BI213: Exam III
1/304
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
305 Terms
endoplasmic reticulum
What is a network of membranes throughout the cytoplasm that makes up 10% of the cell volume?
movement of proteins:
1. Rough ER
2. Golgi
3. Secretory Vesicles
4. Cell Exterior
What is the secretory pathway?
Pulse-Chase: labeling with radioactive amino acids, fixing cells, and exposing to X-ray film
- leave for a certain amount of time and view where the labeled amino acid is in the cell (ER, vesicles, etc.)
How was the secretory pathway initially characterized?
GFP protein - keeps the cells alive
Without the radioactive labeling and fixing cells, how else can we view the secretory pathway experimentally?
nucleus and mitochondria
Where do proteins translated in free ribosomes (in cytosol) end up?
plasma membrane, nuclear membrane, secretory vesicles
Where do proteins translated in the membrane-bound ribosomes (endoplasmic reticulum) end up?
around 12 hydrophobic amino acids
What are N-terminal signal sequences on secretory proteins made of?
- N-terminal signal sequence is recognized by Signal Recognition pArticles (SRP - proteins and RNA)
- allows association with membrane of the endoplasmic reticulum
- the SRP receptor binds everything, allowing the translocon to open and protein enters the lumen
- ribosome remains associated and continues translation
- Signal peptidase removes the sequence
- Protein is released into the ER LUMEN, chaperones in the lumen facilitate folding
Describe the cotranslational targeting of secretory proteins to the ER
in the endoplasmic reticulum, initially inserted into the ER membrane
Where are plasma membrane proteins synthesized?
the presence or absence of signal sequences
What does the orientation of membrane proteins generally depend on?
- some proteins have a signal sequence AND a transmembrane alpha helix in the middle of the protein
- when the signal sequence is removed and the transmembrane alpha helix is inserted laterally, translocon becomes closed and translocation is halted
- alpha helices exit the translocon laterally (C-terminus in cytosol) and transcription continues in the cytosol
Describe the insertion/orientation of a membrane protein with a cleavable signal sequence?
- without a signal sequence, translation will occur in the cytosol until the alpha helix
- SRP recognizes and associates with alpha helix
- NO signal sequence so transmembrane sequences recognized by SRP (binds to alpha helix region) (not cleaved)
- N terminal remains in cytosol and the rest is fed through the translocon
Describe the insertion/orientation of membrane proteins with internal transmembrane sequences? No Signal Sequence
orientation of transmembrane sequence
What also determines the N- and C-terminal location?
function (DO NOT ASSUME THAT AMINO SIDE CAN'T be in lumen without the sequence)
What does the directionality of proteins that span the membrane multiple times depend on?
- SRP recognizes the helix, translocon opens laterally, second alpha helix will be inserted into the membrane
- alternating regions between the cytosol and lumen between helices
- after the third helix, additional polypeptides are inserted into the translocon
Describe the multi-pass orientation of transmembrane proteins
An Hsp70 chaperone that facilitates protein folding in the endoplasmic reticulum
What is BiP?
protein-folding sensory proteins (eg. chapersones)
they are removed from the ER and targeted back to the cystol where they are degraded by the ubiquitin-proteasome system
In ER associated degredation (ERAD) what are misfolded proteins identified by?
What happens to them?
signal initiates at endoplasmic reticulum (3 receptors in ER membrane: ATF6, IRE1, and PERK)
three transcription factors: XBP1, ATF6, ATF4 regulate Unfolded Protein Response genes inc. in
chaperones (they enter the nucleus to do this)
DIFFERENT mechanisms
What occurs in the cell when there is an excess of unfolded proteins?
What are the factors activated by?
some are refolded and some are degraded
What are possible results of the unfolded protein response?
membrane lipids (phoshpholipids, glycolipids, cholesterol)
What are synthesized in the smooth endoplasmic reticulum?
chemical reactions with a series of specific enzymes on the CYSTOLIC FACE synthesizes lipids on outside (not in lumen)
Where does synthesis in the smooth ER occur? How?
flippase
What enzymes flips phoshpholipids on cytosolic face to lumen face?
the way in which parts are arranged
What is topology?
proteins or protein domains inside the ER wind up outside the cell
What is the topology of the secretory pathway?
ERGIC - distinct complex/structure between the ER and golgi
What is the ER-Golgi intermedate complex?
proteins are marked by "retrieval sequences" (KDEL) that signal their return to the ER
What happens when ER proteins are "accidentally" incorporated into vesicles and transported to the Golgi?
KDEL receptor (in the golgi)
What is the recycling receptor? Where is it located?
- Proteins are further processed (additional modifications through O or N linkages) of sugars
- synthesis of glycolipids
- sorting for transport
What happens during transport through the golgi apparatus?
YES - the cis side is closest to the ER and the trans side is closer to the plasma membrane
Does the Golgi have directionality?
regulated secretion - secretion occurs in response to environmental stimulation
continuous secretion - unregulated
Describe the two forms of transport FROM the golgi apparatus?
function of proteins
What do the two forms of golgi secretion depend on?
digestive system of the cell (break down macromolecule)
What are lysosomes?
Golgi vesicles (bud from transgolgi) fusing with endosomes (endocytosis)
Extreme acid environment (pH 5) for enzymatic breakdown of macromolecules
How are lysosomes formed?
What conditions do they have?
turnover of cellular components (recycling to be more efficient/expend less energy under cell stress conditions or normally)
lysosomes
What is autophagy?
What is involved?
proteins move from ER through secretory pathway in vesicles that need to recognize and fuse only with the appropriate target membrane
What is the mechanism of vesicular transport?
1. Cargo selection
2. Coating (proteins cover modified proteins)
3. Budding (associated with microtubulues on cytoskeleton)
4. Uncoating
5. Fusion with correct target (secretion into the new membrane)
What are the steps in the formation and fusion of a transport vesicle?
COPII
What proteins coat vesicles moving from ER to the Golgi?
COPI
What proteins coat vesicles going from golgi to the ER?
Clatharin
What proteins coat vesicles going between Golgi to lysosomes and plasma membranes (both forward and reverse)
Arf (a small GTP binding protein)
What is the formation of a clathrin-coated veiscle regulated by?
GEF
What facilitates the exchange of GDP to GTP in Arf?
- arf is specifically activated in Golgi membrane
- Arf/GTP recruits adaptor proteins which bind to transmembrane receptor (specific and in lumen) + cargo
- Clathrin (coat protein) binds to adaptor proteins
Describe the formation of a clathrin coated vesicle?
distorts membrane and drives vesicle budding
What does clathrin do?
Rab family small GTP binding proteins
What directs vesicle fusion with the correct target membrane?
- initial protein interaction between tethering factors and Rab-GTP
- Tethering factors stimulate coilings interaction between SNAREs (transmembrane proteins)
- SNARE-SNARE interactions provides energy for fusion
Describe the process of vesicle fusion?
hundreds-thousands/cell
metabolic reactions (citric acid cycle) in matrix
Oxidative phoshporylation (generation of ATP) in inner membrane
How many mitochondrion are in a cell?
What occurs in the mitochondria?
- bound by two membranes (outer,inner)
- High surface area - cristae
- extremely impermeable to protons
- usually phospholipid cardiolipin
- high protein content
Describe the physical structure of a mitochondrion
- endosymbiosis (some circular genome remained and wasn't integrated)
What is the human mitochondrial genome the result of?
-1500 proteins
encoded by nuclear genes and transfered to nucleus from ancestral bacterial genome
- human mitochondrial only encode 13 proteins (1%) which are transcribed by RNA polymerase II
About how many proteins do mitochondria contain?
How are 99% of mitochondrial proteins encoded?
free cytosolic ribosomes
N-terminal "presequence" of positively charged amino acids (ends up on the outer membrane)
Where are most mitochondrial proteins translated?
How are they targeted to mitochondria?
Matrix processing peptidase (MPP)
What enzyme performs proteolytic cleavage of the mictochondrial presequence?
TOM (Translocase of the outer membrane) and TIM (translocase of the inner membrane)
In the import of mitochondrial matrix proteins, what must the proteins go through
4 ATP molecules
What does glycolysis + the citric acid cycle yield?
breaks down acetyl coA (from glucose) to CO2 + high energy electrons in NADH and FADH2 (electron carriers)
What does the citric acid cycle do?
high energy electrons from NADH (10 molecules) and FADH2 (2 molecules) are passed through a series of carriers coupled to formation of ATP at energy yielding steps
How does the cell generate 38 ATPs from the output of the citric acid cycle?
3 ATP per pair of electrons (30 ATP total)
What can each NADH molecule generate?
2 ATPs per pair of electrons (4 ATP)
What can each FADH2 molecule generate?
energy from electron transport is used to pump protons across inner mitochondrial membrane at each complex (this energy is then used to drive ATP synthesis when protons flow back into the matrix)
How does the electron transport chain function?
each complex transports 4 protons per pair of electrons so 12 protons per molecule of NADH - note only 10 protons in intermembrane space per NADH molecule
What can each complex do in the electron transport chain?
1. Pairs of electrons enter the electron transport chain from NADH in complex I
2. Electrons are transferred to coenzyme Q which carries electrons to complex III
3. Electrons are transferred from cytochrome b to cytochrome c, which carries electrons to complex IV
4. Complex IV transfers electrons to molecular oxygen
5. Proton gradient is formed, and it is used to drive ATP synthesis as the protons flow back to the matrix through complex V
Describe the process of electron transport from NADH
1. Outer membrane is permeable
2. Inner membrane is impermeable
only route back is coupled to ATP synthesis
Describe the permeability of the mitochondrial membrane
ATP Synthase
What enzyme is complex V and used to generate 1 ATP from minimum of 3 protons
flow of protons down electrochemical gradient through channel drives ATP synthesis by F1 subunit - this is the reverse of a pump that uses ATP to drive flow against a gradient
F1 Subunit
Describe how ATP synthase functions?
What subunit drives ATP synthesis?
3
4
2; 10
How many protons are needed to activate ATP synthase to synthesize each ATP?
So transport of 12 protons from NADH yields how many ATPs?
How many protons are used for H20 in complex IV? How many protons go through ATP synthase?
electric and chemical
What is the nature of the proton gradient
used to drive flow of metabolites (eg. ATP) in and out of mitochondria
What is the energy of the electrochemical gradient used for?
small organelles; single membrane; -500 per cell
DO NOT have their own genomes
What is the structure of a peroxisome?
sites for a variety of biochemical reactions, including oxidation reactions leading to a production of hydrogen peroxide
What do peroxisomes function as?
catalase
What do peroxisomes contain that allows them to decompose hydrogen peroxide into water?
cell shape and movement
- actin filaments, microtubules, intermediate filaments
What does the cytoskeleton effect?
What are the three types of protein filaments that make up the cytoskeleton?
support plasma membrane and determine cell shape
muscle contraction and a variety of cell movements such as cell migration
What are the functions of actin filaments?
globlular (G) actin
What is the monomer of actin filaments?
YES, they have two distinct ends (plus and minus) that is based on the tertiary structure of globular actin
Do actin filaments have distinct ends? What is this based on?
turnover by loss (disassembly) or addition (polymerization) of actin monomers occurs in a process caused tredmilling
What does it mean that actin filaments are dynamic?
- association of actin/ATP is favored (high affinity for actin/ATP) at the plus end so the addition is very efficient
- at some point ATP is hydrolyzed, and the actin/ADP does not have a high affinity so it is lost at the minus end
ENERGY IS NOT REQUIRED
Yes, the stability and remoldeling of actin filaments is tightly controlled in cells
Describe the process of treadmilling
Is energy required at this point? Is this process regulated?
- initiation of a filament (nucleation)
specific proteins (Arp or Formin)
What is the rate-limiting step: the initiation or branching of actin filaments?
What is it mediated by?
Formin
What protein creates the linear filament structure of actin (is a dimer that gets initial actins together)
Profilin
Keeps lengthening at plus end more efficient
What protein stimulates the ADP/ATP exchange in treadmilling?
What does this do?
actin-related protein (Arp)
What protein is responsible for branching actin filaments?
cross-linking proteins
What proteins determine the structure of actin filaments in cells?
a-actinin
rigid and parallel
What proteins help form parallel actin bundles?
What describes this structure?
filamin
flexible and perpendicular
What proteins help form perpendicular structures?
What describes this structure?
association of actin cytoskeleton with plasma membrane: spectrin-actin network
What determines cell shape?
red blood cells
How was the spectrin-actin network discovered?
Integrin-based attachment
What actin cytoskeleton mediates attachment of cells to a surface or extracellular matrix
Focal adhesions
What are the sites of attachment in cytosol near membrane (where you find actin associated with integrin)
Cadherin-based attachment
What occurs when actin filaments mediate stable attachments between cells in tissues?
1. Integrin proteins are attaching cell to surface
2. Extension of leading edge occurs
3. Attachment to substratum by integrin proteins
4. Retraction of trailing edge
*actin filament is undergoing a lot of treadmilling
Describe the process of cell migration
branched actin filaments at leading edge of the plasma membrane (requires an extracellular signal)
What do surface protrusions result from?
Growth factors: Rho family GTP binding, WASP, Arp2/3, Formin (a complex for branching)
What are the growth factor proteins involved in cell migration?
Profilin
What activates ADP-actin monomers in cell migration?
Cofilin
What cleaves existing filaments and provides new plus ends in cell migration?
Myosin motor (a molecular motor that uses ATP to move along actin filaments)
What has movement produced by action of myosin?
ATPase activity (hydrolysis must occur for it to be stimulated)
What activity does the globular head region of Myosin II have?
multiple bundles of actin/myosin filaments (contractile unit)
What do muscle cells contain?
Sarcomere
Contractile unit of muscle
myosin moves toward the PLUS end of actin and Z bands are brought closer together
Myosin is fixed within the unit, but globular head sticks to actin pulling it toward M line
What occurs during muscle contraction in a sarcomere?
How?
Label The Bands of the Sarcomere
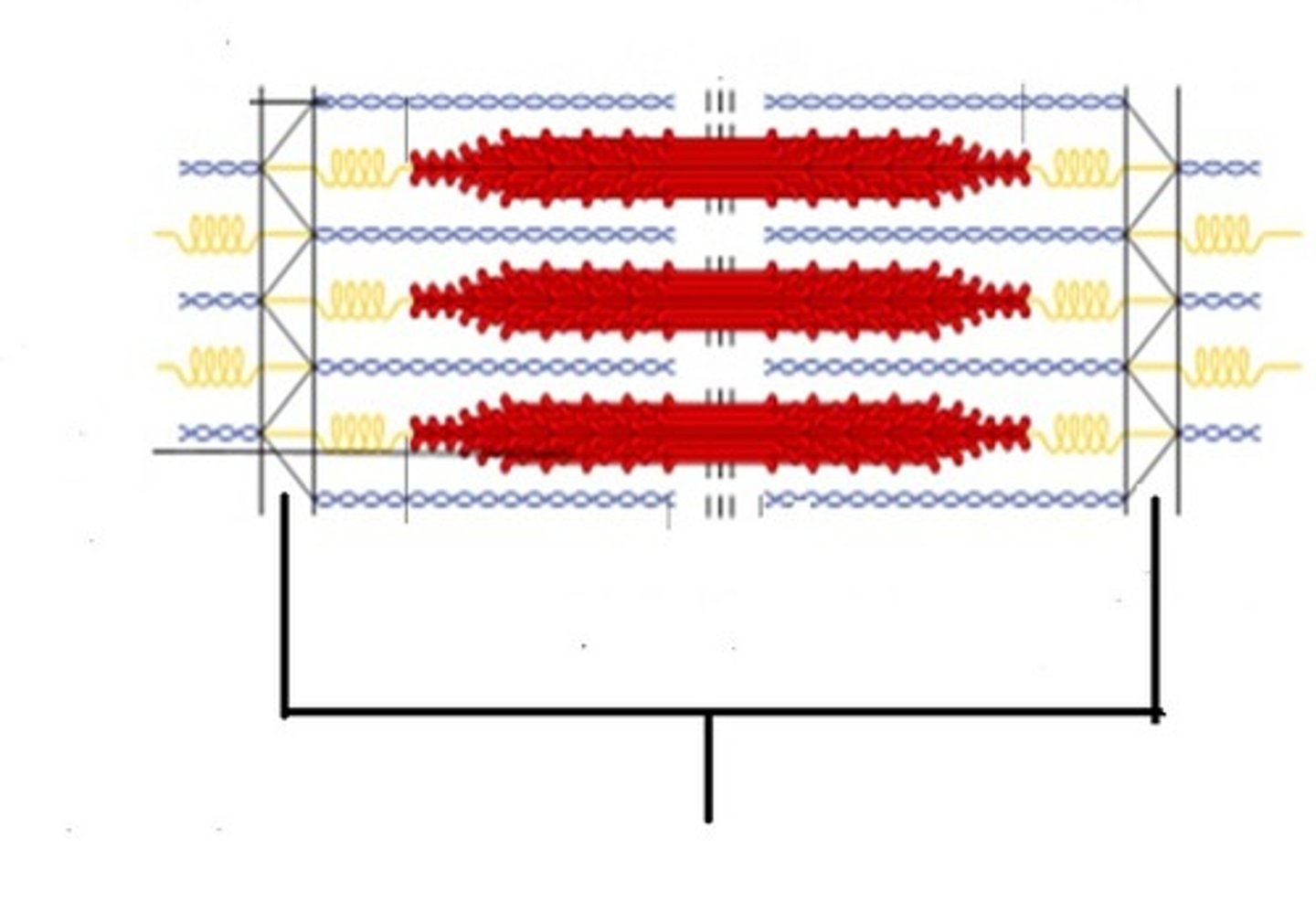
I band
What bands shorten during muscle contraction?
1. Dissociation of actin-myosin complex from binding of ATP
2. ATP hydrolysis conformational change that moves myosin head
3. Myosin head binds to new position on actin
4. Myosin head returns to original position as the release of Pi triggers a "power stroke" followed by release of ADP
Describe the process of myosin actin power stroke