6.2 DNA Replication
1/17
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
18 Terms
DNA Replication
DNA replicates during the S phase of the cell cycle
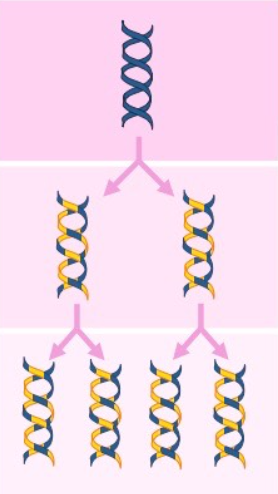
the alternative models of DNA replication are conservative, semi conservative, and dispersive
the correct one for DNA is semi conservative
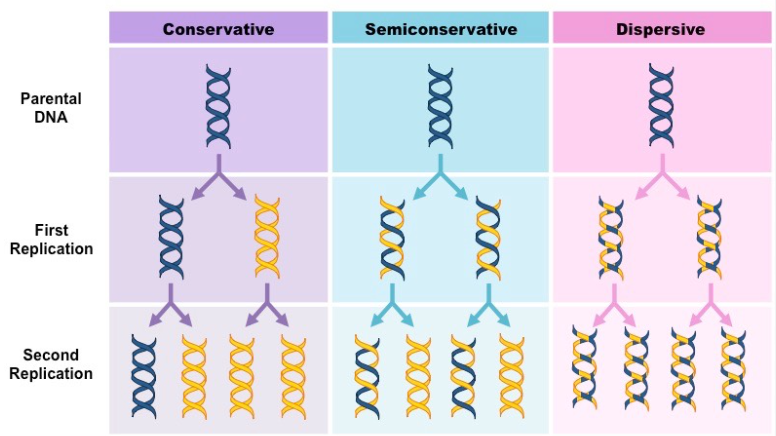
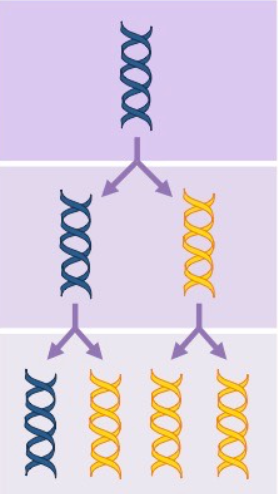
Conservative model
the parental strands direct synthesis of an entirely new double stranded molecule
the parental strands are fully “conserved”
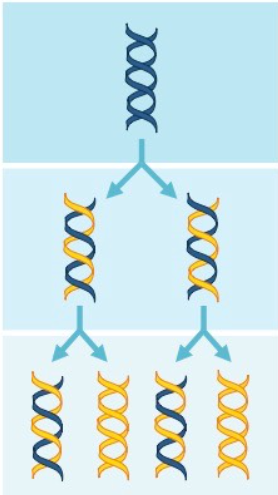
Semi conservative model
the two parental strands each make a copy of itself
after one round of replication, the 2 daughter molecules each have 1 parental and 1 new strand
Meselson and Stahl performed an experiment using bacteria to confirm that this was the correct model
Bacteria was cultural with a heavy isotope
Bacteria was transferred to a medium with a light isotope
DNA was centrifuged and analyzed after each replication
By analyzing samples of DNA after each generation, they found that the parental strands followed the semi-conservative model
Dispersive model
the material in the two parental strands is dispersed randomly between the 2 daughter molecules
after one round of replication, the daughter molecules contain a random mix of parental and new DNA
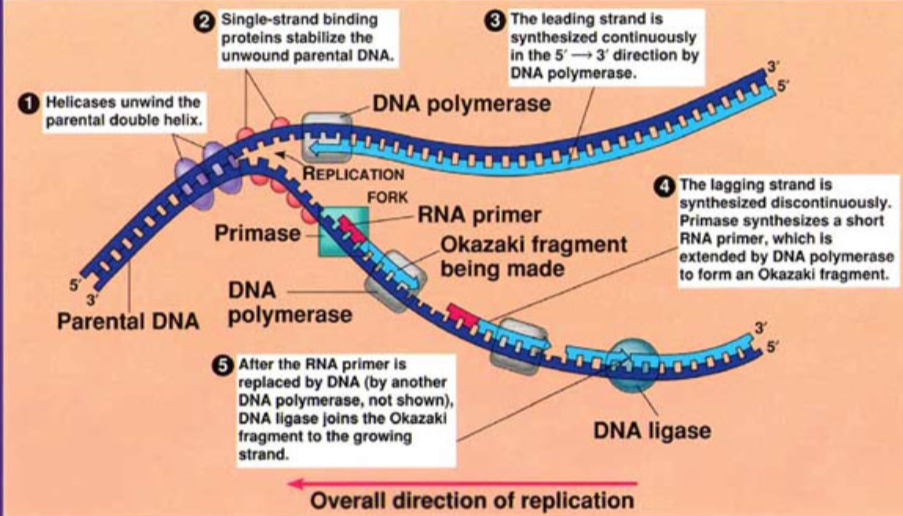
Steps in DNA Replication
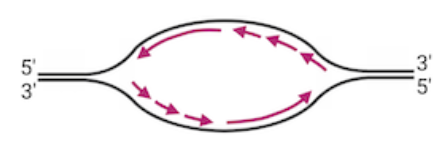
Proteins attach to the origin of replication and open the DNA to form a replication fork
Helicase unwinds the DNA strands at each replication fork
Primase initiates replication by adding primers to the parental DNA strand
DNA polymerase attaches to each primer on the parental strand and reads 3’ to 5’ direction. As it moves, it adds nucleotides to the new strand in the 5’ to 3’ direction
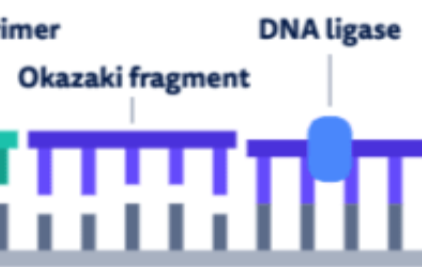
After DNAP III forms an okazaki fragment, a DNAP I replaces RNA nucleotides with DNA nucleotides
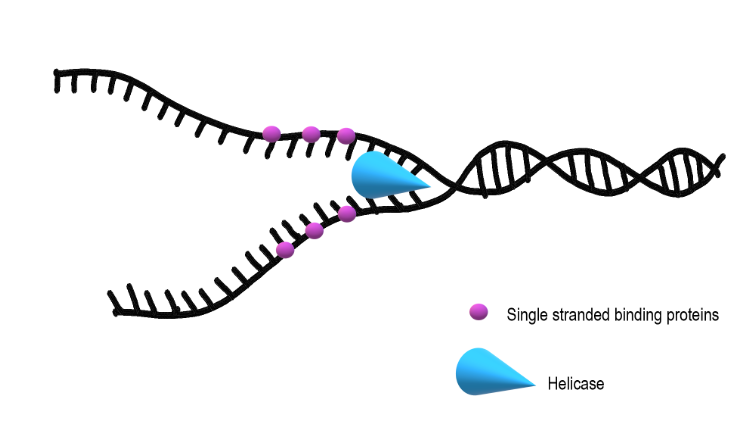
Helicase
unwinds the DNA strands at each replication fork
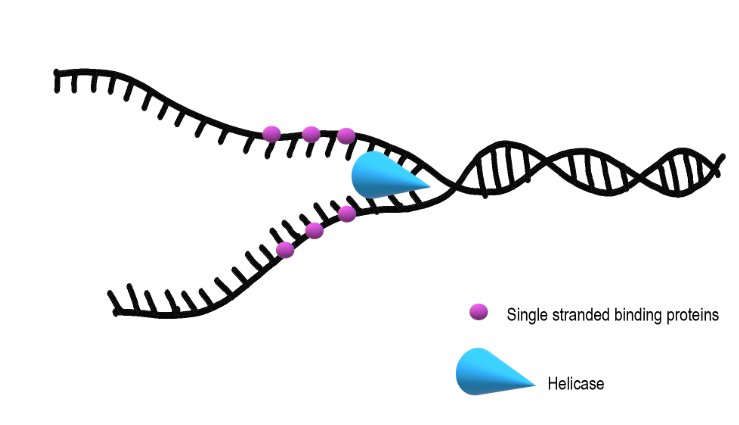
SSBPs
single strand binding proteins
to keep the DNA from re-bonding with itself, proteins called single strand binding proteins (SSBPs) bind to the DNA to keep it open
Topoimerase
helps prevent strain ahead of the replication fork by relaxing supercoiling
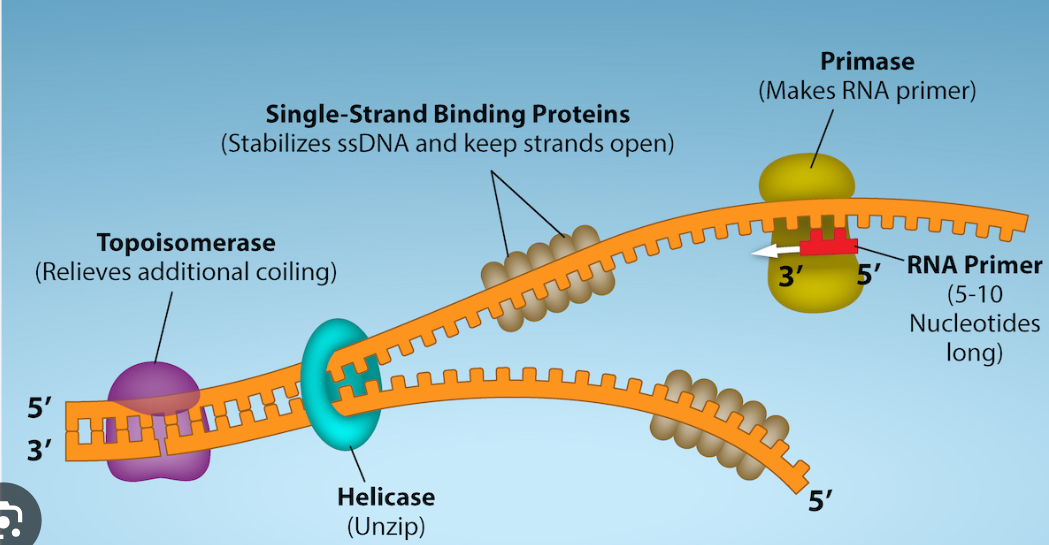
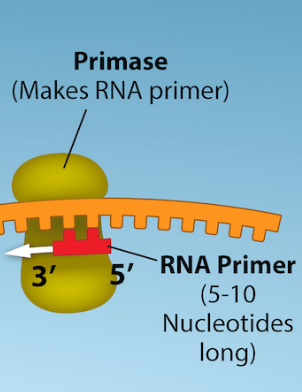
RNA Primase
enzyme that initiates replication by adding RNA primers to the parental DNA strand
the enzymes that synthesize DNA can only attach new DNA nucleotides to an existing strand of nucleotides
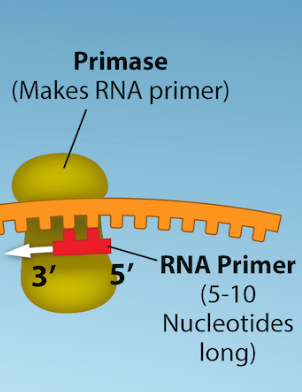
Primers
short segments of RNA
serve as the foundation for DNA synthesis
1 needed for leading strand, multiple needed for lagging strand
DNA Polymerase
antiparallel elongation- (DNAP III in prokaryotes) attaches to each primer on the parental strand and reads 3’ to 5’ directions(can only add nucleotides on 3’ end)
as it moves, it adds nucleotides to the new strand in the 5’ to 3’ direction(now its antiparallel to the parent strand)
the DNAP that moves towards the replication fork synthesizes the leading strand smoothly and continuously, requires only 1 primer
the DNAP on the other template strand moves away from the replication fork, synthesizing the lagging strand discontinuously short segments called okazaki fragments, requires many primers
after DNAP III forms an okazaki fragment, DNAP I replaces RNA nucleotides (from the RNA primers) with DNA nucleotides
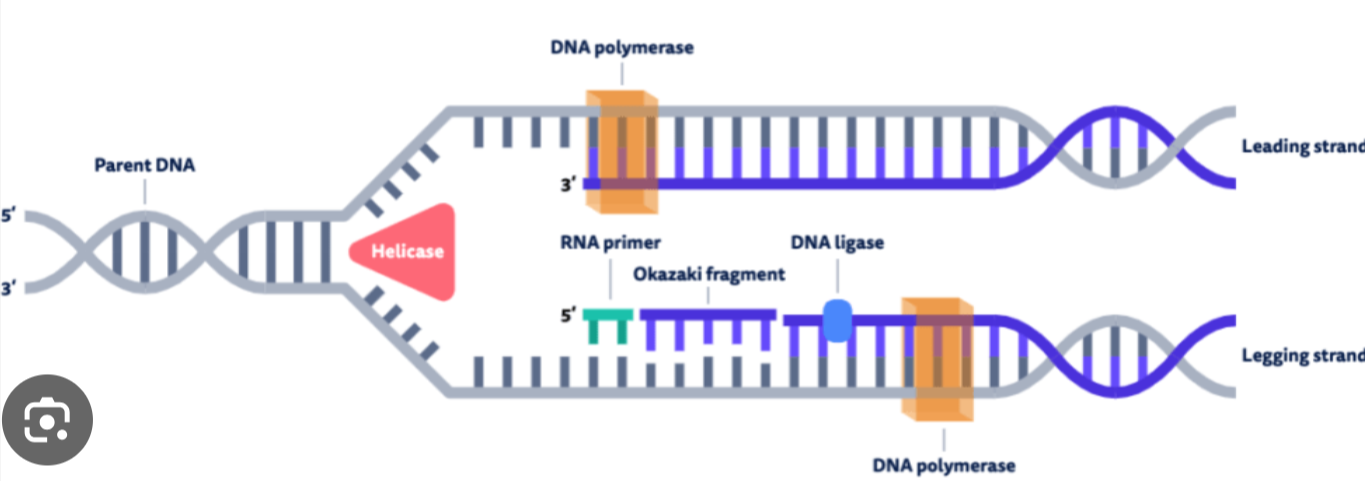
DNA polymerase in prokaryotes
prokaryotes have 2 DNA polymerase important in replication, DNAP III and DNAP I
eukaryotes have 5 distinct DNA polymerases, but the specific names of those aren’t needed
DNA ligase
joins the okazaki fragments, forming a continuous DNA strand
acts like glue for the okazaki fragments
DNA synthesis diagram
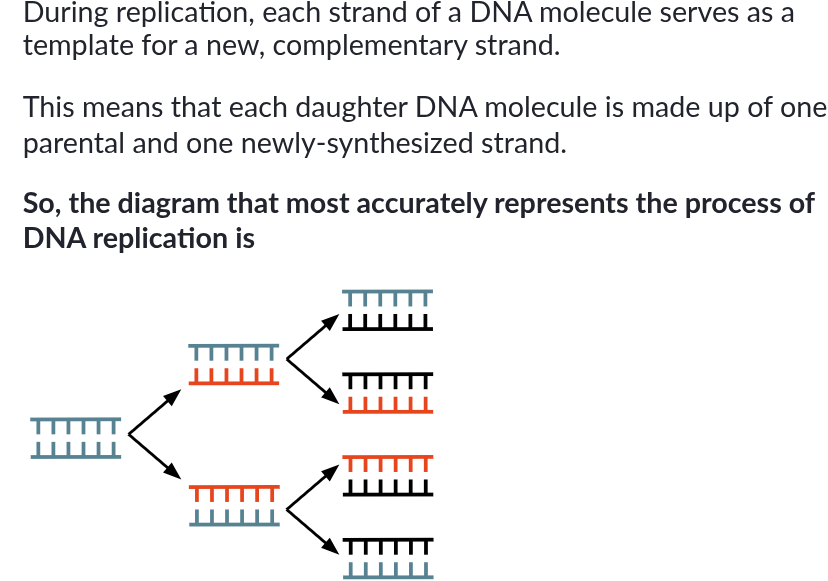
DNA synthesis always occurs antiparallel to the template DNA and in the 5’ to 3’ direction. So, in the correct diagram, each arrow will point towards the 5’ end of its template strand (picture is an example)
Eukaryotes: Problems at the 5’ End
linear chromosomes present a problem at the 5’ end
since DNAP can only add nucleotides to a 3’ end, there is no way to finish replication on the 5’ end of a lagging strand
-over many replications this would mean that the DNA would become shorter and shorter
genes on DNA are protected from this due to telomeres
Telomeres
repeating units of short nucleotide sequences that do not code for genes
form a cap at the end of DNA to help postpone erosion
some cells have the enzyme telomerase, which adds telomeres to DNA
Proofreading and Repair
as DNAP adds nucleotides to the new DNA strand, it proofreads the bases added
if errors still occurs, mismatch repair will take place
Mismatch Repair
enzymes remove and replace the incorrectly paired nucleotide
if segments of DNA are damaged, nuclease can remove segments of nucleotides, and DNA polymerase and ligase can replace the segments