DNA Biotechnology (1)
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
what are the applications of the Human Genome Project?
development of methods for the diagnosis of genetic diseases
- success in gene therapy approaches!
describe the content of the human genome
~3 billion bps that encode 20,000-25,000 protein-coding genes located on 23 chromosomes
describe the 3 primary tools used to complete the human genome sequence
- restriction endonucleases that permit the cleavage of huge DNA molecules into defined fragments
- cloning techniques that provide a mechanism for amplification of specific nucleotide sequences
- the ability to synthesize specific probes, which has allowed for the identification and manipulation of nucleotide sequences of interest
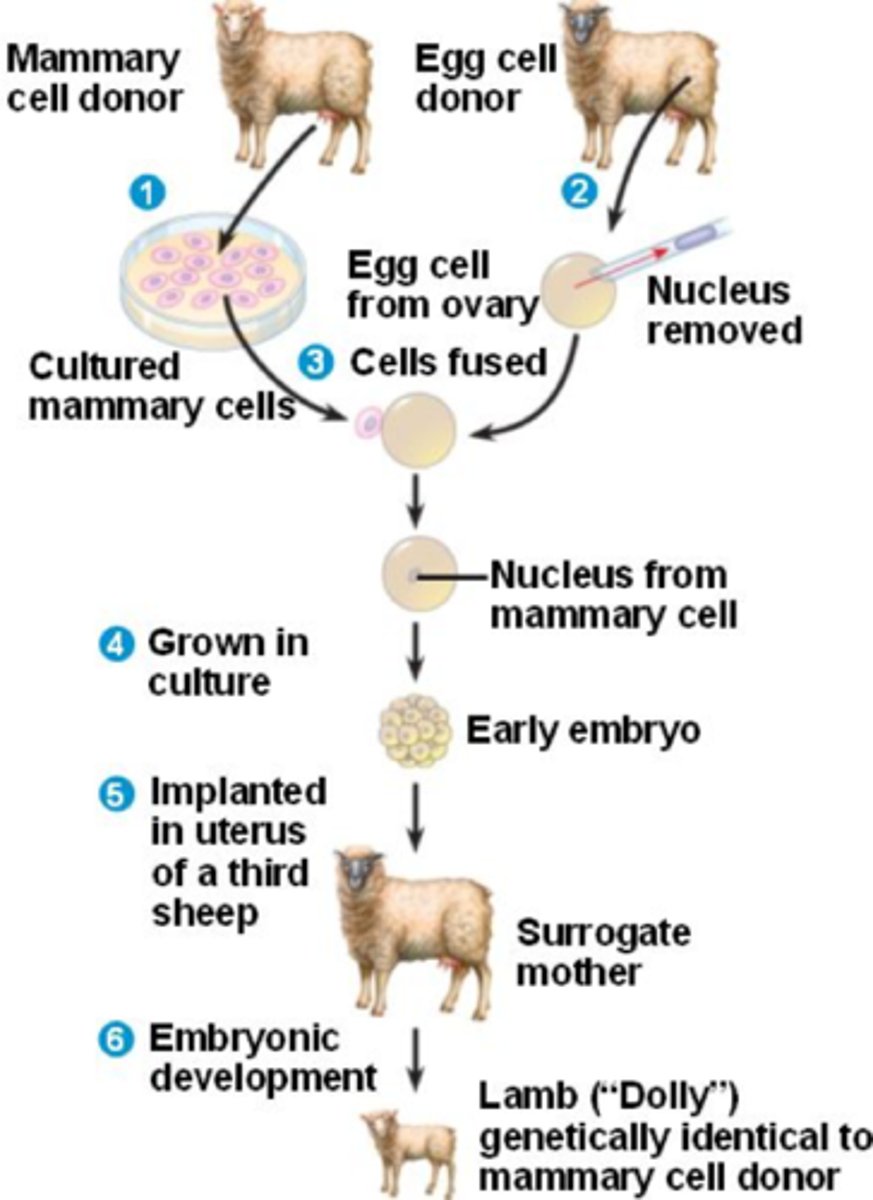
likely FYI - describe the gene cloning breakthroughs that occurred in 1975, 1984, and 1996
- 1975: first mammalian embryo created by nuclear transfer from a rabbit embryo to an enucleated rabbit egg cell
- 1984: first mammal created by nuclear transfer via in vitro fertilization techniques (created a lamb embryo)
- 1996: first mammal created by somatic cell nuclear transfer from an adult sheep's udder into an enucleated egg (Dolly the sheep)
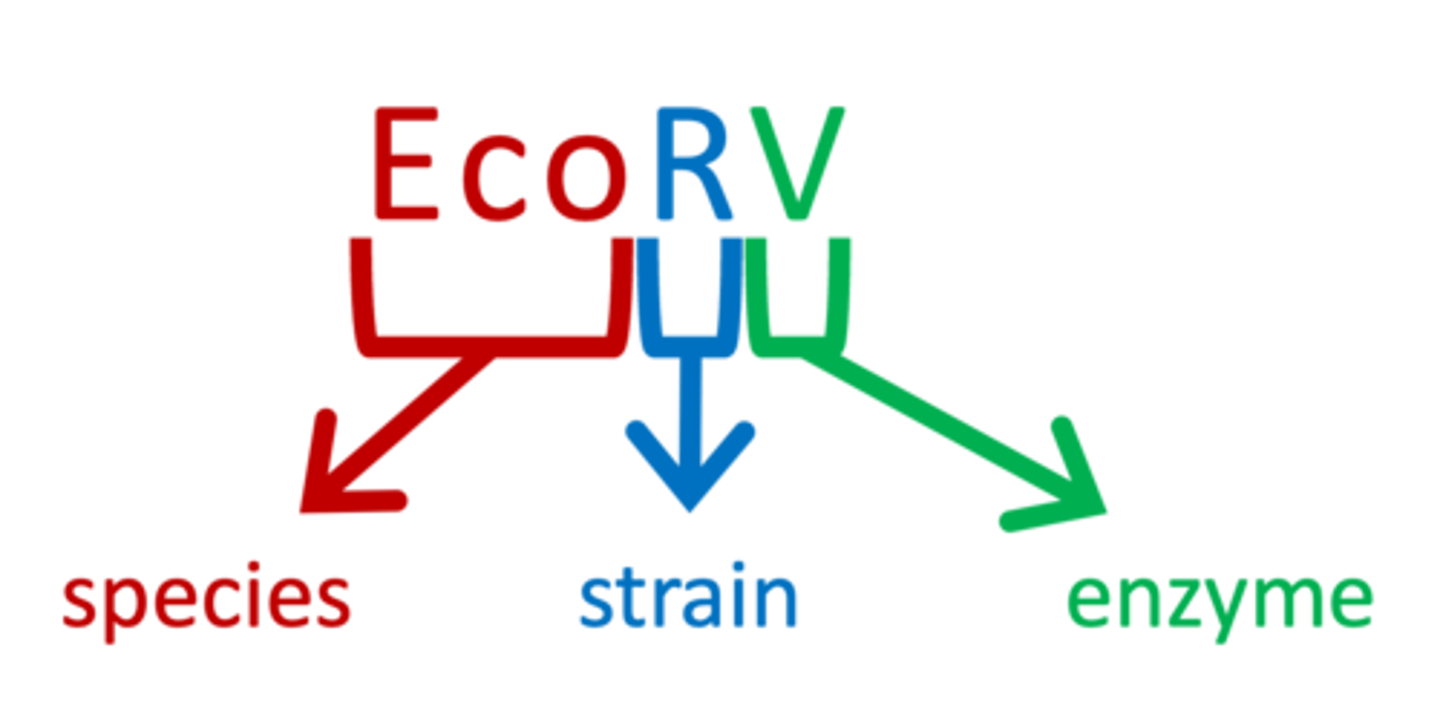
how are restriction enzymes named?
- the first letter is from the genus of the bacterium, and the next 2 are from the name of the species
- an additional letter indicates the type or strain (as needed), and a Roman numeral indicates the order in which the enzyme was discovered in that organism
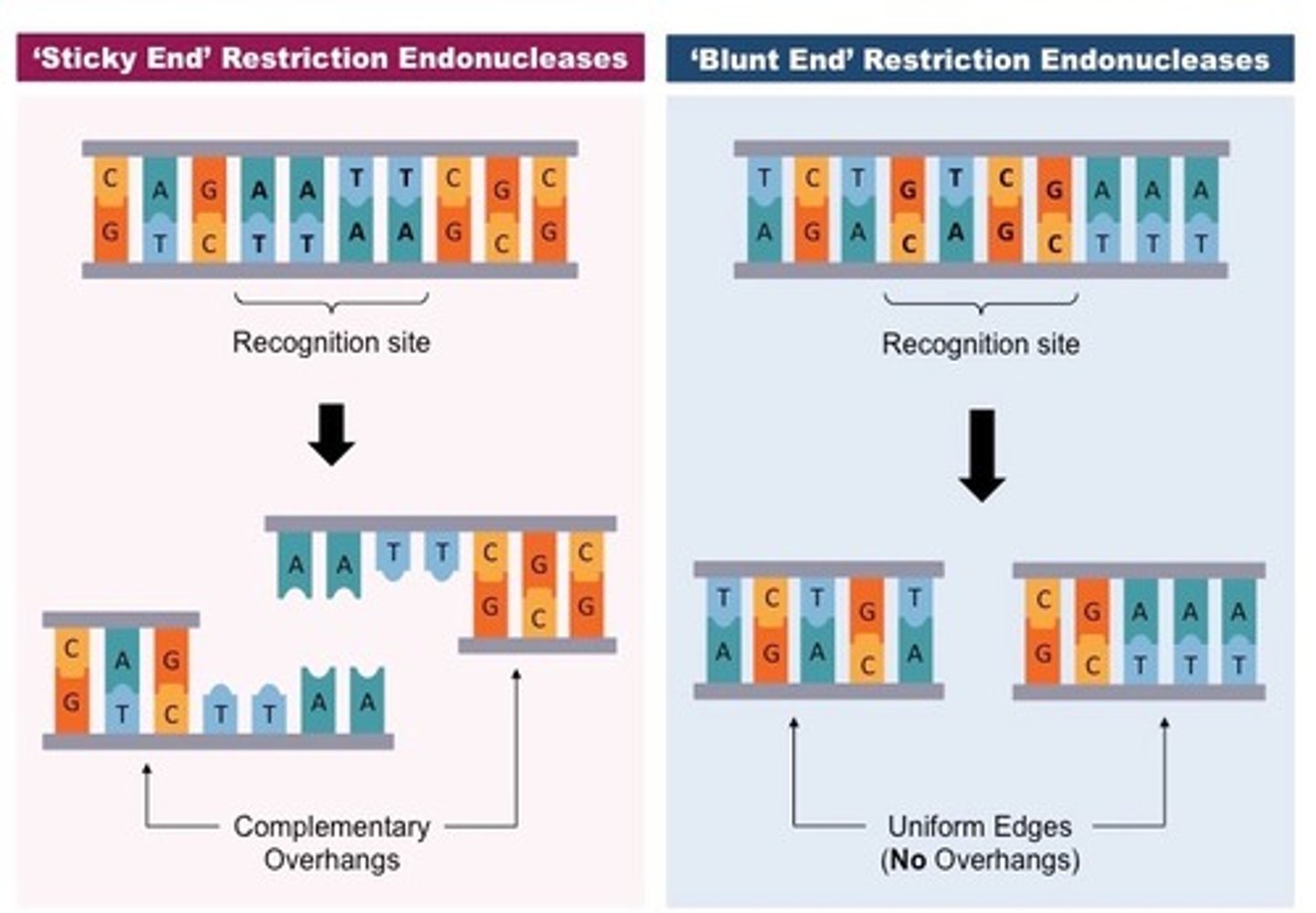
what is the function of restriction enzymes?
cleave double stranded DNA into fragments of different sizes
- restriction fragments are formed based on the side of the restriction site sequence
- recombinant DNA forms from the annealing of restriction fragments with sticky ends!
what's the difference between 'sticky' and 'blunt' ends?
- sticky: produces an overhang of DNA, which allows for more efficient re-annealing (ex: TaqI)
- blunt: DNA is cut with no overhanging DNA, which is less efficient for re-annealing (ex: HaeIII)
what enzyme is responsible for formation of the phosphodiester bond between restriction fragments?
DNA ligase
how is foreign DNA amplified within a host cell?
introduction of foreign DNA into a host cell allows it to be replicated by host machinery
- cellular DNA is cleaved with a specific restriction enzyme
- the DNA fragment is then joined to a DNA vector molecule (cloning vector) to form a recombinant using DNA ligase
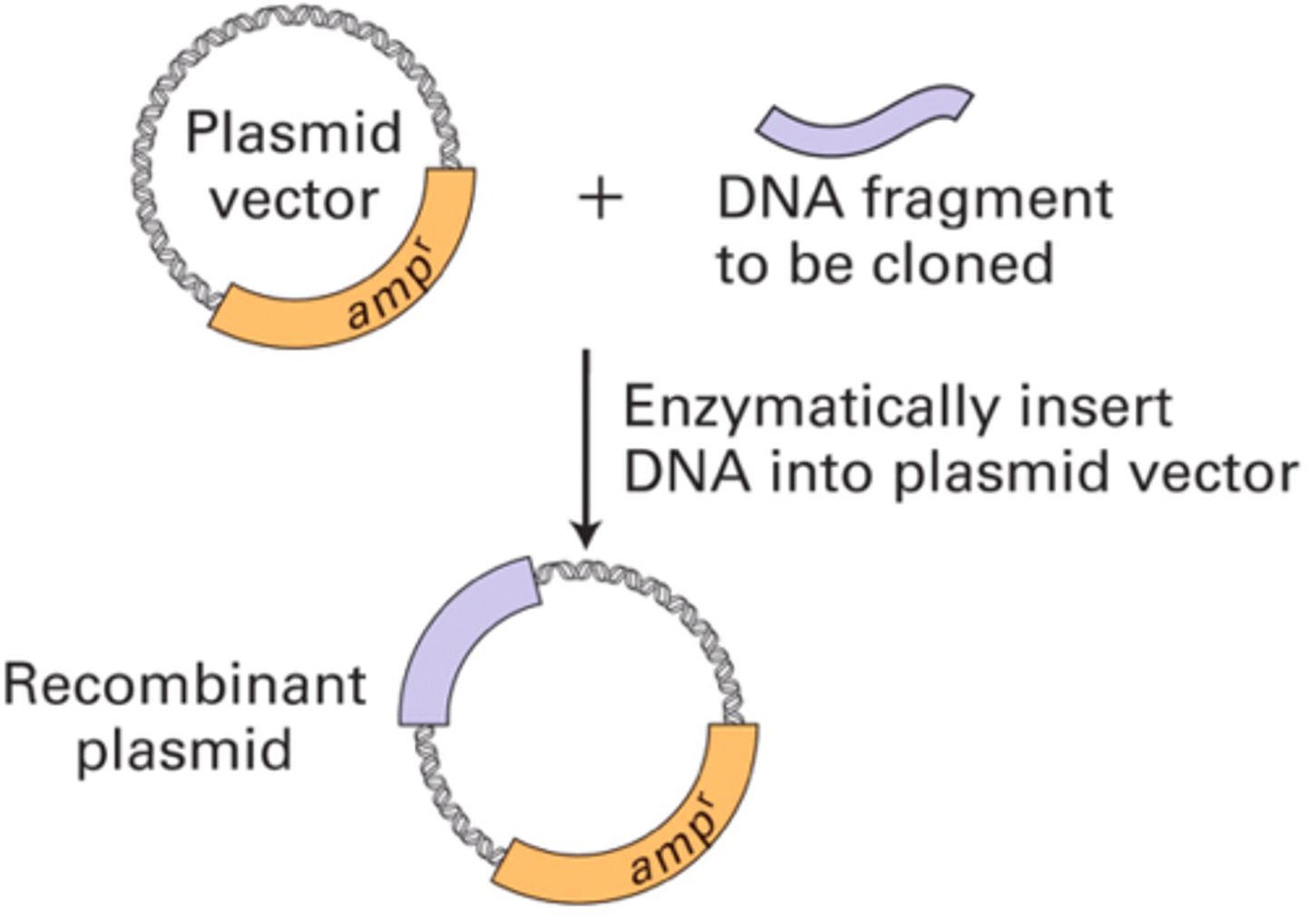
what is a cloning vector? describe the effects of vector pBR322 on the host cell
a vector is a molecule to which the fragment of DNA to be cloned is joined
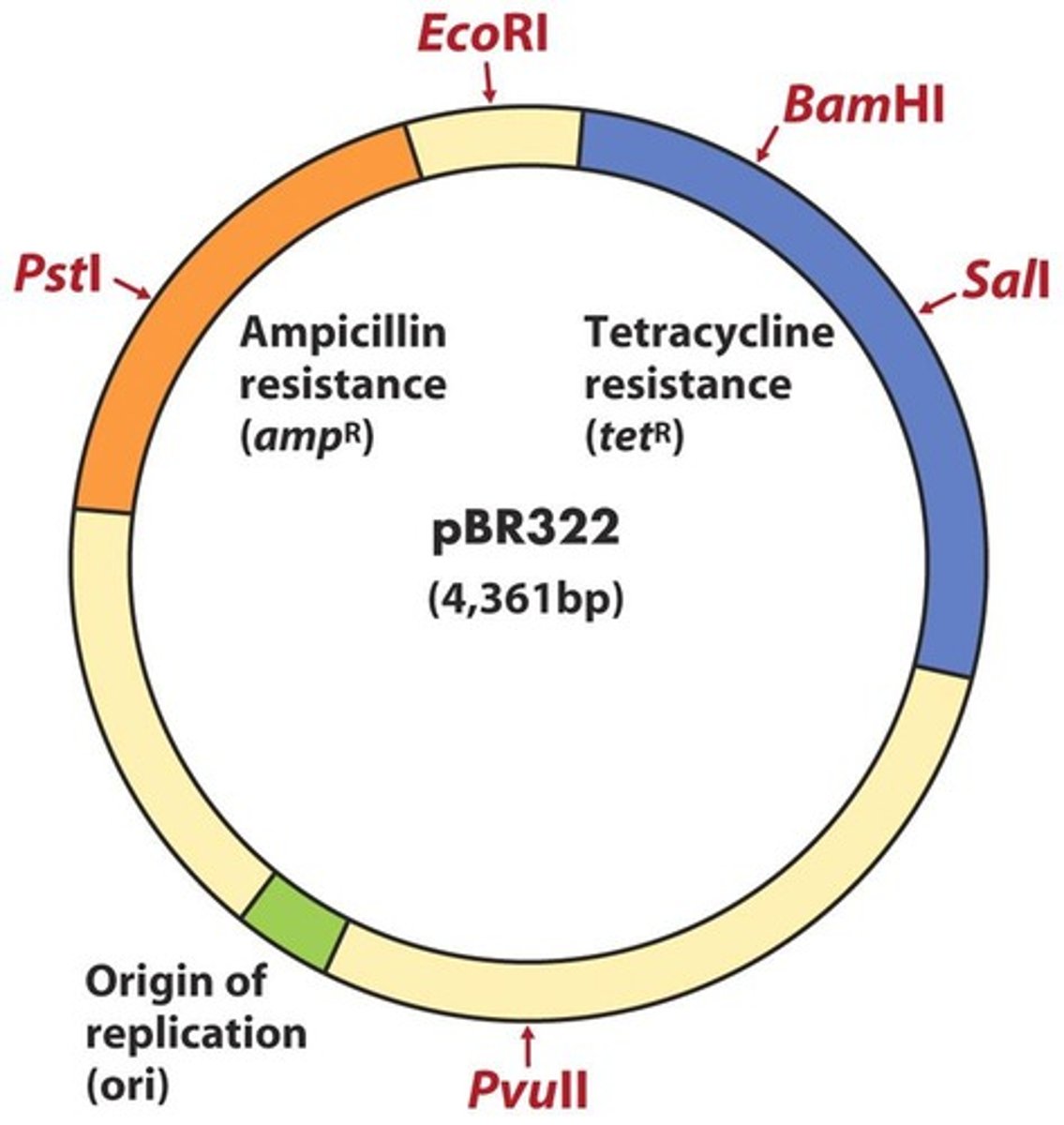
- pBR322 contains an origin of replication, 2 ABX-resistance genes (ampicillin and tetracycline), and >40 unique restriction sites; provides ABX resistance
- testing can then be done to isolate the bacteria that successfully integrated the pBR322 vector
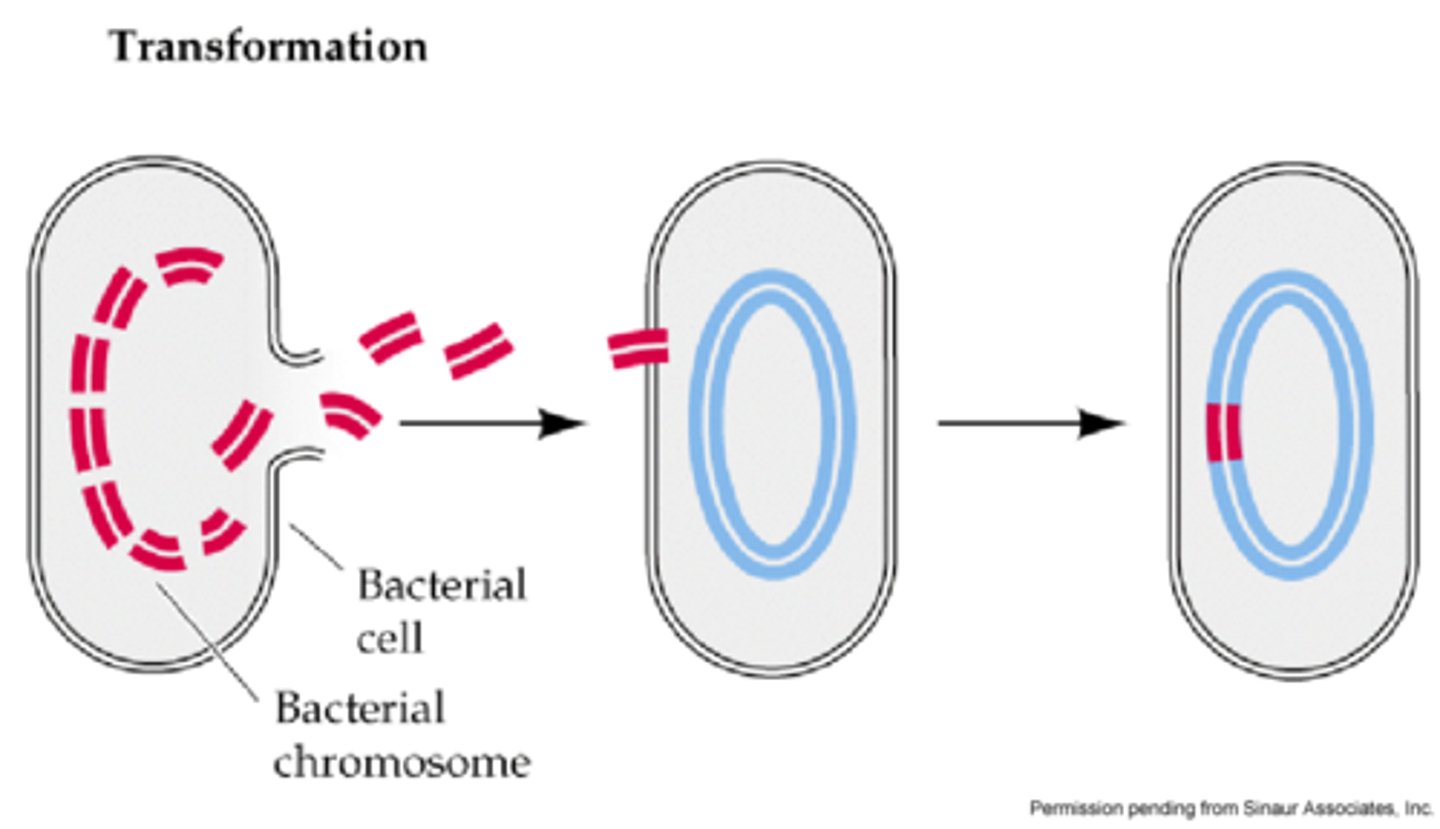
what is transformation?
integration of recombinant DNA into a bacterial cell from the environment
describe the general steps of gene cloning, from the introduction of foreign DNA to isolation
① introduction of foreign DNA into a host cell (often bacteria or yeast)
② host cell multiplies and creates a colony of clones containing the same inserted DNA fragment
③ recombinant molecules are released from host cells via membrane lysis
④ cloned DNA fragments can be cleaved from their vectors using restriction endonucleases and then isolated using purification techniques
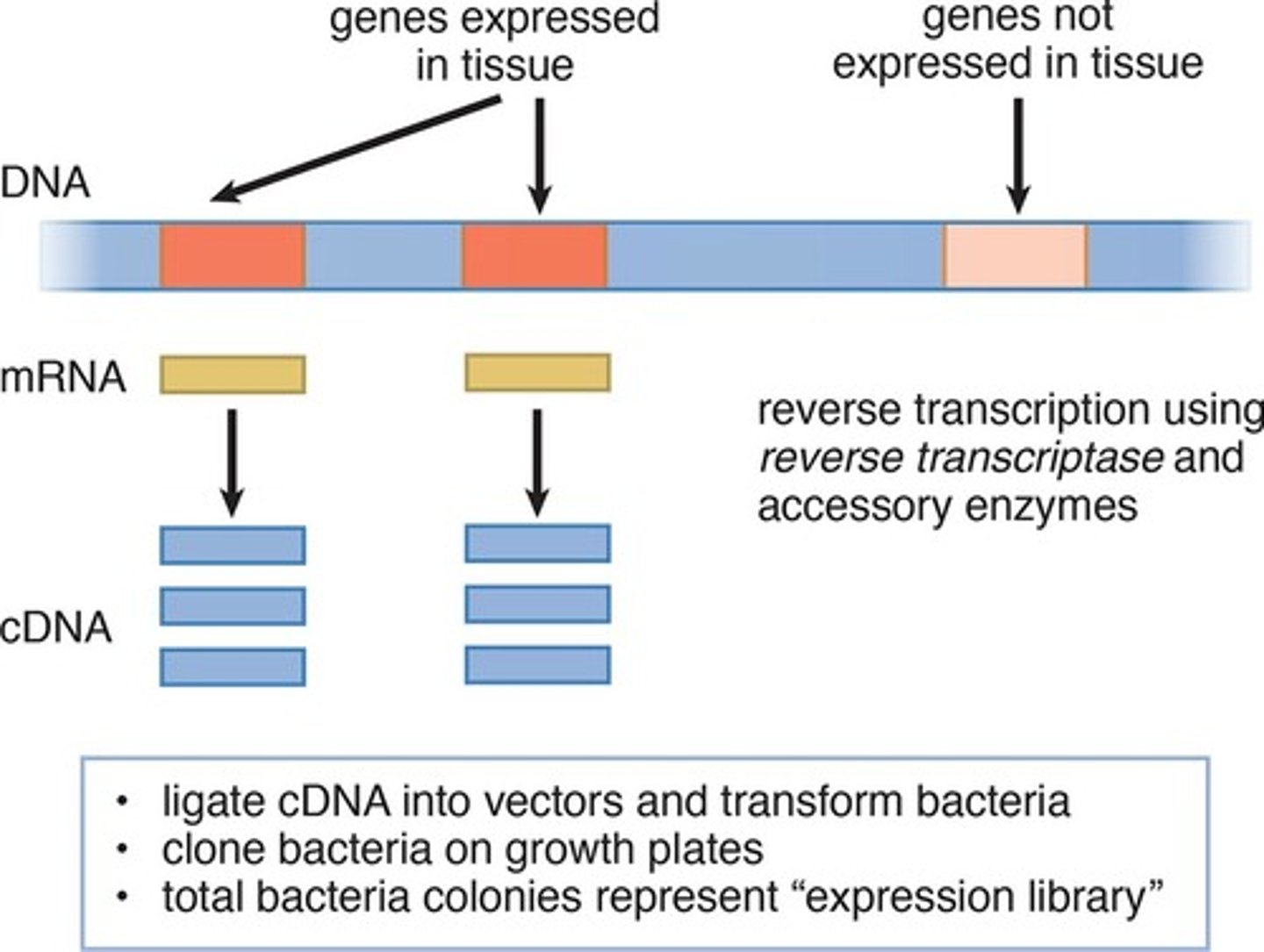
what are DNA libraries and what are the 2 kinds?
a collection of cloned restriction fragments of the DNA of an organism
- 2 kinds: genomic and complementary DNA (cDNA)
- ideally contain a copy of every DNA nucleotide sequence in the genome
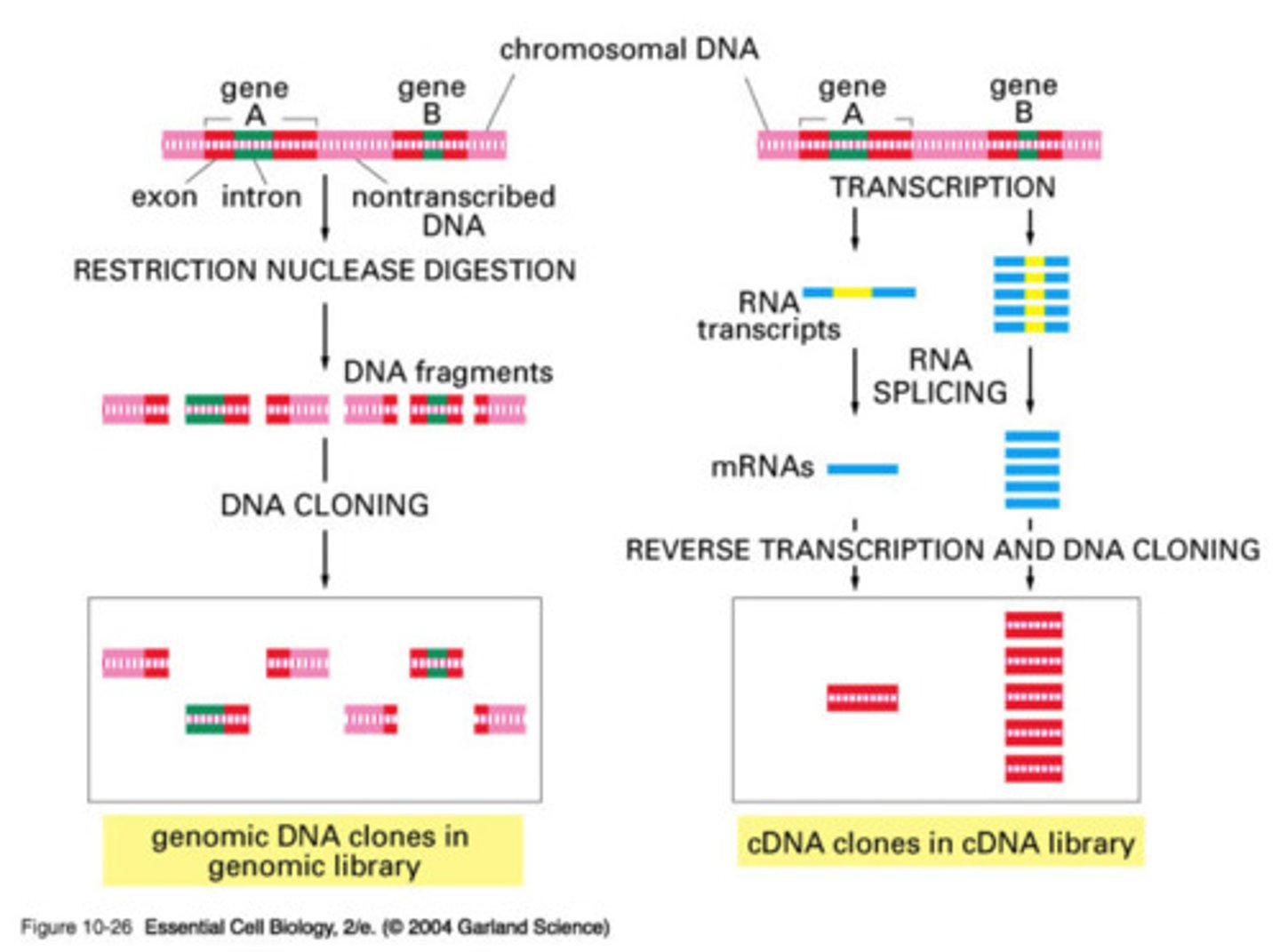
what is kept in cDNA libraries?
contain only DNA sequences that have a corresponding mRNA molecule in the cell
- mRNA can be used as a template to make a cDNA molecules using reverse transcriptase
- cDNA can then be amplified by biological cloning or PCR
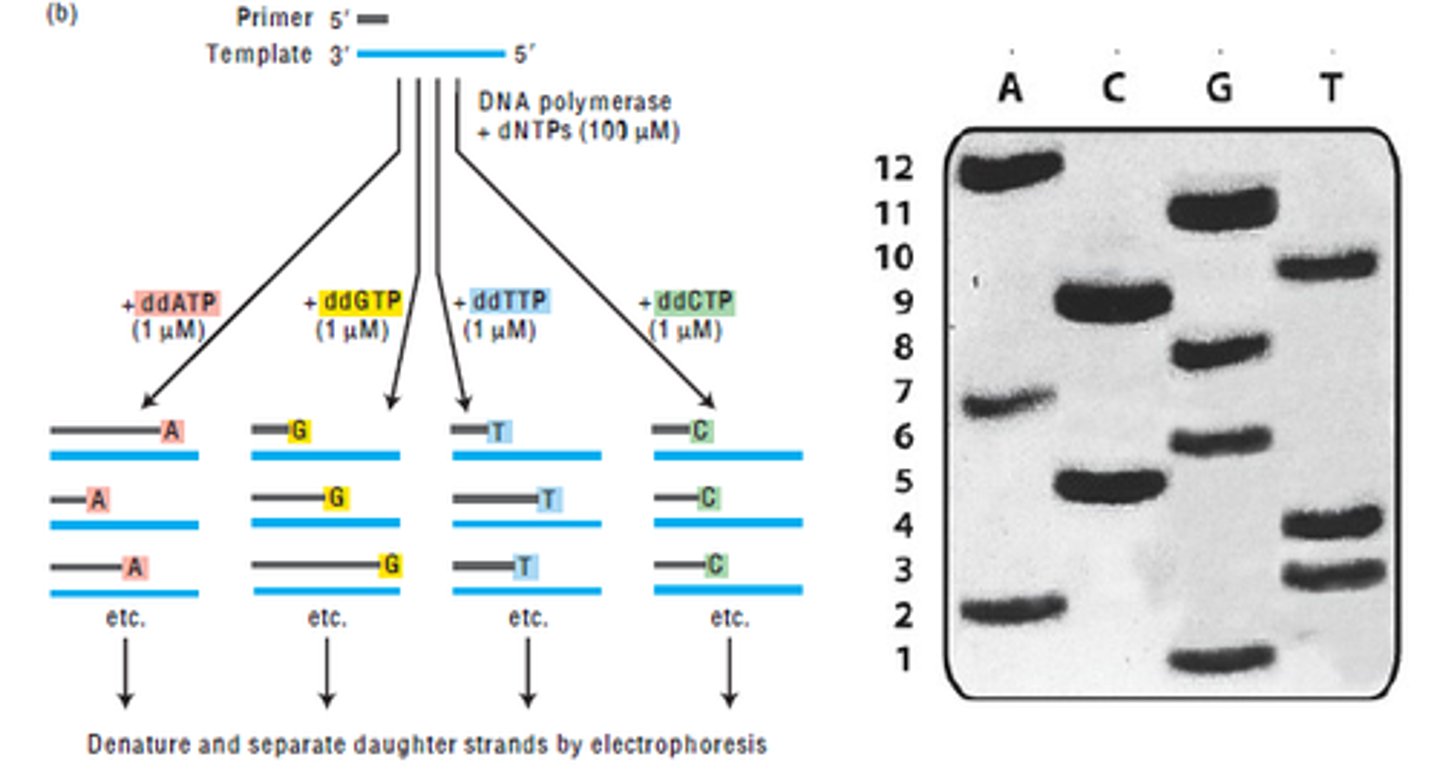
what is the Sanger method? how does it work?
sequences a target DNA fragment
- 4 di-deoxyribonucleoside-triphosphates (ddNTPs), each linked to a different fluorescent dye, are mixed with ssDNA
- the sample is then separated by capillary electrophoresis, and the fluorescent labels are detected and read to generate the sequence
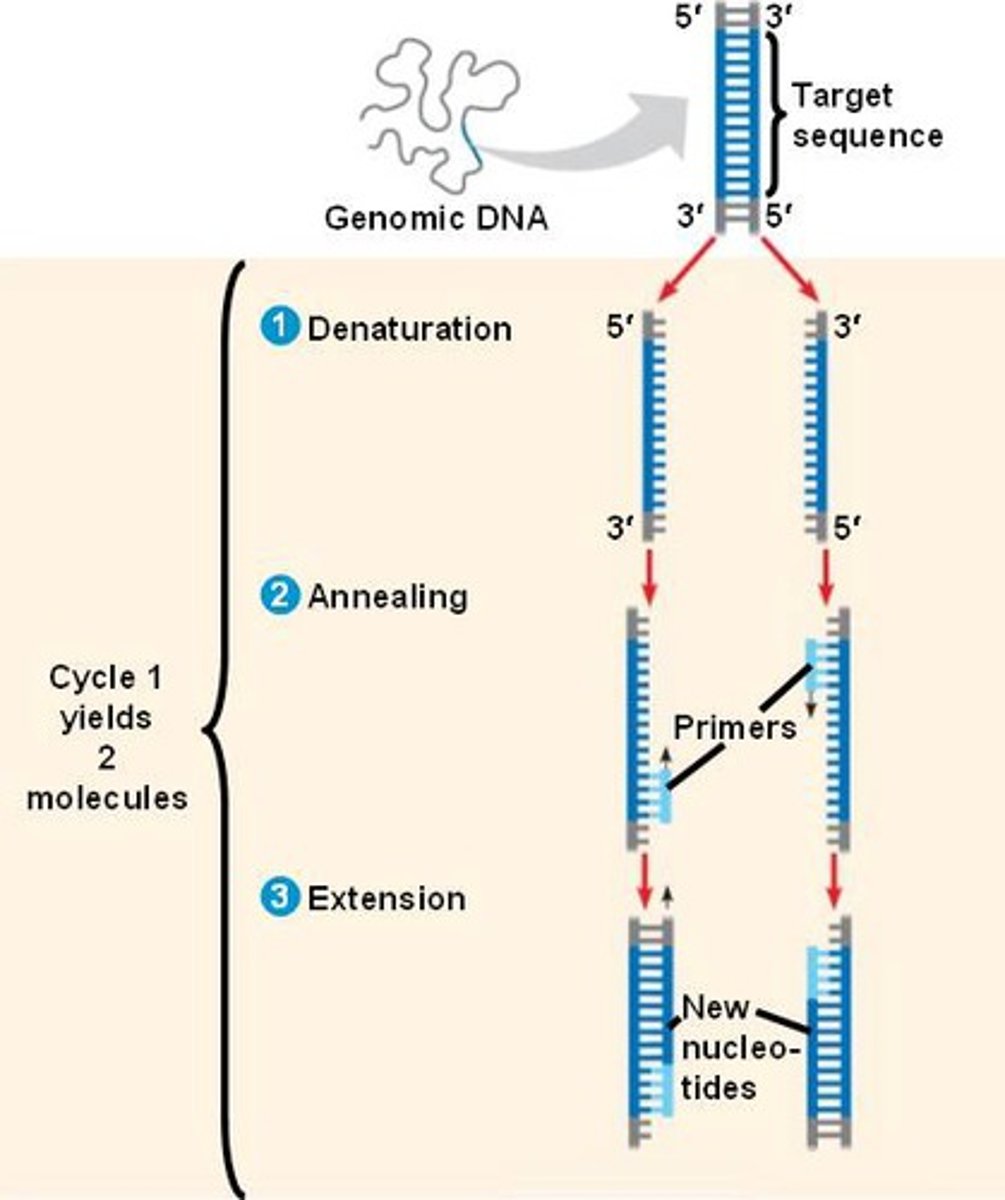
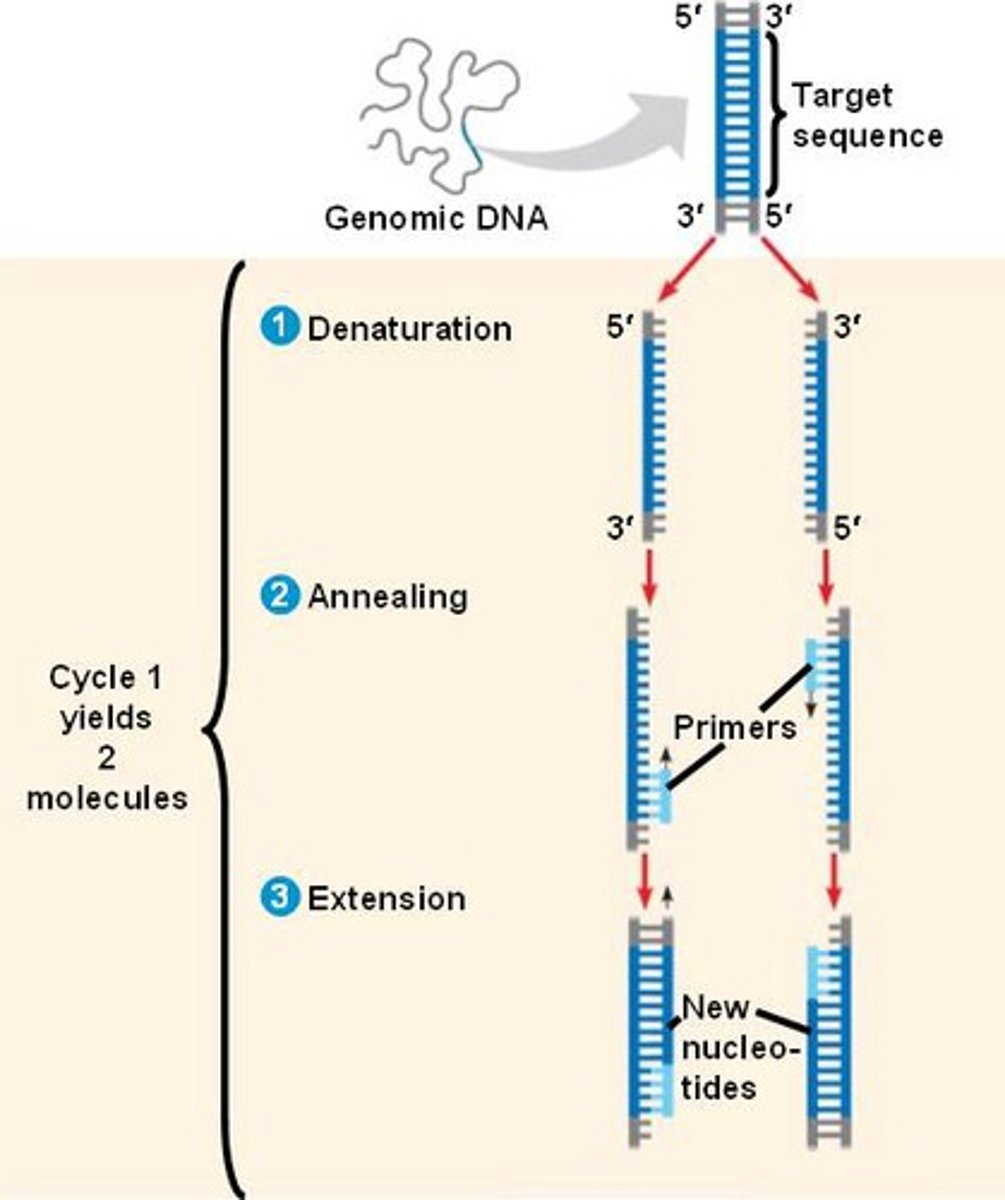
what is PCR? how does it work?
amplifies a target DNA fragment to produce millions of copies
- requires a DNA sample, primers, nucleotides, Taq polymerase, a mix buffer, PCR tubes, and a thermal cycler
- the dsDNA strands are denatured, annealed, and extended
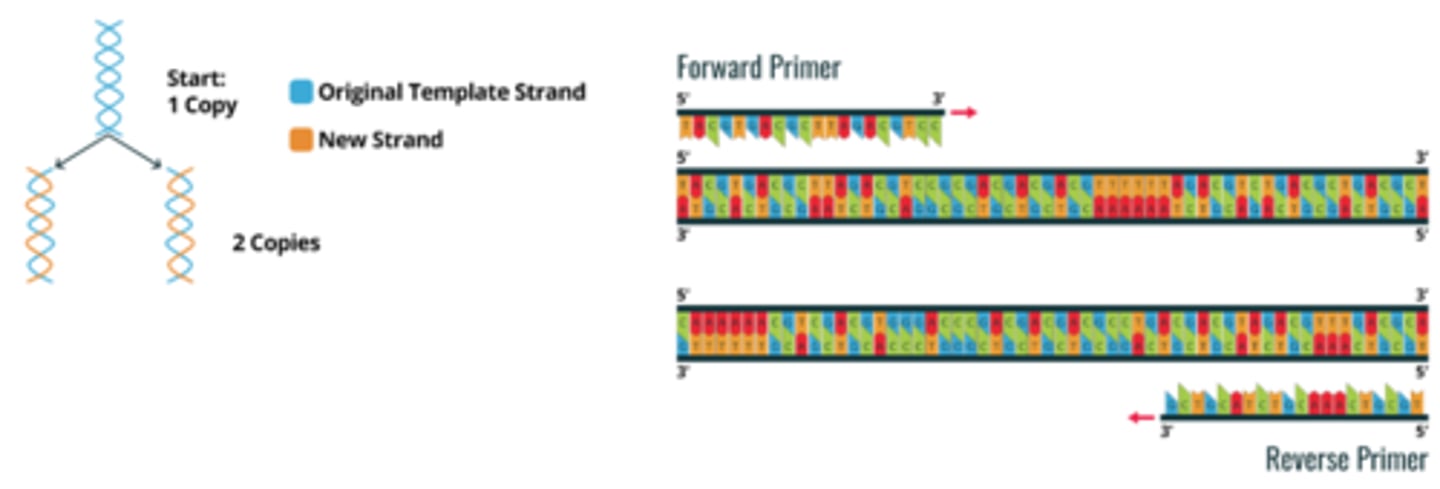
PCR - what's the difference between forward and reverse primers?
forward primers bind to the 3' end of the antisense (template) strand, while the reverse primers bind to the 3' end of the sense (coding) strand, allowing DNA polymerase to synthesize new strands in the 5' to 3' direction on both templates
describe the 6 main steps of PCR
① initial denaturation for 2 mins at 94 C → H bonds are broken between the 2 DNA strands
② denature for 30 secs at 94 C → continued denaturation
③ anneal primers for 30 sec at 55 C → forward and reverse primers anneal to the ssDNA templates
④ extend DNA for 1 min at 72 C → DNAP synthesizes and elongates the new target DNA strand rapidly
⑤ REPEAT steps 2-4 25-30x
⑥ final extension for 5 mins at 72 C → final extension to fill in any protruding ends of the new DNA strands; after 30 cycles, >1 billion copies are generated!
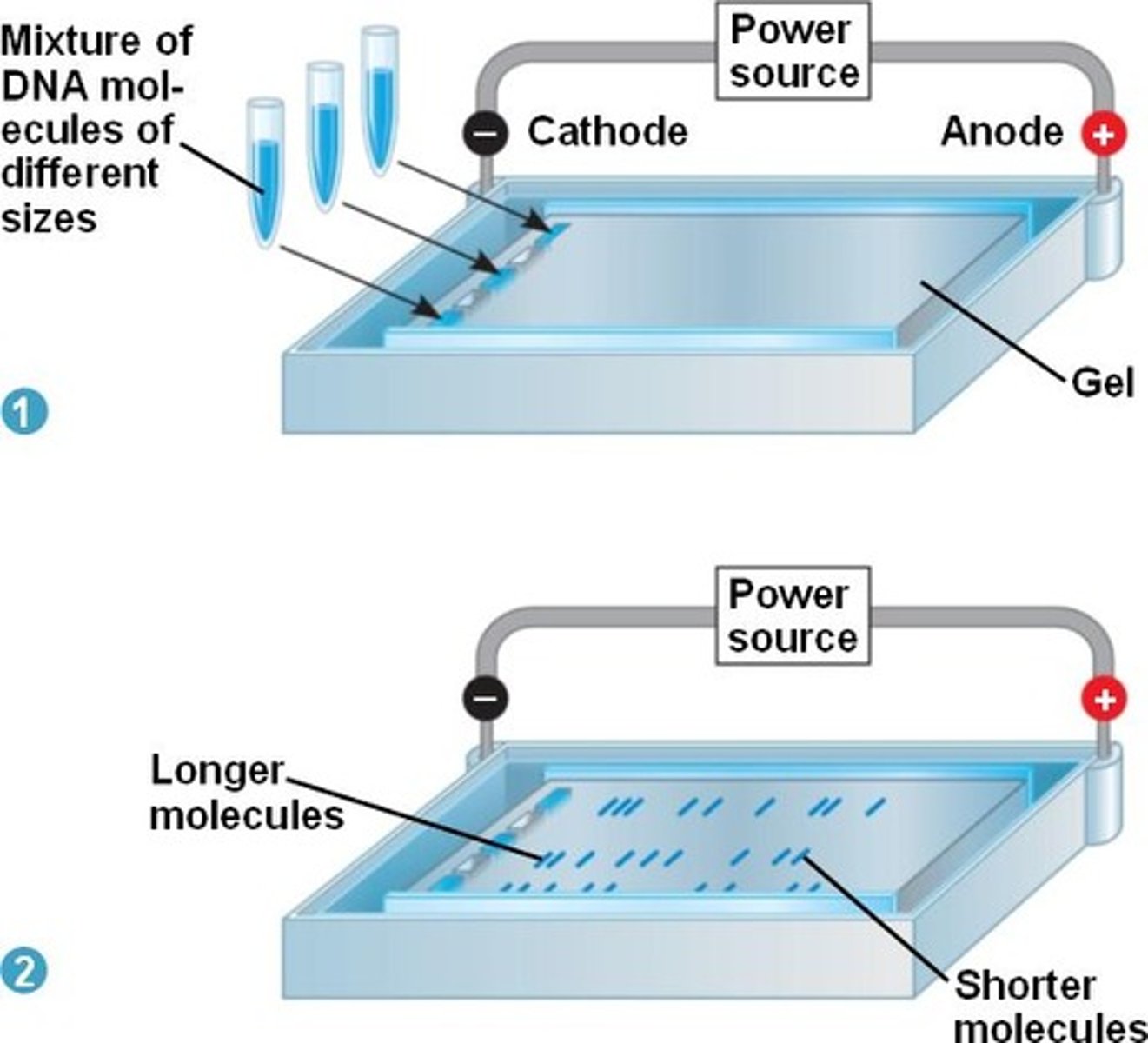
what is gel electrophoresis?
a laboratory method used to separate mixtures of DNA, RNA, or proteins according to electrical charge or molecular size
- the results of PCR are usually visualized using this approach
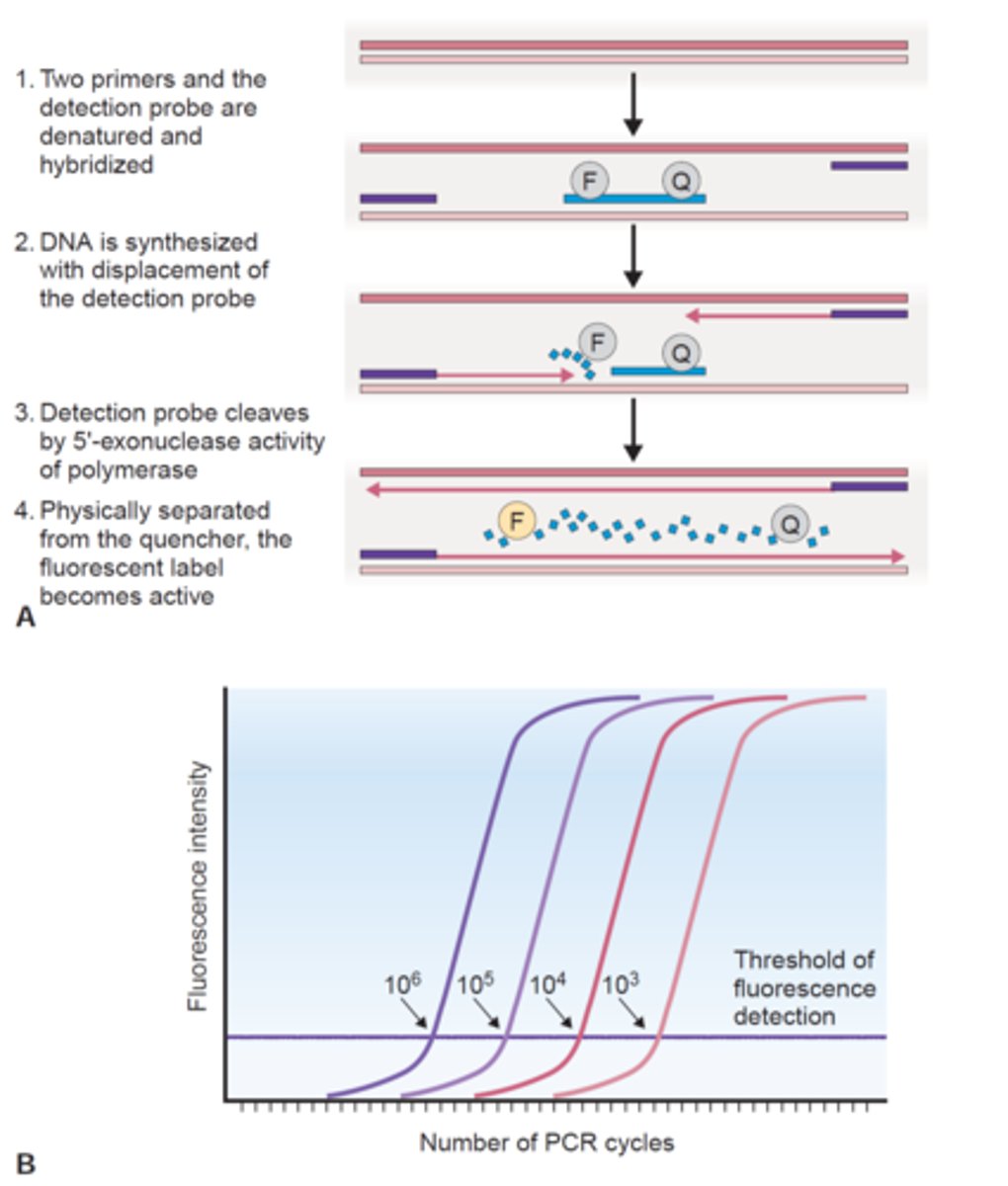
what is quantitative PCR (qPCR)?
quantitative version of regular PCR; aka real time PCR
- enables determination of the starting number of DNA copies in a sample
- the amount of product is monitored at each cycle in 'real time'
- ex. probes: SYBR green, TaqMan
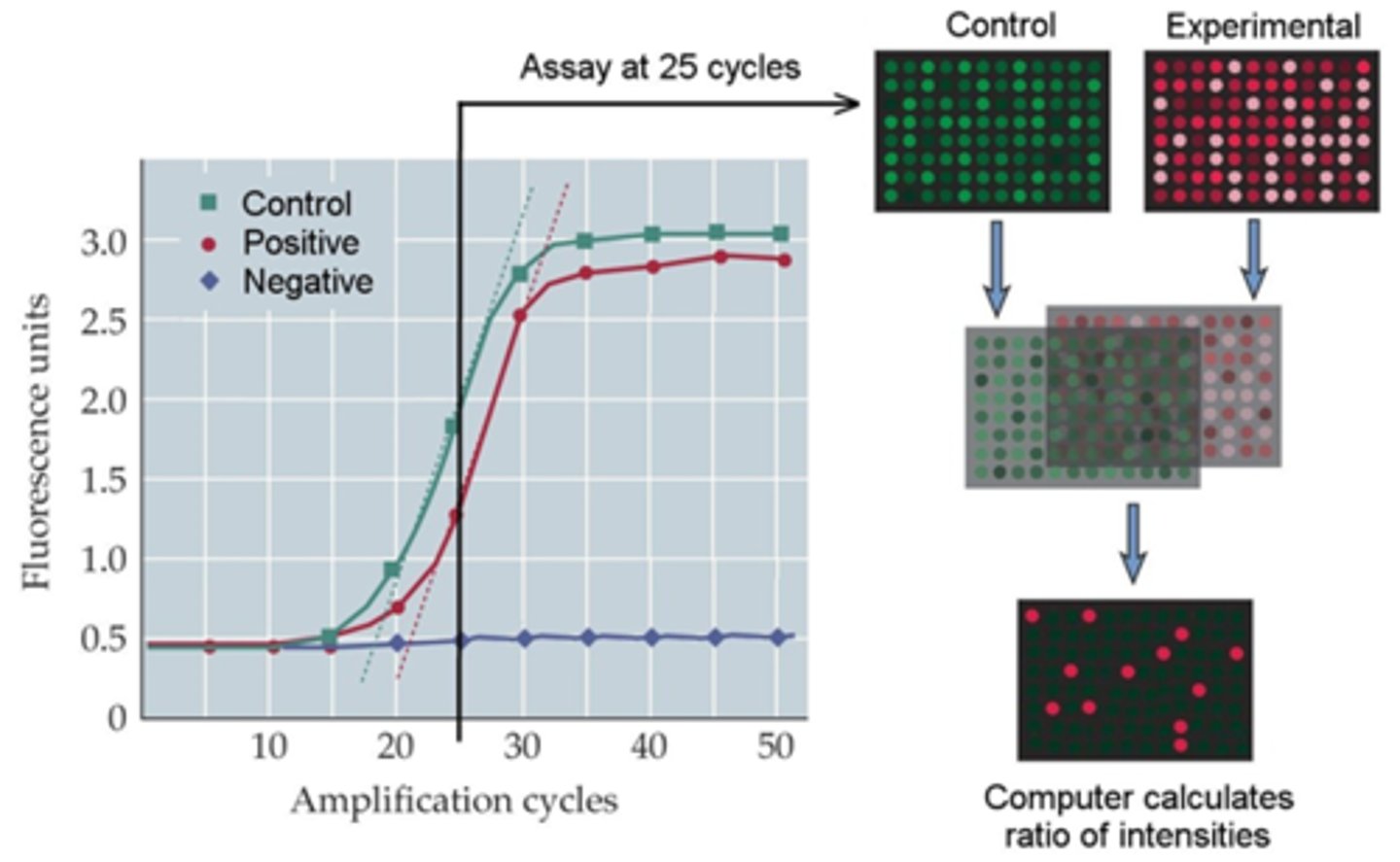
what do real time PCR instruments monitor?
fluorescence
- monitored as the PCR cycles progress