Cancer cells: excessive birth rate
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
15 Terms
What are the 4 types of genetic changes to a gene? (pathogenic variants) which do they all lead to?
Amplification/Deletion (large scale aneuploidy - chromosomal)
Single gene mutation (point/ small insertion -deletion )
Chromosomal translocation
Epigenetic methylation
All lead to aberrant activation or inactivation of specific genes so they function incorrectly (at the wrong time)
Amplification / Deletion
can be caused by
increased number of chromosomes (>2) from errors in cell division/ mitosis
or
Formation of “double minute“ chromosomes - extra fragment of DNA carrying specific (onco)genes outside of the nuclear genome. Can be reincorporated into cell chromosomes
Leads to production of MORE proteins by having extra copies of genes
Deletion can also occur during mitosis/ aberrant DNA repair leading to loss of specific (tumor supressor genes) = less protein
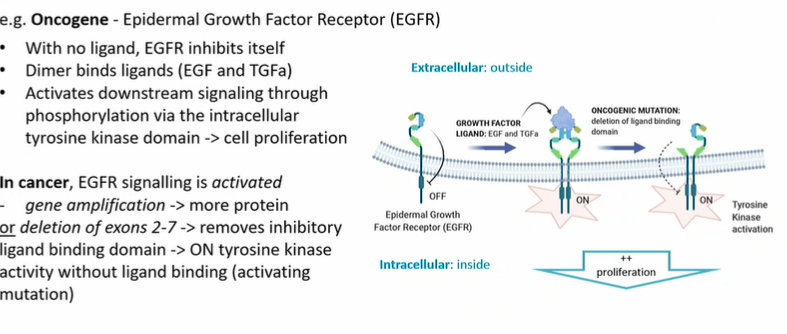
Partial gene deletion - case of oncogene Epidermal Growth Factor receptor
due to deletion tyrosine kinase always dimerised which leads to phosphorylation which leads to constant proliferation
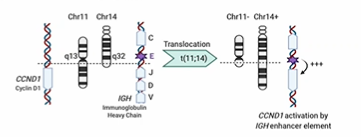
Translocation - description and case of CCND1 oncogene
Rearrangements of genetic material to juxtapose gene promoters/regulatory elements which drives inappropriate gene expression (activating mutation)
Translocation of CCND1 gene from Chr 11 to CHr 14 leading to IGH enhancer element having an effect by switching on CCND1 gene leading to excess Cyclin D1 protein which results in deregulation of cell cycle at R point
Seen frequently in Mantle cell lymphoma
Evidence of effects of excess oncogenes
Transfection of immortilise mouse fibroblast with cancer mutation (eg DNA that carry oncogen RAS) leading to fully transformed cells that are:
- growth factor independenthave 3D growth (disorganised)
lose Anoikis (an apoptotic death triggered by the detachment from the ECM/BM) → allow anchorage independent growth
Form tumours in vivo (invasive/ metastatic)
Inactivating point mutations and epigenetic regulation - two hit hypothesis - case of retinoblastoma
retinoblastoma often caused by inheritance of one mutation in RB1 gene and acquisition of a second mutation which switches off its function (regulating cell cycle and proliferation)
Alfred Knudson’s TWO-HIT hypothesis for the inactivation of tumour suppressor gene
First hit- acquired inheritance or somatic mutation of one allel
Second hit - somatic mution/ deletion or epigenetic inactivation of second organelle
Causes total loss of RB1 encoded protein (pRb). pRb function is lost in 25% of all cancers through inactivating mutation or epigenetic changes (promoter methylation)
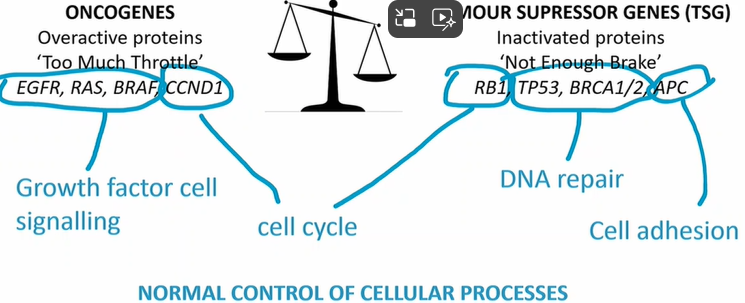
Oncogenes - overactive proteins
EGFR, RAS, BRAF, CCND1
Tumour supressor genes - inactivated proteins
RB1, TP53, BRCA1/2, APC
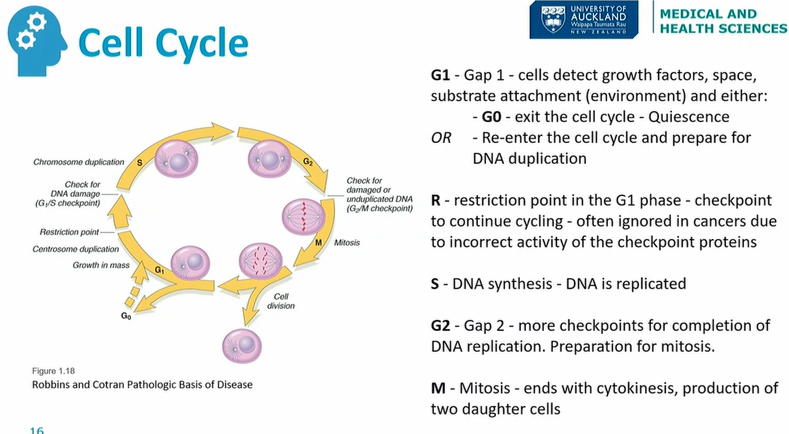
Cell cycle
Normal growth factor signalling
Promotes entry into cell cycle
Relieves blocks on cell cycle progression (promote replication)
Prevent apoptosis
Enhances synthesis of components (nucleic acids, proteins, lipids, carbs) required for cell division
EG, EGF - epidermal growth factor - mitogenic, stimulates epithelial cell migration, stimulate formation of granulation tissue
Oncogenic growth factor signalling
Increased levels of growth factors (sometimes different ones in metastases)
Permanent activation of growth factor receptor
Increased levels of growth factor receptor
Abnormal signal transduction (eg active RAS)
Abnormal behavior = advances cells through the cell cycle
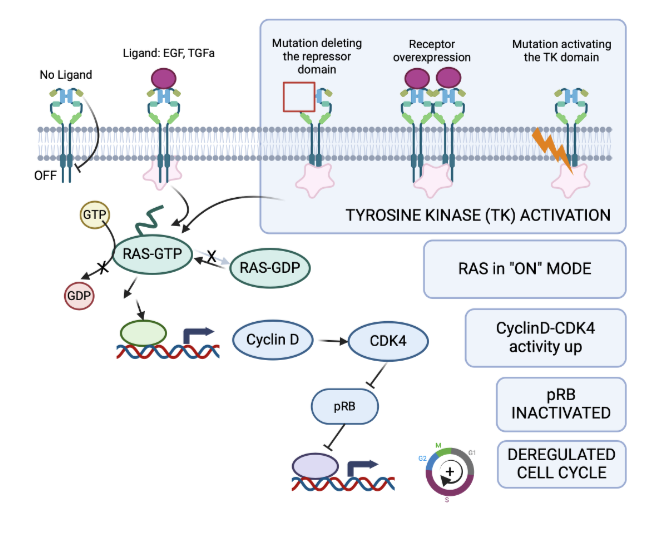
Growth factor signalling pathway
growth factor receptor binds to the receptor (EGF, TGFa ) to EGFR which induces receptor dimerization, tyrosine kinase activation, phosphyrylation of the receptor
some oncogenic muatation (partial gene deletion, point mutation) lead to constitutive activation of the tyrosine kinase receptor signalling, even in the absence of growth factor
Ras pathway
Ras is an intracellular protein that is central to integrating GFR signals
GRF activation → Ras binds GTP and recruits and activates signalling protein (eg Raf, PI3K)
Signal stops when Ras convert GTP to GDP+Pi via its GTPase activity
Cancers with activating mutant Ras have reduced GTPase activity → signalling is ON
Ras activates transcription of CCND1, cyclin D which activates CDK4 to stimulate cell cycle progression. Excess oncogenic cyclin D or CDK4 (by activating mutation, amplification) occur in some cancers)
Retinoblastoma protein (pRB)
The RB exists as two forms during the cell cycle
Weakly phosphorylated at G0 and G) - ACTIVE - halt cell cycle by suppressing transcription factor signalling
Strongly phosphorylated (ppRb) during rest of the cell cycle - INACTIVE protein, therefore cell cycle progresses
Inactivating phosphorylation of pRB is by (oncogene) CyclinD- CKD4)
Loss of pRB (eg by gene deletion/ inactivating mutation/ methylation) leads to uncontrolled cell cycle
By skipping the cell cycle checkpoints, mutation can be passed onto daughter cells