you retain the anions more. This means positive proteins elute first, and negative proteins elute last
In cation-exchange chromatography, the beads are negative, so you retain the cations more. So negative proteins elute first, and positive proteins elute last
Affinity Chromatography
Separates a protein of interest by coating the beads with ligands that the specific protein can bind to. Then to remove the isolated proteins, you put the solution in a very high concentration of free ligands, which displaces the protein
Immunoprecipitation (primary vs secondary antibodies)
Used to detect and separate a protein of interest from a solution of proteins by adding a y that is specific to the protein of interest to a bead. Once the bead and antibody are bound to the protein, it can be removed from the solution (it’s basically affinity chromatography, but we use antibodies instead of any ligand)
Primary antibodies directly bind to the protein, while secondary antibodies are first bound to a fluorescent tag (for visualization), then bind to the primary antibody
Radioimmunoassay
Used to monitor the concentration and amount of a certain protein in a solution by binding to the proteins of interest to antibodies and iodine so their relative light emission can be measured
Flow cytometry
Fluorescent tags are added to specific proteins on cells which are then passed under light so that the cell size or number of cells can be measured
Bradford Quantification
A quantitative lab technique that measures the concentration of protein by binding Coomassie blue (a dye) to proteins and measuring the relative absorbance of the solution (A = \epsilon bc)
Edman Degradation
Used to determine the amino acid sequence of small proteins by removing amino acids one at a time from the N terminus
Salting Out/Dialysis (no not what you give to patients who have kidney failure)
This is used to purify proteins
Salting out is when a high concentration of salt is added to a solution of proteins which causes salt to compete with the protein to interact with water, causing the proteins to precipitate/fall out of the solution to be collected
Dialysis is when molecules of different sizes separate from each other using a semipermeable membrane that allows ions to leave the solution and enter the surrounding water, leaving behinds the proteins alone
In the Ramachandran plot, where do you see right-handed alpha helices, left-handed alpha helices, and beta sheets?
Right-handed alpha helices are in the bottom left corner
Left-handed alpha helices (again, not natural) are in the top right corner
Beta sheets will appear in the top left corner
Describe the steps of PCR.
You heat the sample to denature the DNA
You cool and add primers to allow them to bind to target sequences
You hadd Taq polymerase with dNTPs to extend/synthesize new strands
Repeat the process as many times as you like (number of copies is x 2^N, where x is the number of copies at the start of PCR, and N is the number of times you do this process)
qPCR vs RT-PCR
qPCR measures the amount of concentration of a gene of interest. It does this by using a fluorescent dye that will increase as the number of strands increases and can be measured
RT-PCR identifies if a specific mRNA is present and determine when the genes are being expressed (you add reverse transcriptase to convert mRNA into cDNA)
Fluorescence in-situ hybridization (FISH)
Identifies whether a specific DNA or RNA sequence is present (if it is, it binds to the probe, which will be complementary to the gene of interest)
Hybridization is when two complementary strands bind to each other
In Vitro Mutagenesis
Identifies the functioning of a gene (random mutations are introduced into a gene to see how the functioning of the gene changes)
Restriction Fragment Length Polymorphism (RFLP)
Identify mutations or polymorphisms (you add restriction enzymes to cut gene sequences at palindromic sequences, and if the lengths are different than expected, mutations are present
Tollen’s Test vs Benedict’s Test
Both detect the presence of reducing sugars, which will have a free hydroxyl (hemiacetal or hemiketal) in ring form on the anomeric carbon
In Tollen’s Test, if a reducing sugar is present, Ag+ is reduced to Ag (S) forming a silver mirror
In Benedict’s Test, if a reducing sugar is present, Cu2+ is reduced to a red precipitate form of copper (goes from blue to red)
Gram Stain
Identify the type of bacteria present by staining it with a purple dye, washing it, and then dying it again with a pink dye
If it’s gram negative, the purple dye will be washed away, and therefore they appear pink due to the last stain
If it’s gram positive, the purple dye is not washed away, so it appeaks purple
Reporters
Any measurable signal (color, fluorescence, radioactivity, etc.)
Supernatant
The liquid that remains above a pellet/bead after centrifugation
In Vitro vs In Vivo
In Vitro - anything done outside of a liquid organism, commonly in a petri dish or test tube (it’s artificial)
In Vivo - anything done inside of a living organism (natural)
Mediating vs Moderating variable
Mediating - a variable that helps explain the relationship between the independent and dependent variables
Moderating - A variable that changes the strength or direction of the relationship between the independent and dependent variables
Vehicular Control vs Negative Control vs Placebo Control vs Positive Control
Negative Control groups don’t get anything
Vehicular Control groups receive an inert treatment (like if you are testing a vaccine, the vehicular control group would receive a shot that contains saline)
Placebo Control groups receive a placebo (fake treatment) to control for psychological effects
Positive control groups receive an effective treatment with an already understood response
Potentiation
The factor or degree with which a variable has changed, determining how significantly the variable was changed (like if it increases from 0.1 M to 0.5 M, the potentiation factor is 5)