Genetic - Extensions to Mendel: Incomplete dominance and multiple alleles
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
22 Terms
Define and be able to identify the following: loss-of-function mutation, null allele, gain-of-function mutation, neutral mutation
loss-of-function mutation: A mutant allele that decreases the amount of mRNA produced or the protein has less activity
null allele: A mutant allele that has a special case where the protein produced has no activity or functionality. Usually inherited as a recessive trait
gain-of-function mutation: A mutant allele that increases the amount of mRNA produced OR the protein has more activity—> can gene can express themselves in new tissue or has a new function. Usually the dominant allele
neutral mutation: A mutant allele that has a change in the DNA sequences that doesn’t impact function or expression
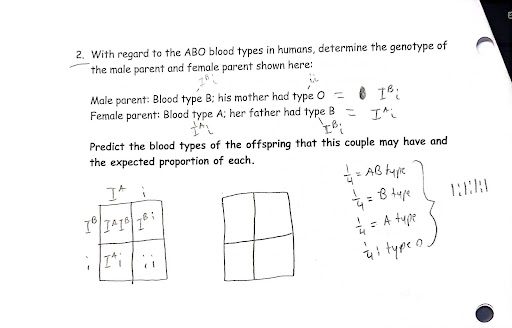
Solve genetic problems (parent genotypes and phenotypes, proportion/ratios of child genotypes and phenotypes) using complete dominance, incomplete dominance, codominance, and different dominant relationships for alleles in a single gene.
complete dominance: a form of dominance wherein the dominant allele completely masks the effect of the recessive allele in heterozygous conditions.
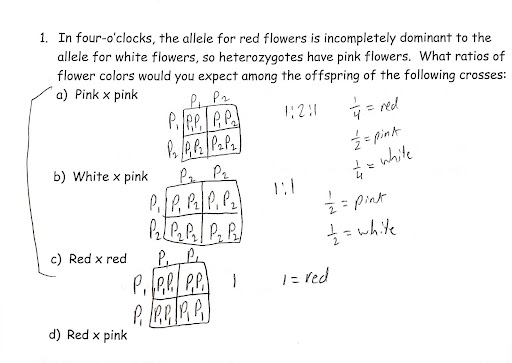
incomplete dominance: When heterozygous individuals display intermediate phenotypes between either homozygous types —> usually more similar to one of the homozygous types than the other.
codominance: When heterozygotes with a different phenotype then that of either homozygotes —> detectable expression of both alleles in the heterozygotes
Solve genetic problems (parent genotypes and phenotypes, proportion/ratios of child genotypes and phenotypes) using multiple (>2) alleles for a single gene causing a single trait.

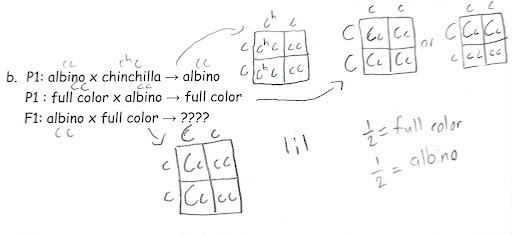
Full Color = C, C_
Chinchilla = cch, cch or cch,c
Himalayan = ch,ch or ch,c
Albino = cc
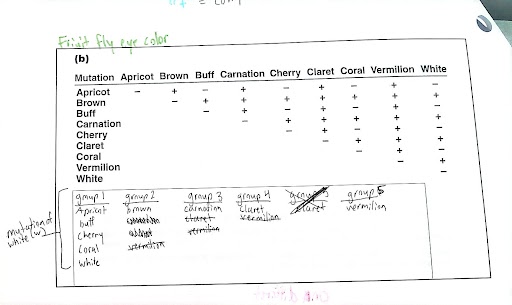
Use complementation analysis to determine if a trait is impacted by 1 gene or 2+ genes.
Infer from the results of genetic crosses the existence of interactions between alleles of different genes including: additivity, epistasis, redundancy, and complementation.
Gene interaction: The production of novel phenotypes by the interaction of alleles of different genes —> Several genes influence a particular characteristic , not necessarily interact with each other—> can lead to altered phenotypic ratio
Additivity: Genotypes at multiple loci act additively to determine phenotype (9:3:3:1)
epistasis: The expression of one gene masks or modifies the effects of a second gene—>alleles of one gene modify or prevent expression of alleles of another gene
redundancy: (15:1)
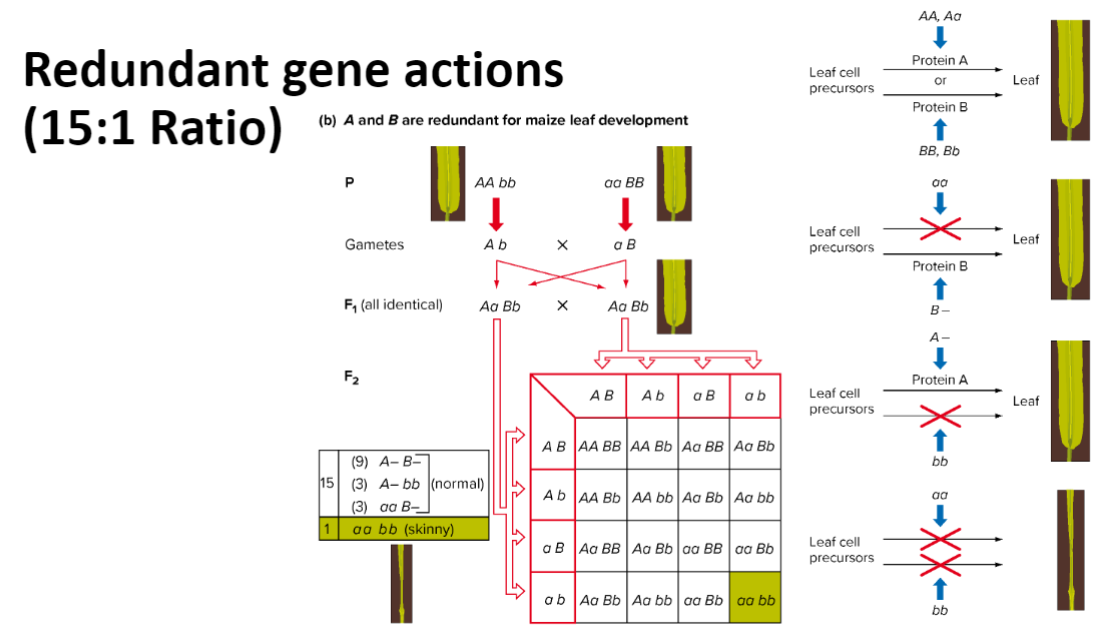
complementation: to determine wheater two independently isolated mutations are in the same gene or wheather they represent mutation in separate genes —> complimentation occurs on seprate genes, and visa versa—→ Such an analysis may reveal that only a single gene is involved or that two or more genes are involved

Solve genetic problems (parent genotypes and phenotypes, proportion/ratios of child genotypes and phenotypes) using gene interactions.
Define pleiotropy, and summarize how and why this occurs.
pleiotropy: A condition in which a signal mutation causes multiple phenotypic effects
It occurs because there c an be multiple genes/traits on a single allele on one loci
2. Recognize progeny ratios that imply the existence of recessive lethal alleles.
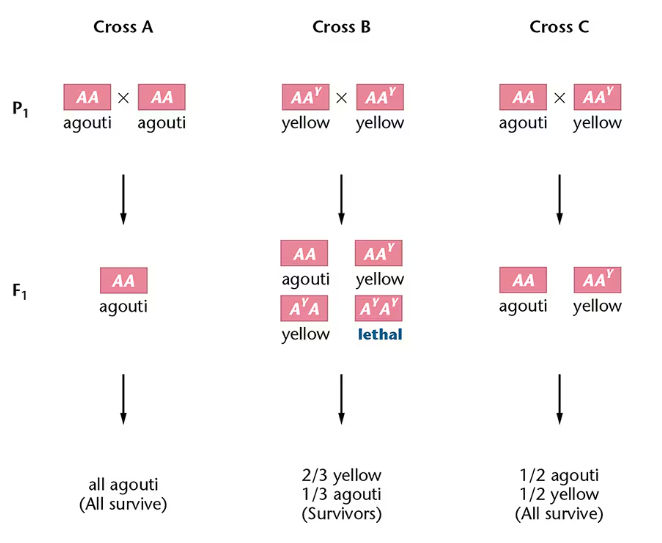
AY AY is lethal —> gain of function with the new coat color but loss of function of embryotic development
3. Differentiate between sex-linked, sex-limited, and sex-influenced traits.
sex-linked: A trait that tends to be associated with one sex or the other.—> sex chromosome
sex-limited: A trait that is expressed in only 1 sex even though the trait may not be X-linked or Y-linked —> sex chromosome
sex-influenced traits: A phenotypic expression conditioned by the sex of the individual. A heterozygote may express one phenotype in one sex and an alternative phenotype in the other sex (e.g, Pattern of baldness) —> autosomal
4. Solve genetic problems (parent genotypes and phenotypes, proportion/ratios of child genotypes and phenotypes) when there is incomplete penetrance and/or variable expressivity.
5. Define temperature-sensitive mutations, and summarize the impacts of these on a phenotype
temperature-sensitive mutations: A conditional mutation that produces a mutant phenotype at one temperature and a wild-type phenotype at another
caused by conditional mutations (A mutation expressed only under a certain condition
Its determined by environmental conditions —> can help induced and isolate in viruses and have added immensely to the study of viral genetics
Define linkage with respect to gene loci.
Linkage: Two genes on a signal pair of homologs— linked genes: Genes that are located close together on a chromosome, and segregate together together (travel together)
MUST BE SYNTENIC GENE AND THEY DO NOT ASSORT INDEPENDENTLY
Complete linkage or linkage without crossing over: Produces only Parental or noncrossover gametes —> equal proportion of parental—> observed only when genes are very close together and # of progeny is relatively small—>1:2:1 ratio
linkage with crossing over: crossover involve only two nonsister chromatids of the four chromatids present in tetrad, results in recombinant or crossover gametes
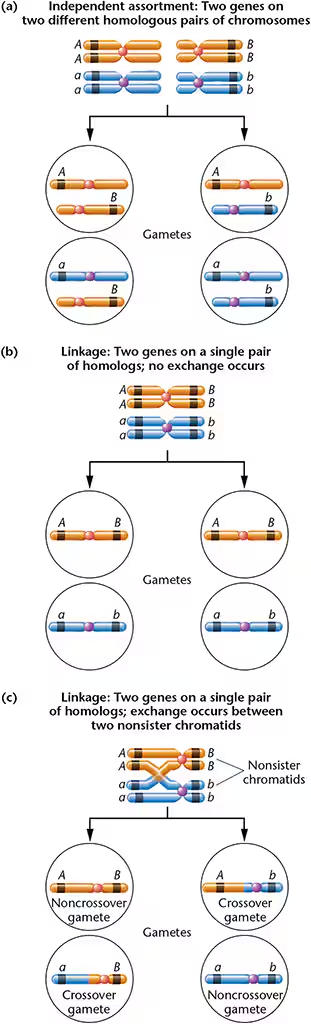
2. Identify and differentiate between between parental and recombinant gametes.
Parental gamete: Alleles of syntenic gene that Homologs that CAN NOT be reshuffled; no crossing over occurs
Recombinant gametes: Alleles of syntenic genes that can be reshuffled when crossing over occurs between homologs
3. Conclude whether two genes are linked from ratios in progeny in a dihybrid cross and using a chi-squared test.
You use a Chi-square of independence instead of chi-square goodness of fit

4. Discuss the relationship between the recombination frequency and the map distance separating two loci on a chromosome.
As distance between the two gene increases, the proportion of recombinant gametes increases and that of the parental gametes decreases
Recombination frequency is likely a reflection of the physical distance between two genes
linked genes with higher recombination frequency are more distant from one another than genes with lower frequencies.
the farther away the gene are from each other, the more likely that recombination will occur
5. Explain why the value of the recombination frequency between any two genes is limited to 50%
in a single crossover(occurs between two nonsister chromatids), recombination is only be seen in 50% of the gametes because
Single crossover (but not between loci) —> goes undeteched (all parental)
single crossover (loci are farther apart)—> recombinantion occurs
depends on where the loci are, because with the four chromatids, there will always result with atleast half of them being parental gametes, so the other half will result in recombinant gametes, but only less than 50% because of the signal cross over
Differentiate between coupling (cis) and repulsion (trans) configuration.
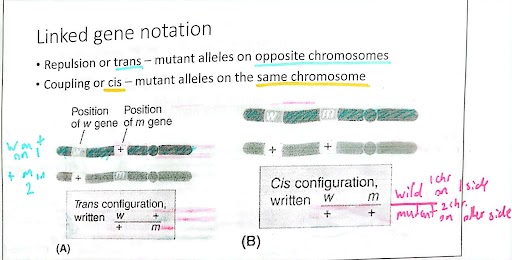
Coupling (cis): mutant alleles on same chromosomes
Repulsion (trans): mutant alleles on the opposite chromosomes
Differentiate between genetic maps (linkage map) and physical maps.
Genetic (linkage) mapping: plots the position of genes on chromosomes by using the frequency of recombination between syntenic genes (measured in centiMorgans)
Physical mapping: Produced through DNA sequencing, are measured in base pairs
Given recombination frequencies, calculate the expected number of parental and recombinant types.
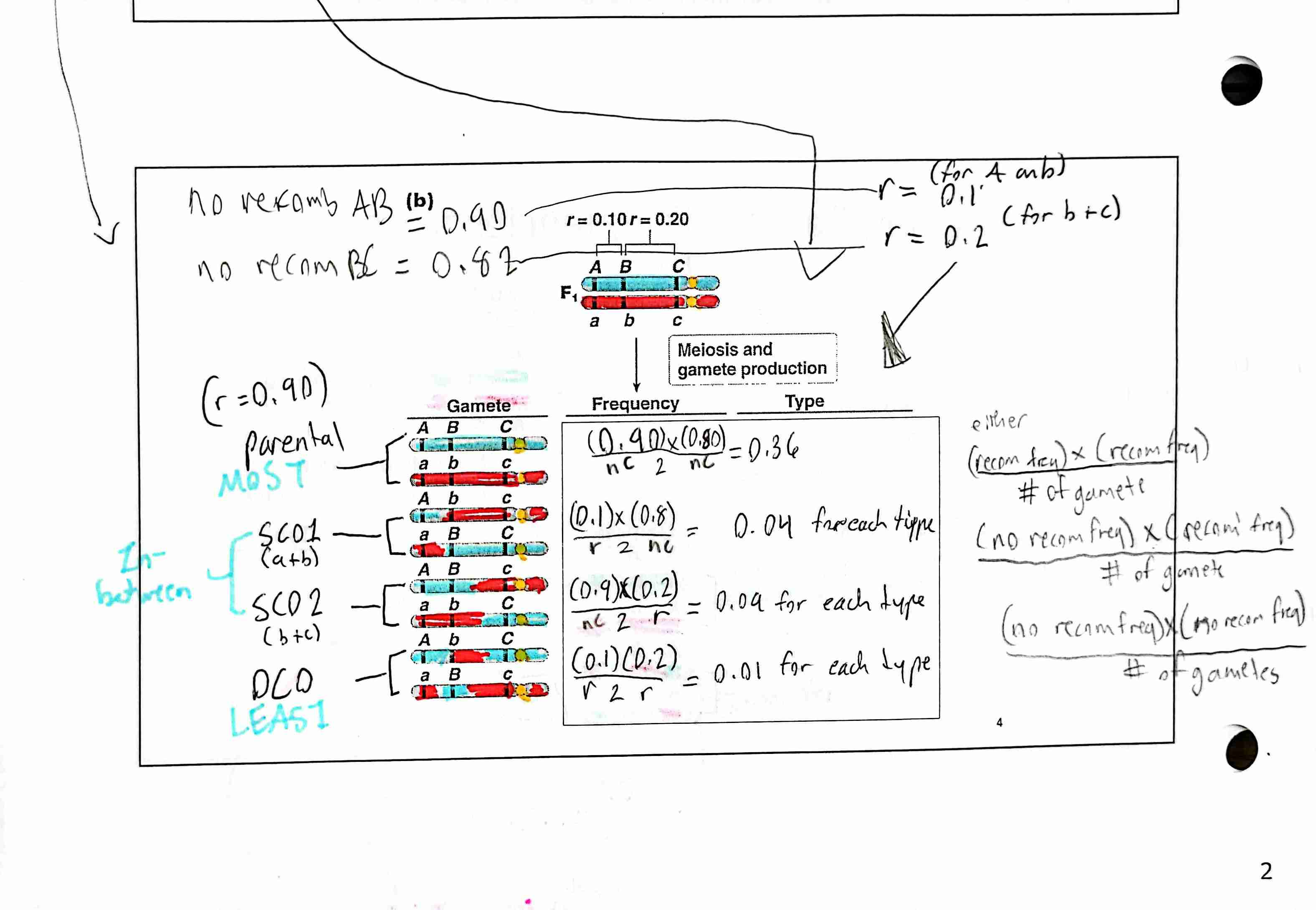
For two-point mapping of genes, identify parental and recombinant types, calculate the recombination frequency between pairs of genes, and draw a genetic map
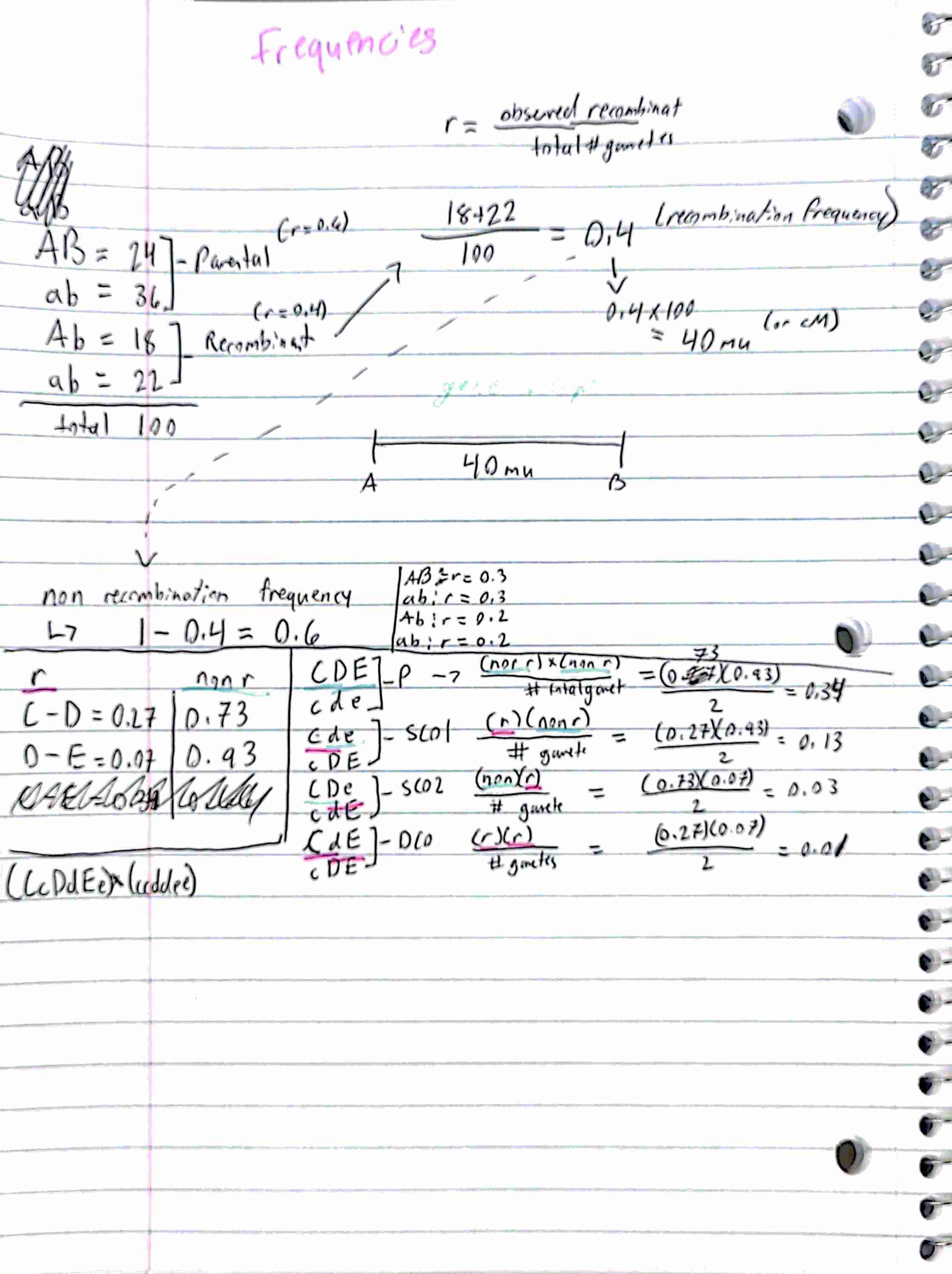
For three-point mapping of genes, identify parental and recombinant types, calculate the recombination frequency between pairs of genes, calculate and explain the results from the coefficient of coincidence and interference, and draw a genetic map.

Compare and contrast SNPs, RFLPs, and microsatellites.
ALL ARE GENETIC MARKERS/DECTECTION
SNPs (single-nucleotide polymorphism): A variation in one nucleotide pair in DNA, as detected during genomic analysis. Present in at least 1% of a population, a SNP is useful as a genetic marker
RFLPs (Restriction fragment length polymorphism): Variation in the length of DNA fragments generated by restriction endonucleases. These Variation are caused by mutation that creat or abolish cutting sites for restriction enzymes. RFLP are inherited in a codominant fashion and are extrememly useful as genetic markers
microsatellites: Short repetitive sequences that are found throughout the genome and they vary in the number of repeats at ant given site—-Short tandem repeats 2-9 bases long found within minisatelities DNA sequence. Used in preparation of profiles in DNA forensics and other applications
Polymorphism Type: SNPs involve single base changes, RFLPs involve changes in restriction enzyme sites, and microsatellites involve variation in the number of short sequence repeats.
Detection Methods: SNPs are detected by sequencing or SNP arrays, RFLPs by restriction digestion and gel electrophoresis, and microsatellites by PCR and electrophoresis.
Frequency & Information Content: SNPs are the most abundant but provide low individual informativeness, while microsatellites are highly polymorphic and informative. RFLPs are less common and less informative in comparison.
Application Areas: SNPs are mainly used in GWAS and personalized medicine, microsatellites in forensic science and population genetics, and RFLPs are largely of historical interest in genetic studies.