DNA Replication
1/51
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
52 Terms
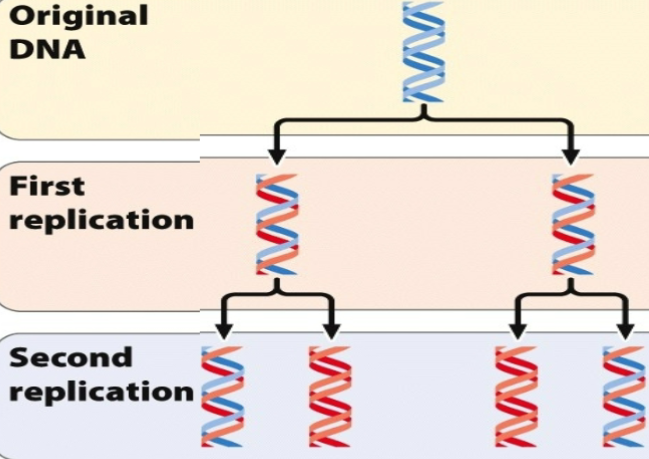
Proposed model of DNA replication - Conservative
The original double strand serves as a template for a new molecule of DNA
ORIGINAL DNA: It opens up and splits → each strand used as a template to generate new strand
FIRST REPLICATION: there is a old strand that came together and a new strand that came together
SECOND REPLICATION: The old strand splits up → there is a new entity in second replication. The new strand splits → there are new entities
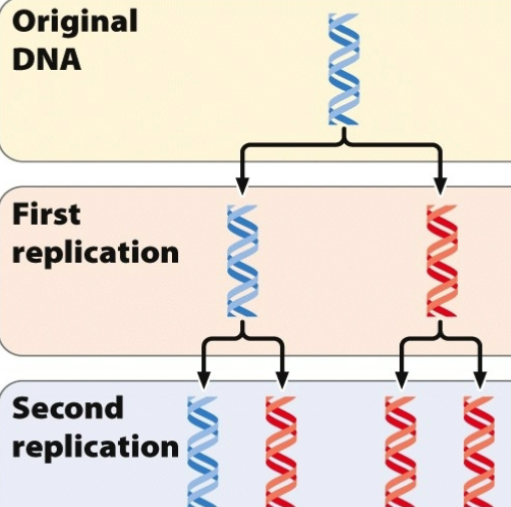
Proposed Model of DNA Replication - Dispersive
DNA Strand both double stranded would break up to multiple fragment and each fragment serves as a template → fragments come together → combination of old and new in both entities
Both DNA molecules break down into fragment and serve as a template for the new fragment
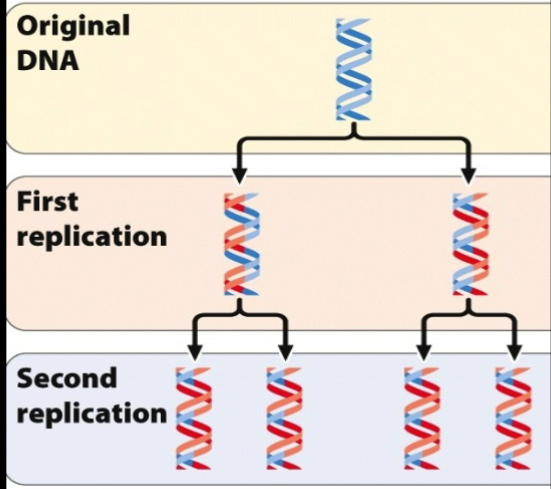
Proposed Model of DNA replication - Semi conservative
The original strand unwinds and is used as a template to generate the new strand
ORIGINAL DNA: Original strand breaks apart → as they break apart → serve as a template to get new strand
FIRST REPLICATION: No breaking apart, just a combination of old and new strand
SECOND REPLICATION: New strands just used red as a template
Meselson and Stahl’s experiment
Needed a way to distinguish the old from new strand. They used 2 isotopes of Nitrogen: 14N and 15N (this was used since bases that make up rungs of ladder are nitrogenous bases)
There are heavy and light form of Nitrogen
Steps of Meselson and Stahl’s experiment
1st: grow bacteria in heavy 15N for many generations. This made the DNA HEAVY.
2nd: Took a sample of those bacteria in 15N and switched the rest to 14N. The rest left out was left in 15N. New DNA contained 14N which was lighter.
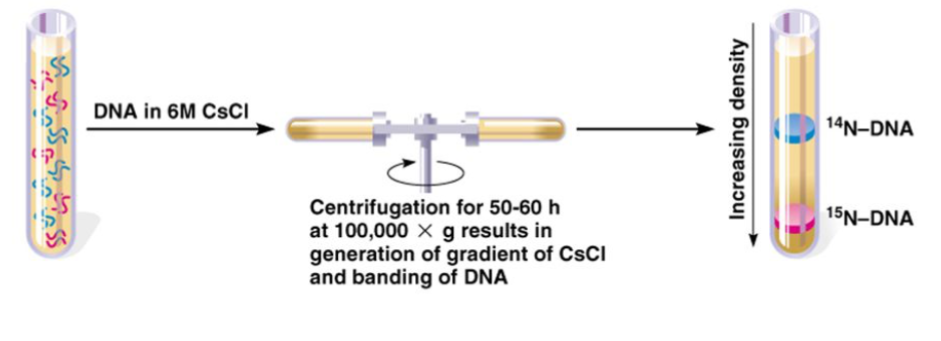
3rd: Ran the collected DNA on an equilibrium density gradient centrifugation to distinguish between heavy and light strands.
Meselson and Stahl’s Equilibrium Density Gradient Centrifugation
Take acquired DNA and put in tube of different levels of density as you move down the tube
If you put DNA on top and spin it in centrifuge → DNA travels down and stops where it meets identical density → establish density gradient using different amounts of sugar (create increased density in solution)
At the bottom, it is heavier material (more sugar)
As you go up, it is slightly lighter and there is less sugar
This allows it to separate DNA molecule
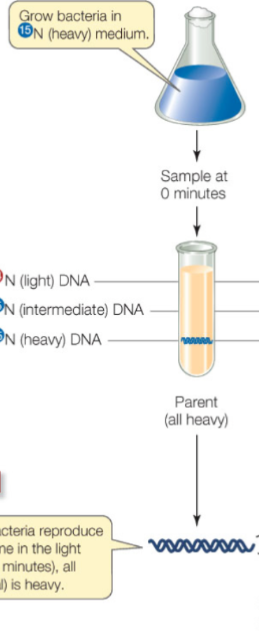
Details of Meselson and Stahl’s experiment (0 mins)
One sample was kept at 15N and the other was switched to 14N
Sample at 0 mins: found double stranded molecule with 15N and sunk down further (from sample with 15N)
Interpretation: All bacteria is heavy
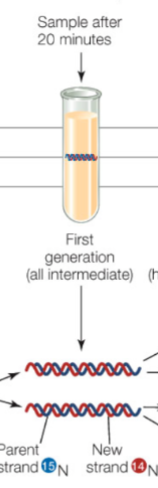
Details of Meselson and Stahl’s experiment (20 mins)
Sample that was transferred to 14N was rested in intermediate zone (not too light nor heavy)
This is slightly slower and lighter
Interpretation: it contained both parent 15N and 14N in strands
Details of Meselson and Stahl’s experiment (40 mins)
Found strand in intermediate zone that had both 14N and 15N
Found band with just 14N which is lighter
It is semi conservative since there is a band that is light meaning there were 2 strands that were generated using 15N and strands that had a single copy of 14N
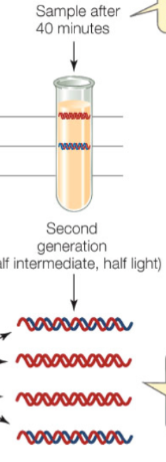
Meselson and Stahl demonstrated DNA replication is semiconservative
After 2 rounds of replication, if replication were dispersive, you expect the weight of DNA to be: all in intermediate range where every strand has a mixture of old and new DNA
The fact that some of the DNA was light and some was of intermediate weight after 2 rounds of replication suggested semi-conservative replication - dispersive replication will produce DNA that contains both light and heavy DNA - all intermediate weight
Several modes of DNA replication
There are several ways that semi conservative replication can take place differing based on whether the DNA is circular or linear
Theta replication
Rolling Circle replication
Linear eukaryotic replication
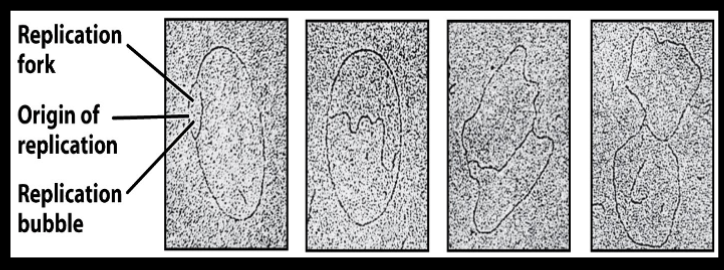
Several modes of DNA replication: Theta replication
Takes place in circular DNA/plasmid such as e. coli, bacteria, and prokaryotes
Expect organisms that undergo replication to have circular plasmids
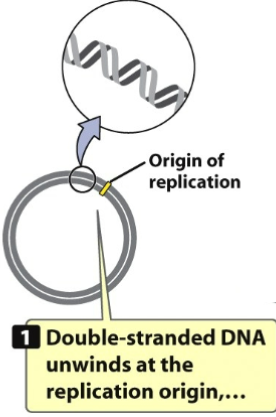
1st: 2 strands that make up plasmid need to melt (unwind/separate)
Unwinding starts at origin of replication where polymerase will start where it will bind and begin to do job of replicating
It needs to unwind before origin of replication so all machinery can get there and ready to do its job
There is a slow unwinding and it’s called a replication bubble
Theta replication (2nd)
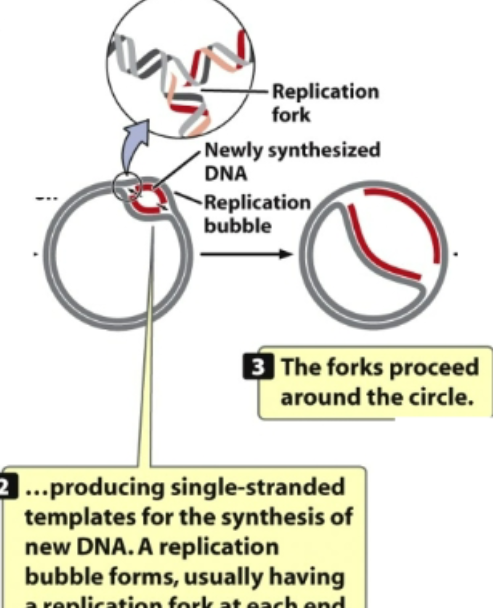
2nd: the unwinding continuous and bubble gets large. Location where there is unwinding is called replication fork.
If there is unwinding on both ends → bidirectional replication
If unwinding is on one side → unidirectional
In this case, it is bidirectional
Replication bubble
proteins come in and do their job
proteins decide whether DNA replication is unidirectional (travels 1 way) or bidirectional (bubble opens and DNA replication is going to take place around)
Theta replication (3rd)
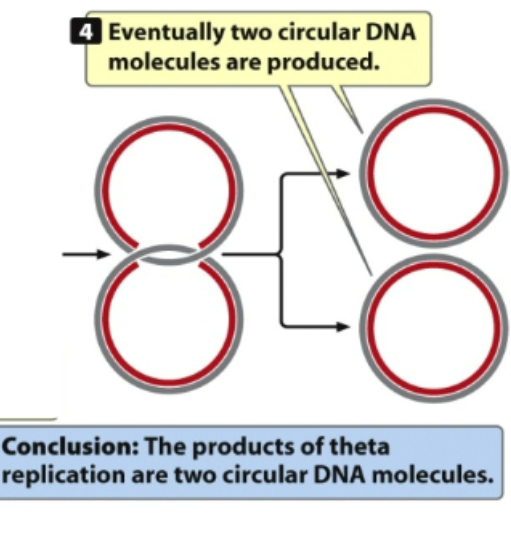
3rd: DNA synthesis proceeds in 5-3’ direction
DNA will be built in where polymerase will actually bind DNA moving in 5’ to 3’ prime direction
physical pieces that the nucleotides are being added in 5’-3’ prime direction so it synthesizes the strand in that direction
DNA is antiparallel
Original template is moving in opposite direction
Theta replication summary
In replication, bubble forms and its bidirectional and need to split from one another
Several Modes of DNA replication: Rolling Circle replication
1st: Single strand break creating a 3’ OH and 5’ Phosphate group
DNA polymerase: move from 5’ to 3’ direction
Next one that attaches to phosphate and C (5’ C) which is attached to another ring
There is a reliance on OH group in order for synthesis to occur
Critical elements required for function of DNA polymerase is 3’ OH (which comes off 3’ C)
When there is a plasmid in circle → no access to 3’ OH group so you create a nic → break DNA to expose 3’ OH
Polymerase can come in and find 3’ OH group → attach and synthesize
Unidirectional
Bacteria uses this
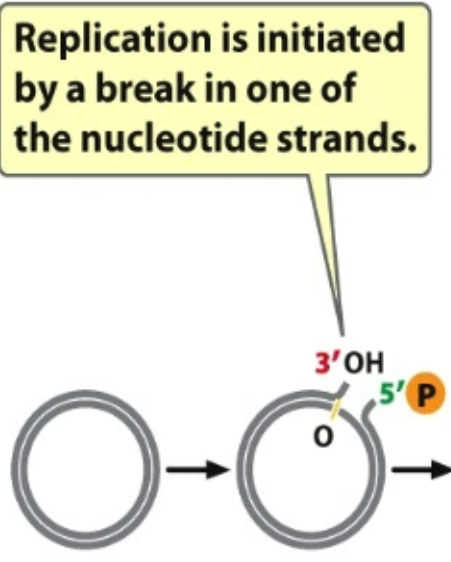
Several Modes of DNA replication: Rolling Circle replication (2nd)
2nd: New nucleotides are added on the 3’ OH break using the inner strand as a template
Now that the 3’ OH group establishes (in yellow area), DNA polymerase synthesizes new strand (red) → displaces original strand that existed → polymerase is a chunky protein moving along old DNA and using it as a template to generate new strand
By displacing old DNA, it can be used as a its own template to generate new strand
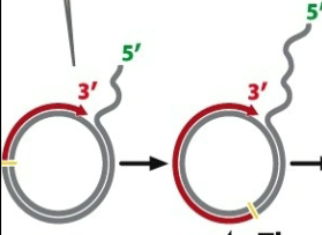
Several Modes of DNA replication: Rolling Circle replication (3rd)
3rd: As the new strand gets elongated, the original strand gets displaced, eventually rolling off the entire plasmid. This can continue for several rounds until many copies are made.
Purpose of rolling circle: repeat and generate new strand and it is a continuous cycle
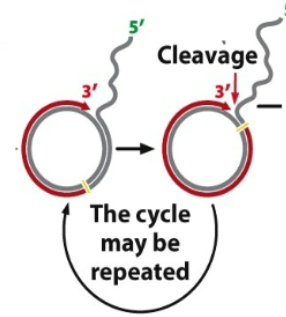
Several Modes of DNA replication: Rolling Circle replication (4th)
4th: the linear fragment that rolls off can be used as a template for a new strand
Run into position where every strand is being displaced every single time and now circularized and used as a template to generate a double strand
Every time it goes around original strand, it gets displaced to make more copies
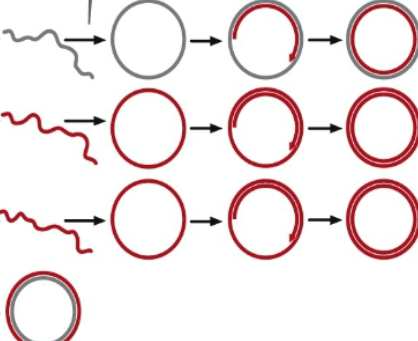
Several Modes of Replication: Linear Chromosomes
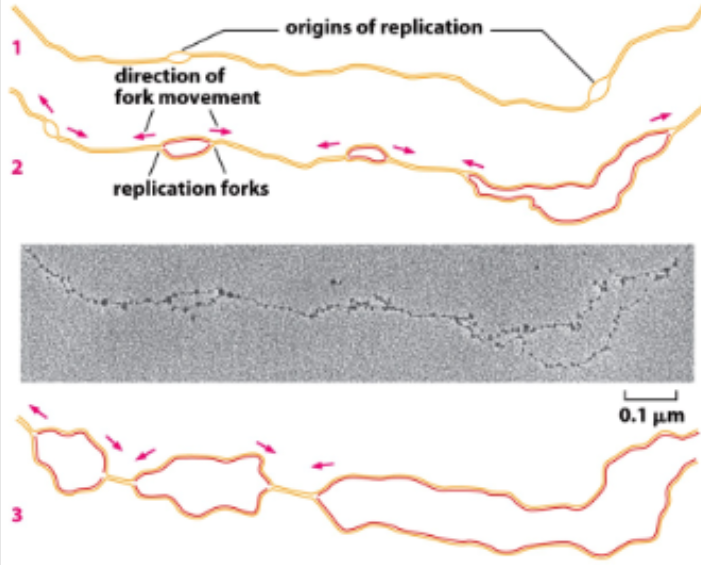
Too large to have a single replication origin, so linear chromosomes in eukaryotes need multiple origins
Linear chromosomes: complex and linear and lots of nucleotides and many sites of replication (unique and need to melt DNA)
Origin 1, 2, 3 initiate replication, meet up when bubbles merge together and have replication of entire strand
Bidirectional
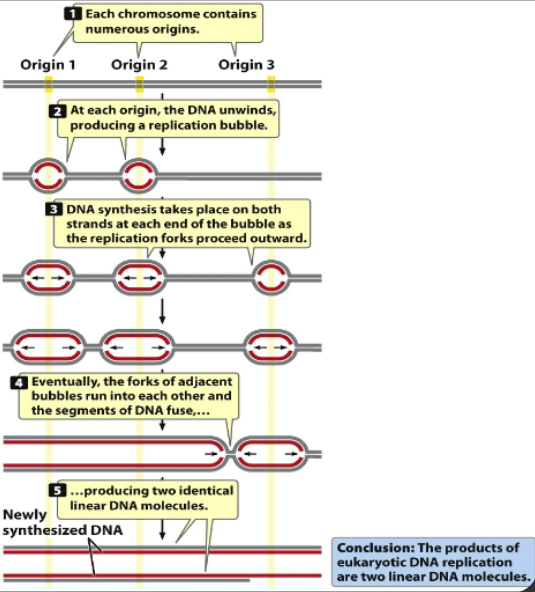
Stages of DNA replication of Eukaryotic and Prokaryotic
Initiation
Unwinding
Elongation
Termination
Stages of DNA replication Prokaryotic: Initiation
Initiation: DnaA protein binds to the origin of replication (oriC)
Initial bubble that forms and refers to unwinding part of DNA (melting)
Opens a small site for Helicase and single-strand binding protein to enter and being unwinding the 2 strands
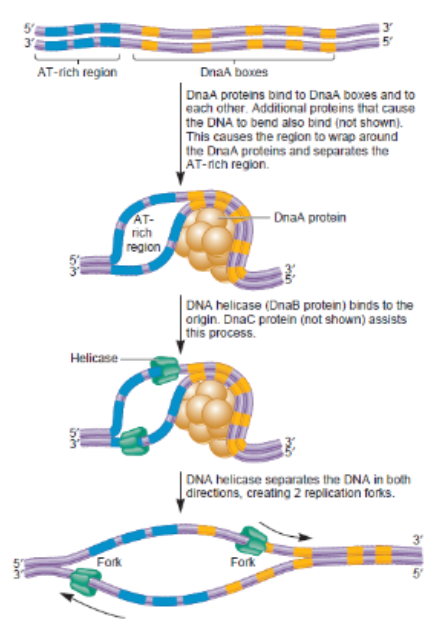
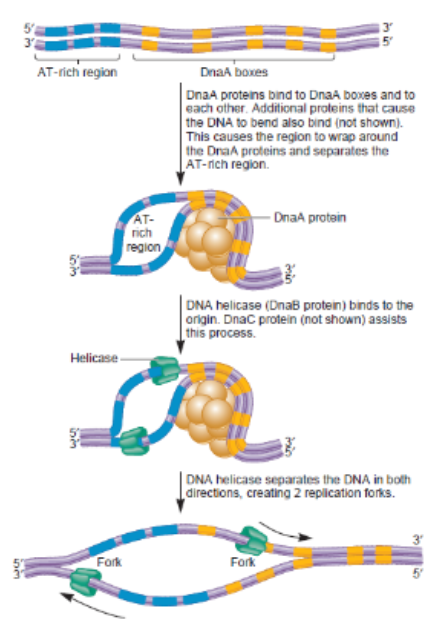
AT rich region
Area where replication, initiation needs to begin so by bending → causes unwinding of location → can throw machinery for work to take place → establish opening of entire strand
Seen in initiation
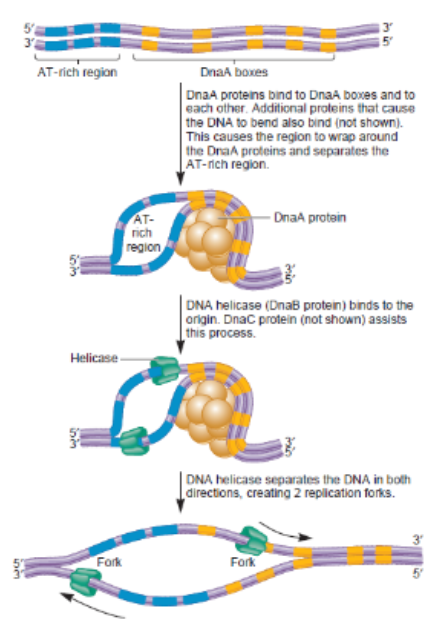
DnaA box
Location where it has high tendency to bind proteins (DnaA proteins) and when binding to it → causes conformational change where DNA twist where releases its hold on upstream region
Seen in initiation
Protein Helicase
responsible for unzipping inner wrongs and undoing hydrogen bonds between base pairs in middle
Seen in initiation
Stages of DNA replication Prokaryotic: Unwinding
Unwinding: DNA replication requires a single strand template. Multiple enzymes
Unwind whole thing
Helicase undo H bonds between base pairs
Strands separate: need something to hold DNA from not collapsing
The second you unwind, it wants to go back → it keeps it open to allow continuous replication by using SSN proteins → stay bound to unwounded DNA so machinery can do their job
Supercoiling is prevented during unwinding with DNA Gyrase
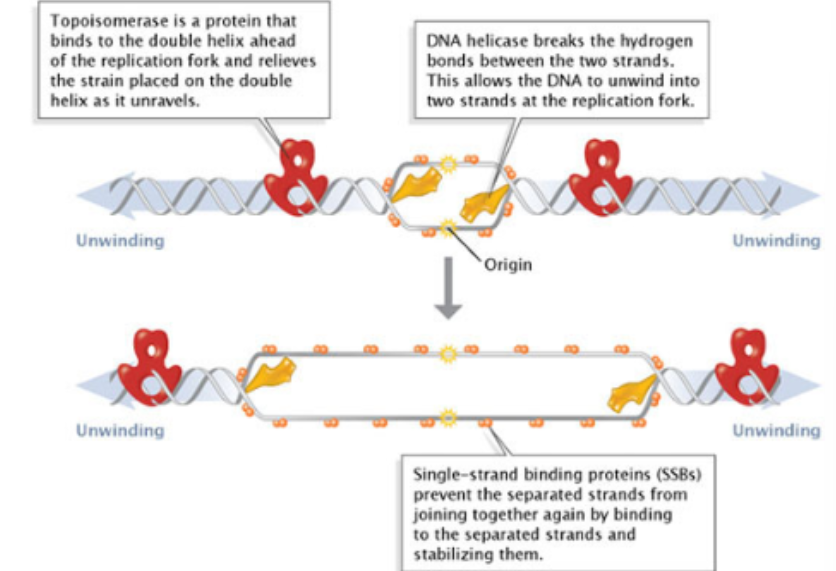

DNA Helicase
breaks hydrogen bonds between bases
involved in unwinding

Single-strand binding proteins
bind to single strands once they form to stabilize the structure
involved in unwinding
DNA gyrase
known as topoisomerase
prevents supercoiling and upstream torsional strain the builds up during unwinding
Forces torsional strain upstream → take wound DNA force apart, push it out → prevent you from going further
Need to unwind a bit so replication can continue

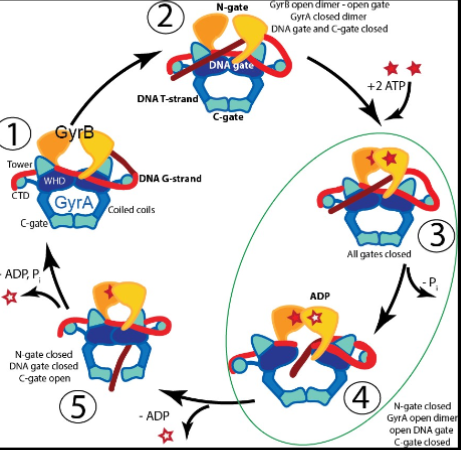
Kinds of DNA Gyrase: Topoisomerase 1
Creates single stranded breaks in DNA
take 2 strands that are there → clip one of them and twist it back to the other one → undoes a single turn to relax DNA
Kinds of DNA Gyrase: Topoisomerase 2
Creates double stranded breaks in DNA
Breaking the double strand, passing the double helix through it and resealing again
Does something similar to 1 but will clip both strands and turn them around each other → undo a single turn → both functioning a way to relax DNA before replication occurs
It is needed since it will seize to continue since it will not be able to unwind and there will be too much strain upstream
Stages of DNA replication Prokaryotic: Elongation
Elongation: single strands of DNA are used as template to make a new strand.
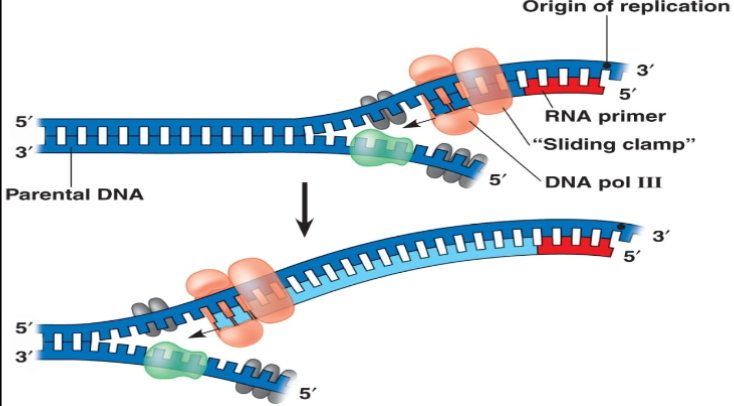
Primers
DNA polymerase can’t start replicating on its own. It needs a free 3’ OH to start adding nucleotides to. To bypass this issue primase is used to generate a short 10bps of RNA which ends in a free 3’-OH group. The RNA is later removed and replaced with DNA.
Elogation
DNA polymerase 3
Replicates DNA using the RNA 3’-OH group. Has very high processivity (holds on to the DNA and doesn’t release until its done). Works on leading strand.
Has 5’ → 3’ activity for replication
Has 3’ → 5’ exonuclease activity for backing up and replacing errors
Elongation
DNA polymerase 1
Removes RNA primer and replaces it with DNA. Working on the lagging strand
Has 5’ → 3’ activity for replication
Has 3’ → 5’ exonuclease (enzyme that removes nucleotide, specifically bad ones) activity for backing up and replacing errors
Has 5’ → 3’ exonuclease activity to remove primers and replace with DNA
Stages of DNA replication Prokaryotic: Elongation (2)
Poly 1 removes the RNA primer and replaces it with DNA
But a single break remains between the DNA generated by Poly 1 (replacing RNA primer) and poly 3.
This break is sealed by the enzyme Ligase
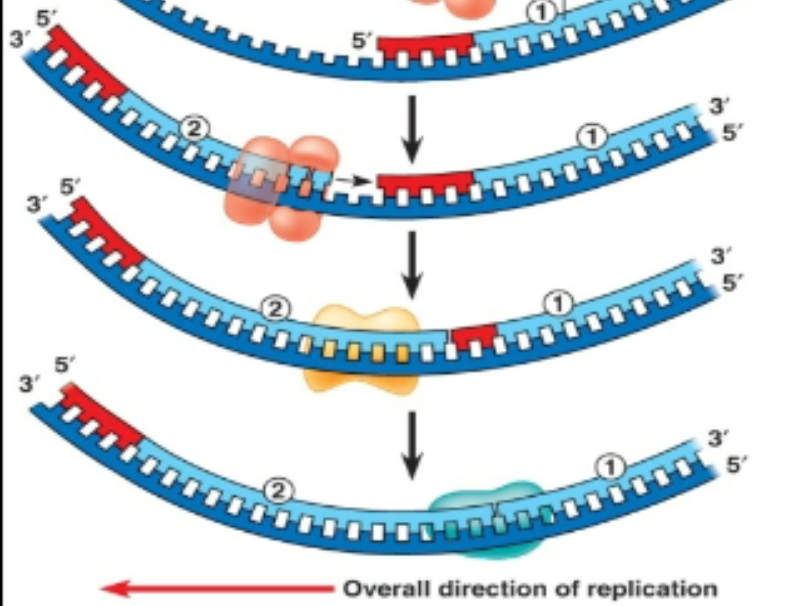
Stages of DNA replication Prokaryotic: Elongation (3)
DNA polymerase 1 can add/remove but not give DNA.
Once there is a new strand, DNA polymerase 1 budges all RNA primers off and its gone.
It cannot stitch together stands before and after primer
Little gap (no stitching) remains unglued (Ligase glues the gap back together)
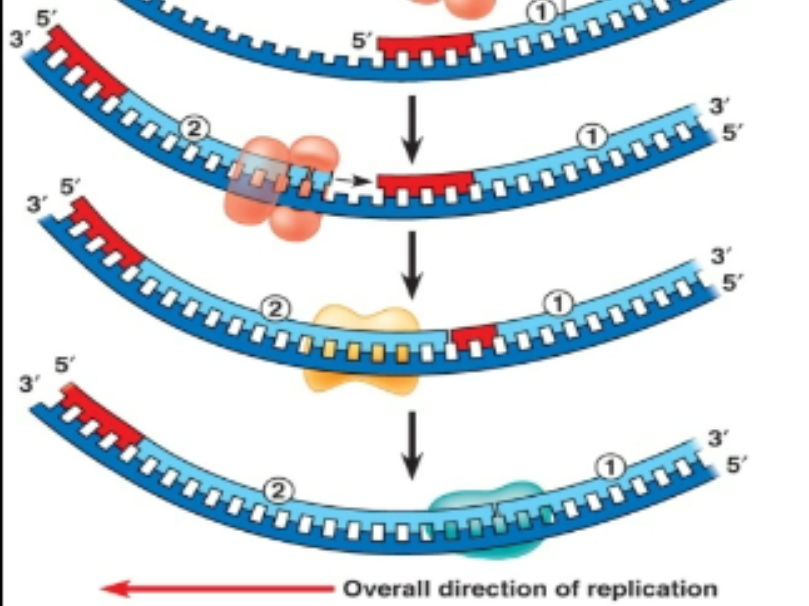
Stages of DNA replication Prokaryotic: Termination
Termination:
1 - Some DNA replication terminate when 2 replication forks meet
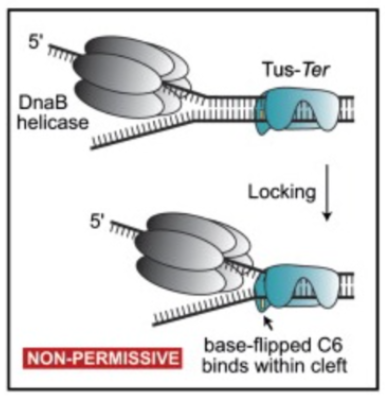
2 - Others need assistance of termination proteins Tus. The Tus protein binds to a sequence known as a Ter sequence. Upon binding, the complex blocks replication.
Tus behaves like a barricade which prevents polymerase/hell case from moving any further past that point. It is like a clamp and acts like a physical barrier.
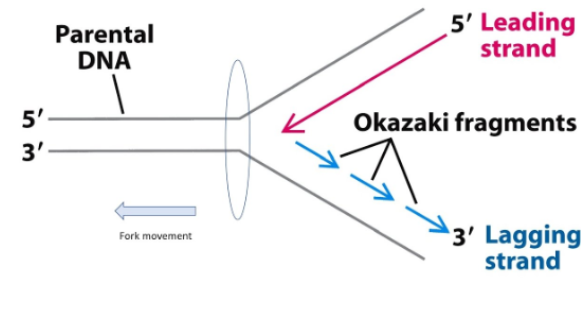
One strand must be synthesized in fragments
Structure is antiparallel - 2 strands run in opposite orientations (5’ to 3’ versus 3’ to 5’)
The new chains are synthesized in what for them is the 5’ to 3’ direction
The template is read in what for it is the 3’ to 5’ direction
Fragments: DNA polymerase can replicate in 5’ to 3’ direction
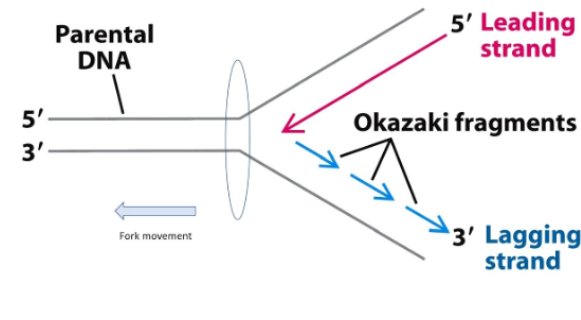
One strand must be synthesized in fragments 2
Polymerase can synthesize on leading strand without problem
The primer is generated on leading strand
Okazaki fragments: this strand is laying down 5’ to 3’ direction → then polymerase can bind and build in this direction
Eukaryotes DNA replication
Origin
Cell cycle
licensing
DNA polymerase
Nucleosome
Origin
multiple origins in eukaryotes in long linear strand
Unique sequences that are associated with yeast called autonomously replicating sequence (every time you see it → it indicates origin of replication. They are AT rich)
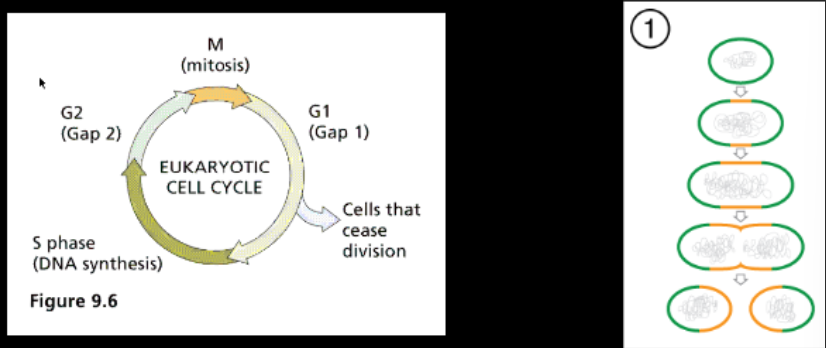
Cell Cycle
Has specific proteins that need to be turned on and off to cycle
There are checkpoints where DNA is checked to make sure it is correct
more dependent on timing
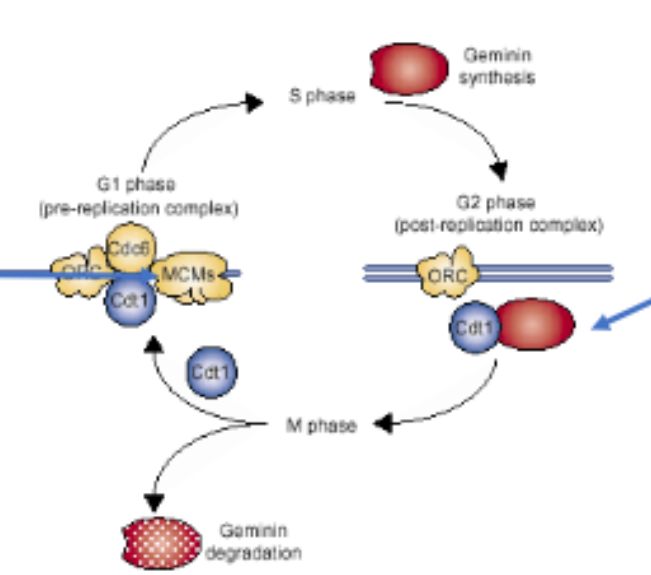
Licensing
Takes place to ensure replication forks initiate at the same time and doesn’t initiate more than once per cycle
Step 1: origins are licensed. Licensing proteins (origin recognition complex - ORC) get attached to the origin.
Step 2: Initiation machinery (MCM2 - helicase) binds and starts replication. Licensing factor is removed as the replication fork moves away from origin. Licensing factor is removed to ensure that replication can’t occur more than once per cycle.
MCM - minichromosomal maintenance. Has helicase activity and unwinds a small stretch of DNA at the beginning of replication.
Geminin Binds to Cdt1, which is used in replication initiation, and degrades it at G1. Therefore, replication can not re-initiate.
DNA polymerase
DNA polymerase in Eukaryotes
Alpha: has primase activity and initiations DNA synthesis by generating a short primer
Delta: takes over after the primer has been laid down (by alpha DNA polymerase) on the lagging strand
Epsilon: functions like Delta but works on leading strand
There is a small binding pocket in these polymerases that doesn’t allow incorrect bases to enter. If there is a lesion and an incorrect base needs to be paired, then these polymerases detach, and a trans lesion polymerase comes in and adds an incorrect base just to bypass the error. Error can always be fixed later.
Nucleosome
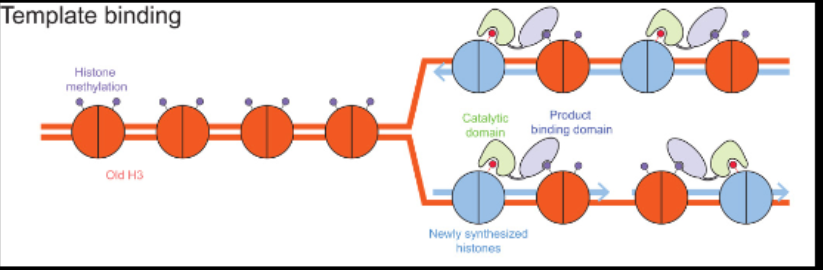
Eukaryotic DNA is complex and packaged for stability with histone proteins. It has 8 histone proteins, with DNA wrapped around it. During replication, the DNA needs to unwrap, and histones need to be removed so that all DNA can be replicated.
1: original nucleosome is removed
2: some old histones find their way to their location on the new strand
3: new histones are made and attach to old histones