BS1040- Bacterial genomes, cloning and AMR
1/56
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
57 Terms
What is the bacterial genome
Term coined by Hans Winker
• Current usage - all DNA within a cell
• Chromosome, plasmids, other genetic elements
What is genomics
• Genomics
• The study of genomes and genome sequences
• Mapping, sequencing and characterising genomes
Bacterial chromosome structure
-Usually one circular chromosome
-condensed into the nucleoid
Size and complexity
• link to structural complexity and biosynthetic capability
The E. Coli chromosome
• E.coli cell:
- ~0.5 μm wide x 1-2 μ m long: ~1-2 μm3 vol
•Chromosome:
- single dsDNA (double stranded DNA) molecule = 430 μm circle (~500x cell
width)
- 4.639 x 10^6 bp
- 1.5mm long! (ie. ~1000x cell length)
The bacterial nucleoid- structure
- occupies ~ 1/5 cell (~ 0.2-0.4um3)
• no membrane boundary -more diffuse in slow growing cells
- Compaction is dynamic, linked to growth rate
• in growing cells, typically in centre & bi-lobed
• Chromosome organised into large supercoiled domain loops.
• DNA has variety of proteins bound to it
- Histone-like protein HU very abundant structural protein
How do bacterial cells divide
• Bacterial cells divide by a process known as BINARY FISSION
• Single cell divides into 2 identical daughter cells
• Bacteria increase in length and mass
• Nucleoid expands - semi-conservative replication of the chromosome
• Synthesis of a septum at the midcell - divides the cell into two compartments
Semi conservative DNA
Semi-conservative DNA replication means that when DNA is copied, each new DNA molecule has one original (parent) strand and one newly made strand.
Origin of replication (oriC) and termination point (terC)
• Replication of the bacterial chromosome is BIDIRECTIONAL (processed in two directions at the same time)
• Starts at oriC - formation of two replication forks (Enzymes like DNA helicase unwind the double helix at oriC, creating an open replication bubble.
• From this point, two replication forks form and move in opposite directions around the circular chromosome.)
• Replisome complexes - replicate DNA at each fork
• Two replisomes approach each other at the terC site
terC is the termination site (the "end" of replication).
• The two replication forks move around the circular chromosome and meet at the terC region on the opposite side of the circle from oriC.
• When they meet, replication stops, and you get two identical circular DNA molecules
Chromosome replication and cell division
• Chromosome is attached by the oriC at the equator of the cell envelope
• Formation of 2 replication forks
• Two new oriC sites move apart at the cell expands
• The terC stays in the middle of the cell
• Completion of replication triggers FtsZ to form the Z-ring complex
• DNA chromosomes are separated and cell divides
Growth rate
• Chromosome replication time independent of growth rate.
• Time to replicate E. coli chromosome is 40 minutes
• But E. coli can divide every 20 minutes!
• Time to replicate chromosome can be longer than time for cell division
• The next round of replication can be initiated before the first is complete
• Faster growing cells have multiple replication forks
• The DNA inheritied by the daughter cells is already partially replcated
Plasmids, structure and function
• Found in most bacteria
• Size varies
• 1000s to 100,000s bp
• Like "mini-chromosome"
• Genes encoding protein and RNA
• Not normally essential for host growth
• Adaption. Selective advantage in specific
niche conditions
• Often contain multiple IS and Tn
Plasmid regulation points
• Autonomous replication: independent of
chromosome;
• fixed origin of replication; oriV
• some encode own initiator protein; oriV-specific
• Many control systems use functional RNAs
• use host DNA-synthesis functions. eg. DNA Pol
• semi conservative replication
• short replication time; 6 sec for 5kb, 3 min for 150kb
• Distributed on cell divisio
Plasmid regulation maintenance
• Self-regulating
• characteristic fixed copy number per cell;
• plasmid determined genetic property
• F @ 1 copy
• R6 @ 5-6 copies
• ColE1 @ 15 copies
• copy-control genes linked to initiation of replication at
oriV; Basic replicon
• related plasmids cannot coexist in same cell
(Incompatibility)
Role of plasmids
• Roles
• Conjugative plasmids eg F & P
• Transmissible
• Sequence transfer- evolution & AMR spread
• Tumour induction
• Agrobacterium Ti plasmids
• Resistance plasmids eg R
• Resistance (antibiotics, metals) and
antimicrobial production
• Metabolic capacity
• Rhizobium N fixation in legumes (symbiosis)
• Virulence
• Recombinant DNA technology
Bacteriophages
• "Eaters of bacteria"
• Twort & d'Herelle, 1920s
• Protein/membrane coat containing phage genome which is a nucleic acid
• Very small
• Presence detected by plaques which are Holes on lawn of bacteria (where they have died)
• causes Cell lysis
• OR insert into the chromosome called prophage
What are Insertion Sequences and Transposons
Insertion sequences (IS) and transposons are types of mobile genetic elements that can move within and between genomes.
Insertion Sequences and Transposons
Move (transpose) around genome
• Moves as unit
• No "free" form
• Transposition mediated by Transposase
• Transposition tightly controlled
• Inverted repeat sequences at ends to be recognised by transposase
• Can insert into chr, b'phage and
plasmid
• Moves genes between genetic elements
in genome
• Cargo genes (additional genes) eg:
• Anti-microbial resistance common
• Spread of AMR
• can have a mutagenic effect eg:
• inactive a gene it is insterted into
Horizontal Gene Transfer (HGT) - how
bacteria acquire new DNA
• Bacterial can exchange DNA within and between species
• Selective advantage
• Rapid acquisition of new DNA sequence and potentially functions
• Important source of genetic diversity
What are the 3 mechanisms of genetic exchange in bacteria
conjugation, transformation, transduction
Conjugation
• Transfer of DNA directly between bacterial cells via cell-cell contact
• Usually plasmid DNA
• Direct contact through pilus formation
Transformation
Transformation
• Bacteria take up DNA from their environment
• Natural transformation - cells must become "competent" to take up
DNA
• We can induce competence in the lab as a tool in cloning
Transduction
• Transfer of DNA between bacteria via bacteriophage
infection
• Phage infect a cell, replicate and produce new phage viruses
• Some host DNA can be packaged inside the virus - this is passed to the next bacteria when it is infected
Why is the size of bacteria important
- Link genome size to adaptive capability
• biosynthetic capability
• synthesis of nutrients
-Stress resistance
• resist environmental insults
-structural complexity
• surface structures, sporogenesis
-Regulation -sensing signals and transcriptional responses
• detect change or requirement and respond appropriately
• transcriptional regulation
Importance of genetic variation in bacteria
• Mutations
• Single nucleotide mutations and other small errors in
replication
• Error rate =10-5/bp with repair =10-10/bp
-Genetic Recombination
• Rearrangements: changes in organisation and deletions
-Genetic elements (plasmids, bacteriophage, Tns
etc)
• Acquisition of new genetic material
• Recombination with genetic elements
• Post-transformation recombination
-Core and non-core genome content
What is core and non core genome
Pangenome is entire set of genes found in all the strains of a species.
• Set of genes found in all strains is the core genome
• Genes found only in some strains is the non- core genome
• Non-core genome are the variable or accessory genes
• High level of horizontal gene transfer(HGT) promotes more non-core genes (open pangenome)
• Low level or no HGT promotes fewer non-core genes (closed pangenome
Lecture 2
DNA cloning
the process of making multiple, identical copes of a particular
piece of DNA
• Core technique that is essential for many areas of biology and medicine
What is a genome library
• Collection of DNA fragments that have been cloned into vectors
• Each DNA fragment can be identified and isolated for further study
• Genomic libraries - collection consists of overlapping DNA fragments that together make up the total genomic DNA of an organism
• cDNA library - collection of cloned DNA sequences that are
complementary to the total mRNA extracted from an organism -
representative of all of the genes that are expressed
Cloning vectors- key features
-Maintenance (replication) in host cell
• plasmid DNA
• Other types of vectors are available...
-Restriction enzyme site
• insertion of foreign DNA fragment
• unique
• multiple cloning site
• create RECOMBINANT DNA molecule
-Method to in2troduce into host cell
• transformation (plasmids)
-Genetic selection marker
• small proportion transformed
• need to recover cells containing vector/clones
• antibiotic resistance -direct selection
- Identify recombinants
• system/screen for recombinants
• Insertional inactivation
How to construct a genome library
-Isolate chromosomal DNA
-Digest with restriction
enzyme
• Digest chromosomal DNA
• Digest vector DNA
-Ligate chromosomal DNA
with vector DNA
• Transform ligated DNA into E. coli
• Select cells containing vector- eg. Antibiotic containing
medium
• Transformation is relatively inefficient
• Not all ligated DNA maintained in E. coli
• Not want all ligated products
Outcome of transformation
• Linear molecules destroyed in E. coli by nucleases
• Transformant will not replicate on antibiotic-containing medium
• Transformant will survive as vector plasmid has antibiotic resistance gene
• Transformant will survive as recombinant=vector plasmid has antibiotic resistance gene
Blue white screening for recombinants
lacZ encodes β-galactosidase
• β-galactosidase converts X-Gal (colourless) to blue compound
• X-Gal
• 5-bromo-4-chloro-3-indolyl β-D- galactopyranoside
• Vector containing lacZ fragment
• lacZα gene
• Insert DNA fragments into sequence encoding lacZα
• Insertional inactivation
• β-galactosidase no longer produced, X-Gal not converted
• SCREEN for recombinants
No lacz activity= white, insertional inactivation
LacZ activity= blue
What can you do with a DNA library
• Each DNA fragment can be identified and isolated for further study
• We can SCREEN the library to identify specific clones that we may be interested in
Protein product based approaches
-Assumes DNA is expressed
-Use antibody specific for target protein product
• Find clone antibody binds to
-Use specific assay for protein product
• Assay for enzyme activity
-Complementation
• Complement a mutant in the gene
DNA sequence based approaches
DNA sequence-based approaches:
-Utilise base sequence complementarity
• Hybridisation to labelled DNA probe
-Utilise known DNA sequence
• Use PCR amplification
How to make a genomic library
• Representative Library
• There is a clone in the library for every part of the genome
• Reality is some parts will be missing- Why?
• Larger the genome the greater number of clones needed to be representative
• Larger the insert the fewer clones needed
• Usability vs Size
Considerations in making library of expressed genes
• Lots of non-coding DNA means many clones will not be of interest
• Not a significant issue with majority of bacteria
• Why not just focus on DNA that is expressed:
• Use the mRNA
-Ability to focus gene library on different expression patterns
• Time, place, response.....
• cDNA library (complementary DNA)
• DNA synthesised from expressed RNA is used to make a library of clones. A cDNA library
Making a cDNA library
1. Grow cells under required conditions or isolate cell type
2. Isolate mRNA
• Release from cells
3. Purify and remove any DNA
• Remove cell debris and contaminating DNA
4. Use Reverse Transcriptase to copy mRNA
• Makes RNA-DNA hybrid
• Needs 3' primer
-Remove RNA strand
• RT activity or RNase
-Synthesise second DNA strand
• Use DNA polymerase and primer
• Use to make a cDNA library
Genome vs cDNA libraries
Complete: Genomic library
-Contains all DNA in genome
• Coding and non-coding DNA
• Abundance of specific sequence matches abundance in genome
Enriched: cDNA (complementary DNA)
-Contains copies of expressed mRNA
-Contains coding sequences only
• No introns, No promoters, No intergenic regions...
-Clones in library reflect mRNA abundance
• Enrichment for highly expressed genes
• Subset of genes can constitute significant % of library
Genome library approach to sequencing a genome
• Clone based library in vector (not now approach of choice)
• Representative collection of fragments to sequence
• Simple guide to sequencing a genome:
• Chop genome up
• Make a library
• Sequence the library inserts
• Assemble the bits of DNA sequence into
contiguous sequence
• Fill in any gaps
• Find the gene
Coding sequences, ORF, and consensus sequences
• Coding sequences (genes):
These are regions of DNA that contain the instructions for making proteins.
• Open Reading Frames (ORFs):
These are stretches of DNA that start with a start codon (AUG) and end with a stop codon (UAA, UAG, or UGA) — they are potential protein-coding regions.
• Consensus sequences:
These are short, common patterns of bases found at important sites (like promoters or splice sites) that help identify where transcription or other processes start.
Example: the TATA box in promoters
How can these be studied
Bioinformatics is the use of computers and software to study biological data — mainly DNA, RNA, and protein sequences.
Scientists use it to analyze, compare, and interpret genetic information.
-Bioinformatics uses computers to scan DNA and protein sequences to find:
• Genes and coding regions (like ORFs and consensus sequences)
• Similarities (homology) between sequences to understand function and evolution.
Homology
• Homology means similarity due to shared ancestry.
• Bioinformatics tools (like BLAST) compare DNA or protein sequences from different organisms to find regions that are similar or identical.
• This helps scientists:
• Find related genes in different species
• Study evolutionary relationships
• Predict functions of unknown genes or proteins based on known ones
Transcriptonomics
The study of RNA and differences in mRNA expression across the whole genome
Profiling differences in patterns of gene expression across the genome
This means studying which genes are turned on or off in different cells, tissues, or conditions — across all genes in an organism's genome.
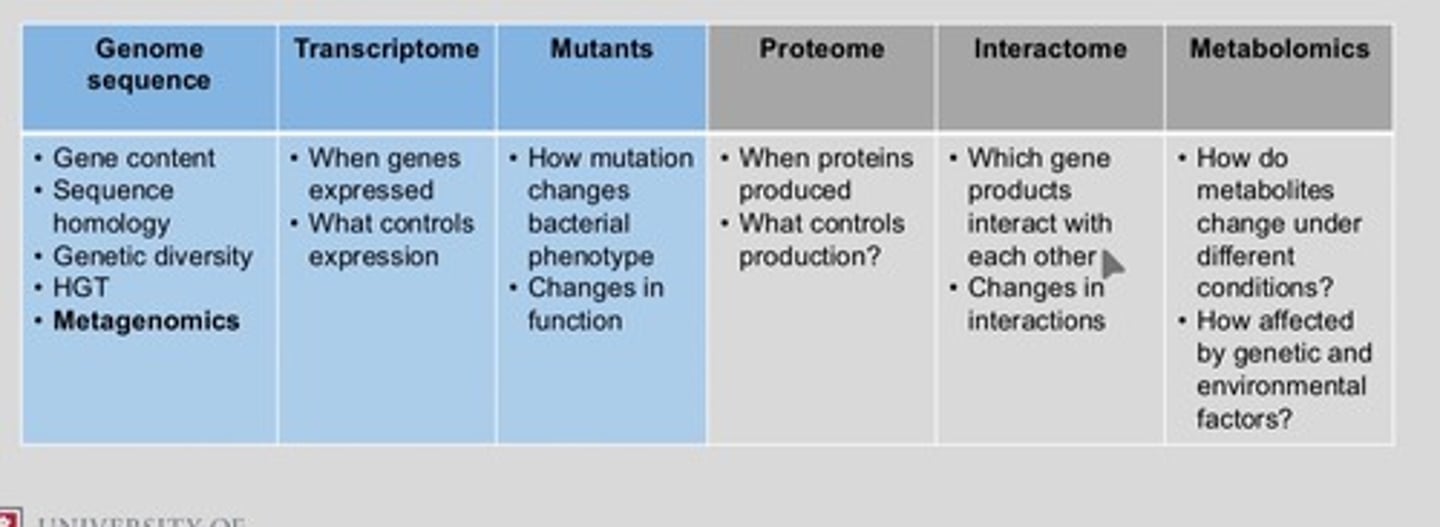
Lecture 3
What is AMR
Antimicrobial Resistance (AMR) occurs when bacteria,
viruses, fungi and parasites change over time and no longer
respond to medicines, making infections harder to treat and
increasing the risk of disease spread, severe illness and
death
Antimicrobials
• Compounds that stop of slow the spread of microorganisms
• Include antibiotics, anti-fungal, anti-parasitic and anti-viral drugs
• Focus for the remainder or this lecture is antibiotics -
specifically target bacteri
What is resistance
• Microorganisms that are no longer controlled or
killed by an antimicrobial
How do antibiotics work
Inhibit protein synthesis
Inhibit cell wall synthesis
Inhibit DNA synthesis
Distrust bacterial cell wall
How do antibiotics work 2
Spectrum of Activity Antibiotics target specific
bacterial structures
Choice of antibiotic depends on the type of infection
Mechanisms of antibiotic resistance
- avoid- modify target of the antibiotic
- attack- destroy the antibiotic
-remove- actively remove the antibiotic from the cell
-limit- reduce the uptake of the antibiotic
How do bacteria become resistant
• Genes that encode the resistance mechanisms
can be inherited or transferred
• Mobile genetic elements - Horizontal gene
transfer
• Spontaneous mutation
• Change in expression level
Factors affecting AMR
- globalisation
- uncontrolled use of antibiotics
Consequences of AMR
- common diseases can return to being major issues
- global pandemics