Module 2.3- DNA Extraction & Assembling DNA Parts
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
34 Terms
DNA extraction
A process that isolates DNA from a biological sample, such as blood, tissue, or saliva, by breaking open the cells and separating the DNA from proteins and other components
Purpose of DNA extraction
Obtain DNA from cells
Isolate and purify DNA from contaminants such as proteins, carbohydrates, lipids or other nucleic acids
Use the purify DNA for further studies or applications
Potential uses of purified DNA
Genetic Engineering Plants and Animals
DNA with desirable genetic traits are extracted and purified and transplanted into the organism’s genome.
Pharmaceutical Products
Pharmaceuticals made via recombinant genetics include the Hepatitis B vaccine and human growth hormone. In addition to a number of other hormones created using DNA extraction, the most common is insulin.
Medical Diagnosis
DNA is extracted from the patient and purified for analysis to diagnose conditions such as cystic fibrosis or Huntington’s disease
Identity Verification
Genetic fingerprinting is a process that matches genetic material from an individual with other genetic material available. One example is paternity testing, to determine someone’s biological father
Harvesting
Routinely isolate DNA from human, fungal, bacterial and viral sources
Some cells may need pretreatment, e.g.:
Whole blood
Tissue samples
Need sufficient samples for adequate yield
3 steps of DNA extraction
Lysis, separation, and purification
Lysis
Release DNA by breaking the cell
Separation
Separate DNA from other cell components (e.g., carbohydrates, proteins)
Purification
Remove any unwanted material from DNA
Types of lysis
Mechanical Disruption
Homogenization
Mortar and pestle
Chemical Lysis
Detergents (e.g. SDS)
Buffering Salts (e.g. Tris-Cl, EDTA)
Methods of separation
Protein Denaturation/Precipitation
RNA denaturation
DNA precipitation
DNA adsorption
Protein denaturation/precipitation in separation
Methods include:
Enzymes (e.g. Proteinase K)
Phenol/Chloroform extraction
DNA in aqueous phase
Proteins in organic phase
Chemical (e.g. Guanidine hydrochloride)
Salting out
High salt concentration (e.g. LiCl, NaCl)
Low pH
RNA denaturation in separation
Via RNase (enzyme)
DNA precipitation in separation
Precipitate DNA using alcohol (i.e., ethanol, isopropanol)
DNA adsorption in separation
Use solid matrix (e.g. membrane) to bind DNA
Wash away contaminants
Elute DNA from column
Purification method
Wash DNA with 70% ethanol
Re-suspend DNA in buffer
Usually in TE buffer (Tris-HCl and EDTA)
Methods of analyzing DNA
Gel electrophoresis and spectrophotometry
Gel electrophoresis
Analyze DNA quality by electrophoresis
Determine the size of DNA isolated
Spectrophotometry
DNA concentration can be determine using absorbance at 260 nm
1 𝐴260 = ~50 𝜇𝑔/𝑚𝑙
DNA purity can be determine by A260/A280 ratio
Traditional molecular cloning workflow
Preparing both the cloning vector and the DNA fragment (insert) to be cloned
Ligating the insert into the vector to create a recombinant molecule
Transforming this recombinant molecule into a host organism like bacteria
Screening the colonies to identify the correct clone
NOTE: Must cut plasmid and gene fragments with the same restriction enzymes (Scissors)
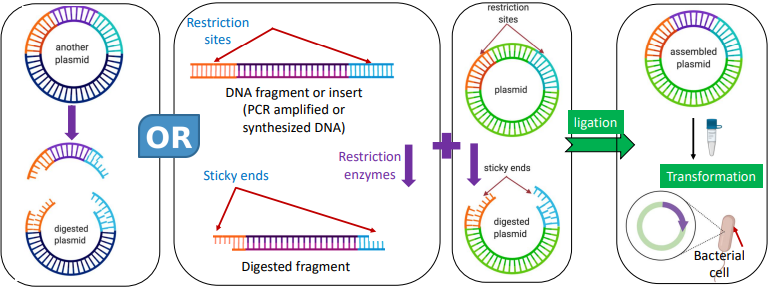
When can we form a recombinant plasmid?
Once we have amplified (and verified) our DNA (biological parts)
What we need to form recombinant plasmid
Amplified and purified biological part(s)
Target plasmid
Reagents
Techniques to form recombinant plasmid
DNA extraction (miniprep)
Restriction/Ligation (cut and paste)
Gel
Plasmid
Circular or linear double-stranded DNA
Replicate separately from the chromosome, uses the same enzymes -
Typically much smaller than chromosomes: 1 kbp to more than 1 Mbp. less than 5% of the size of the chromosome
Usually not “essential” to the cell
Each cell can contain more than one copy or even one type of plasmid
Features of a recombinant plasmid
Origin of replication
A gene that is advantageous for survival, such as an antibiotic resistance gene
Cloning sites
Promoter
Insert, restriction sites, ribosome binding site, primer binding sites
Origin of replication (ori)
DNA sequence that directs initiation of plasmid replication by recruiting the bacteria transcription machinery
Controls the host range and copy number of the plasmid
Not all origins of replication are created equal. Some will produce many plasmid copies and others produce just a few copies (5 -700)
Cloning sites
Sequences that can be cut with restriction enzymes
Ribosome binding site
Various Shine-Dalgarno sequences have been found in prokaryotic mRNAs
These sequences lie about 6-10 nucleotides upstream from the AUG start codon.
Activity of a RBS can be influenced by the length and nucleotide composition of the spacer separating the RBS and the initiator AUG.
Different in prokaryotes and eukaryotes
Primer binding site
Initiation point for PCR amplification or DNA sequencing of the plasmid
Can be utilized to verify the sequence of the insert or other regions of the plasmid.
Why are plasmids with the same ori incompatible?
They cannot be in the same bacteria, because they will compete for the same machinery, creating an unstable and unpredictable environment
The best choice of ori depends on…
How many plasmid copies you want to maintain
Which host or hosts you intend to use
Whether or not you need to consider your plasmid's compatibility with one or more other plasmids.
Promoter
100 to 1000 base pairs long and found upstream of their target genes.
Controls the binding of the RNA polymerase and transcription factors
RNA pol in bacteria is a single enzyme
Eukaryotes have multiple polymerases → the promoter must be compatible with the type of RNA to be made
What is the strength of a promoter important for?
Controlling the level of insert expression (i.e., a strong promoter directs high expression, whereas weaker promoters can direct low/endogenous expression levels)
Constitutive promoter
A DNA sequence that drives the continuous transcription of a gene
The gene is expressed at a constant level regardless of external conditions or regulation.
It is always "on" and binds RNA polymerase to initiate the production of mRNA, leading to the consistent expression of a protein.
Inducible promoter
A DNA sequence that controls the transcription of a gene in response to a specific signal, allowing for gene expression to be turned on or off, or adjusted.
This regulation is often achieved using activators or repressors, where a small molecule inducer can bind to the regulatory protein to either enable or block its interaction with the DNA, thereby controlling the expression of downstream genes.