Graph Algorithms
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
27 Terms
Explain the basic graph concepts: path, cycle, cyclic/acyclic graph
Path
A path from v1 to vn is a sequencev1 v2 ... vn
such that every next vertex is reachable by an edge.
Example:
Edges: a -> b, b -> c
Path: a b c
A simple path means no vertex repeats.
Cycle
A cycle is a simple path that comes back to the start.
Example:a -> b -> c -> a
Cyclic / Acyclic graph
Cyclic: has at least one cycle
Acyclic: no cycles
What is strongly connected graph, strongly connected component (SCC),
For every pair u , v, there is:
a path from
utovand a path from
vtou
Example:a -> b -> c -> a
All nodes reach each other → strongly connected
Connected (directed)
Ignore direction by adding reversed edges.
If the result is strongly connected → graph is connected.
Reversed edges set:ET = {(v, u) | (u, v) in E}
Strongly Connected Component (SCC)
A maximal set of vertices where:
every vertex reaches every other
Think: “islands” of strong connectivity.
What are the basic operations a graph data structure should support?
successor = neighbour
If v = a (Vertex is a) and edges are a -> b, a -> c
Then aE = {b, c}
So aE contains only vertices you can reach in ONE edge from a.
What are adjacency lists and adjacency matrices, and when should each be used?
Adjacency list
For each vertex v, store a list of successors vE.
Example:
0: [1, 3]
1: [2]
2: []
3: [2]
Adjacency matrix
A 2D table where entry (u, v) is:
trueif edge existsfalseotherwise
Example:
0 1 2
0 [ 0 1 0 ]
1 [ 0 0 1 ]
2 [ 0 0 0 ]Adjacency list (usually better)
Memory efficient
Fast iteration over neighbors
Best for sparse graphs
Adjacency matrix preferred if
Graph is dense
(|E|close to|V|^2)Edge existence tests are very frequent
Performance difference doesn’t matter
How does edge testing and edge iteration work using an adjacency matrix?
Data Structure:
The graph stores:
number of vertices
nmatrix
adj_matrix[u][v]
Edge test
Check directly: adj_matrix[u][v] = true
means edge (u, v) exists
Time: O(1)
Iterate over all edges: List all edges (u, v) that actually exist in the graph.
Because every edge is stored as one cell in the matrix, we must:
Look at every row
uLook at every column
vCheck whether
adj[u][v]istrueIf yes →
(u, v)is an edge
How does edge testing and edge iteration work using an adjacency list?
For each vertex u:
store a list
adj_list[u]of neighbors.with its size
Example:
0: [1, 3]
1: [2]
2: []
3: []Edge test
Check if v is inside adj_list[u]:
adj_list[u].contains(v)
Time:
worst case:
O(deg(u))can be improved with binary search or hash table
Iterate over all edges
Steps:
For each
uFor each
vinadj_list[u]Output
(u, v)
Total time:O(|V| + |E|) -> is faster
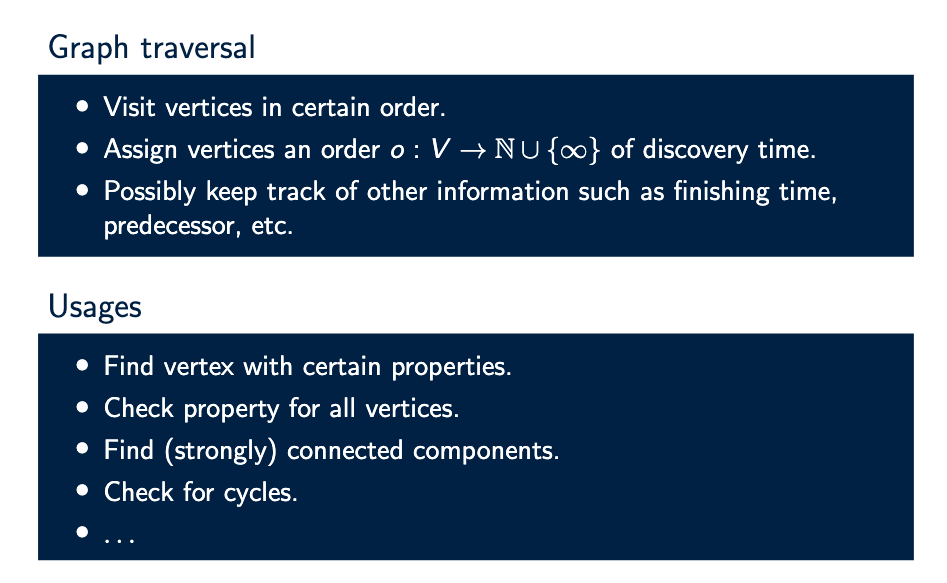
What is graph traversal, and what is it used for?
What is the basic idea of Depth First Search (DFS)?
Depth First Search (DFS) explores the graph:
as deep as possible before backtracking
Key idea:
use a stack to store vertices that still need exploration
Steps (high level):
Start from a vertex
Go to one of its successors
Keep going deeper
If no unvisited successor exists → go back
This is why it’s called depth-first.
DFS algorithm steps
We explain DFS using:
a stack
Sa discovery order
o(v)
Initially:
for all v:
o(v) = ∞
Step 1 — Start
Choose a start vertex (e.g.
a)Push it onto the stack
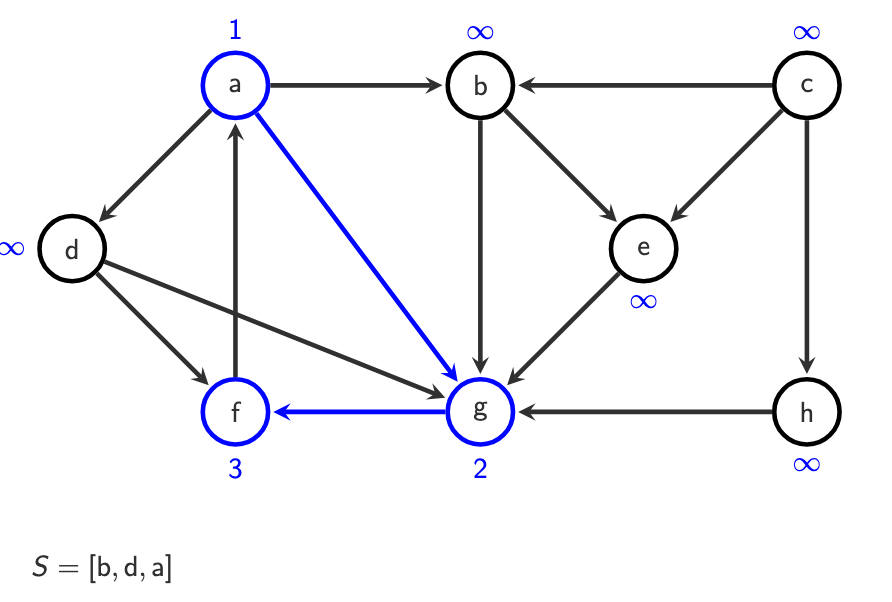
S = [a]
Step 2 — Pop & discover
Pop top of stack
If not visited (
o(v) = ∞), assign next number
Example:
o(a) = 1
Step 3 — Push successors
Push all successors of
aonto the stack
Example:
a -> b, d, g
S = [b, d, g]
(Exact order depends on implementation)
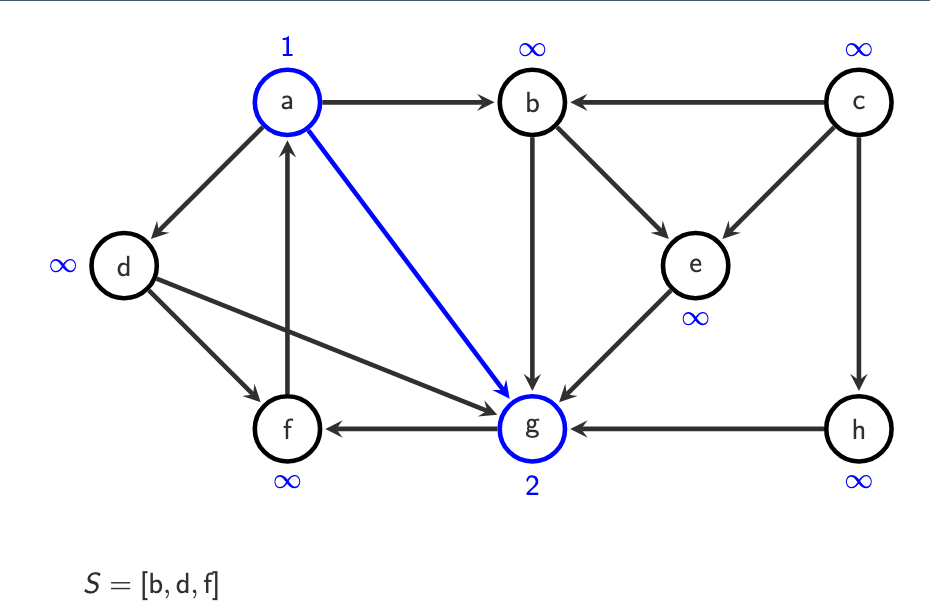
Step 4 — Repeat
Repeat while stack is not empty:
Pop a vertex
vIf
o(v) = ∞, assign next number &Push all successors of
v
Key observation (VERY IMPORTANT)
A vertex can be pushed multiple times
It is only processed once
The check
o(v) = ∞prevents revisiting
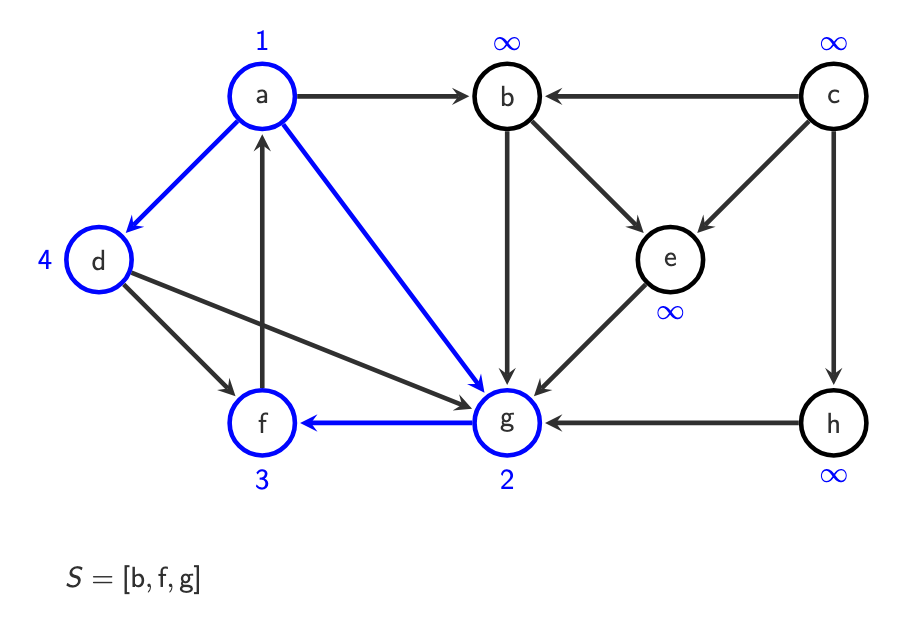
When DFS ends
DFS finishes when
S = []
& only after we tried every vertex in V and its oo(v) ≠ ∞
DFS procedure
Main loop
For each vertex v:
if
o(v) = ∞, start DFS fromv
DFSExplore procedure
push v onto S
while S not empty:
v = pop S
if o(v) = ∞:
o(v) = i
i = i + 1
for each u in vE:
push u onto S
Running time
Each vertex and edge is processed at most once:
O(|V| + |E|)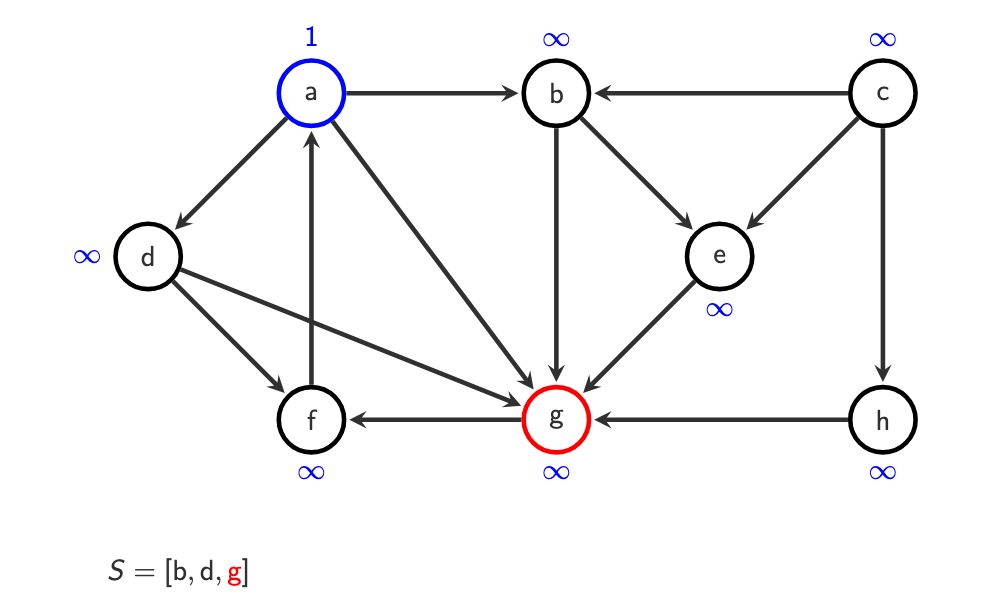
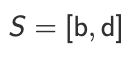
…
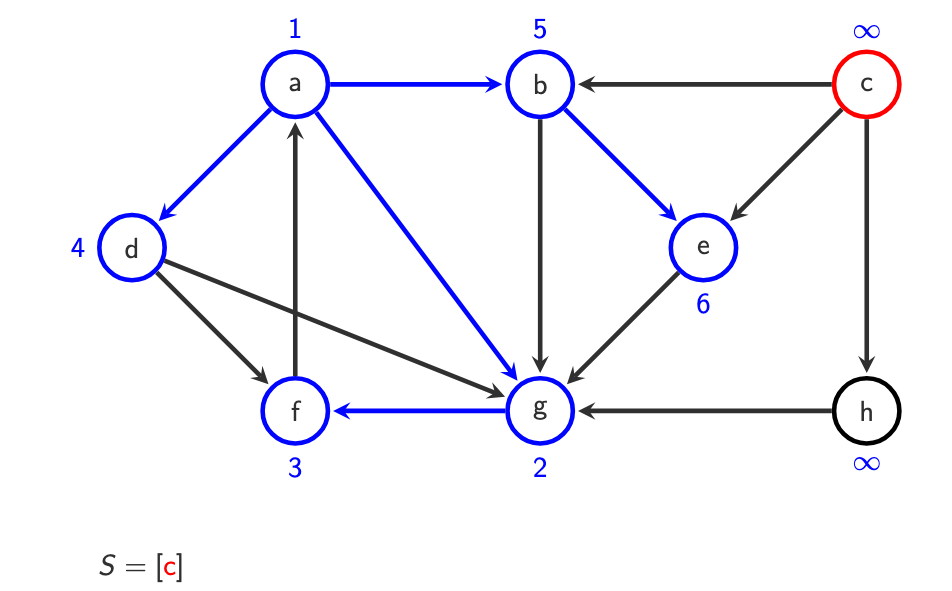
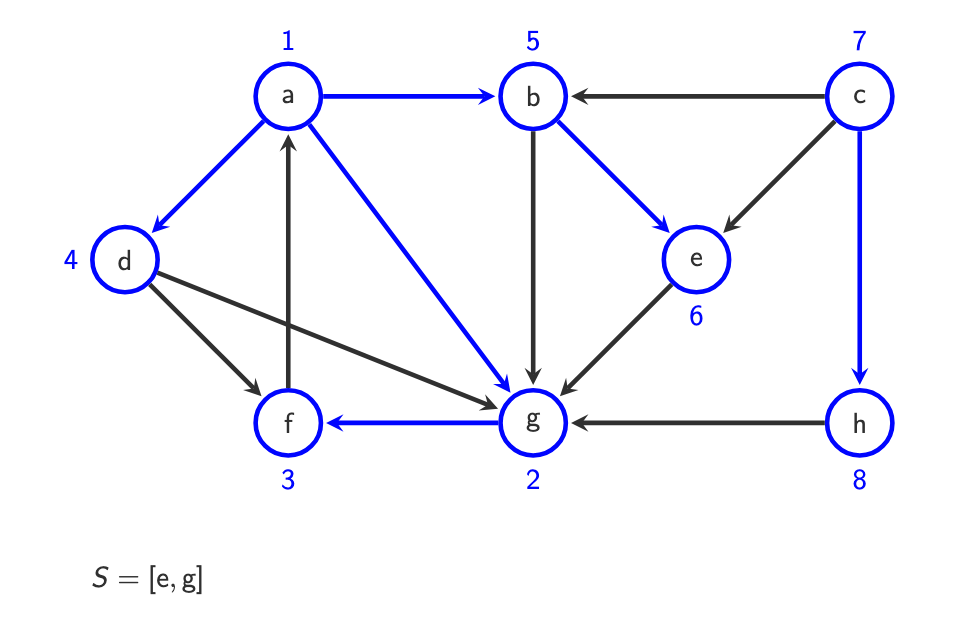
S= [] finished and all vertices visited.
How can DFS be used to detect cycles?
DFS detects cycles by looking for back edges.
A back edge is an edge to a vertex that is already on the current DFS path. (in the stack, waiting)
If such an edge exists, the graph contains a cycle.
In undirected graphs, the edge to the predecessor is ignored.
What is the basic idea of Breadt First Search (BFS)?
Breadth First Search (BFS) explores the graph:
level by level, starting from a source vertex.
Key difference to DFS:
DFS → stack → goes deep
BFS → queue → stays shallow first
So BFS visits:
all vertices at distance 1
then distance 2
then distance 3
etc.
Start from vertex a.
Step 1 — start
S = [a]
o(a) = 1Step 2 — visit neighbors of a
From a, we discover:
b, d, g
They are:
marked as discovered
put into the queue
S = [b, d, g]
Step 3 — process queue in order
Important:
they are all distance 1 from
a
Now BFS continues:
dequeue
benqueue undiscovered neighbors of
b
Then:
dequeue
dthen
gthen their neighbors
So the queue ensures:
vertices discovered earlier are processed first
This guarantees level-by-level traversal.
When a vertex u is discovered:
o(u) = 0
This marks it as:
discovered
but not fully processed yet
BFS procedure
Main procedure
for each v:
o(v) = ∞
S = empty queue
i = 1
for each v:
if o(v) = ∞:
BFSExplore(v)
BFSExplore procedure
enqueue v into S
while S not empty:
v = dequeue S
o(v) = i
i = i + 1
for each u in vE:
if o(u) = ∞:
o(u) = 0
enqueue u into S
Running time
Each vertex and edge is handled a constant number of times:
O(|V| + |E|)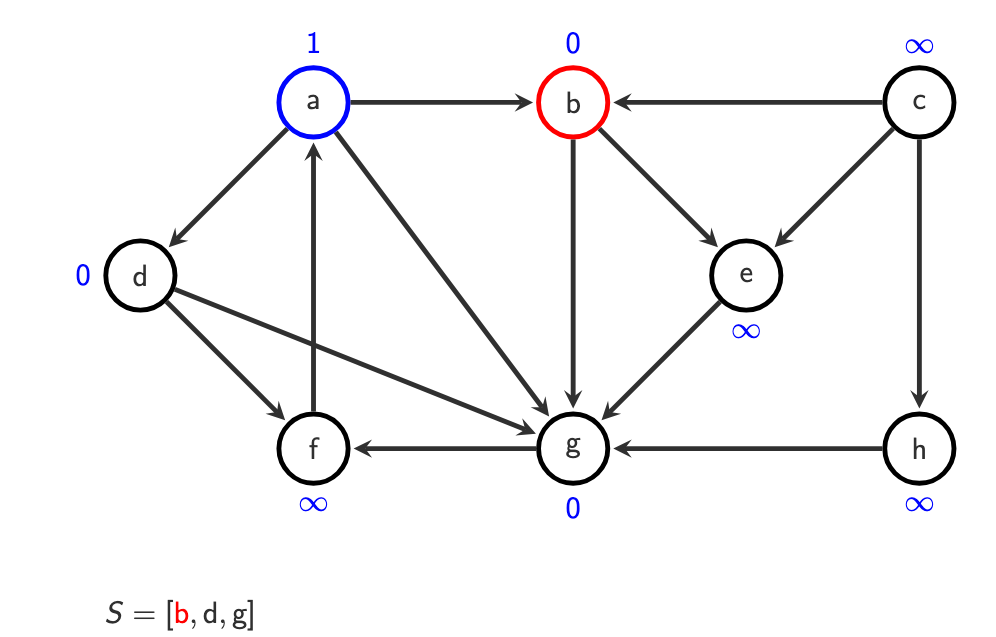
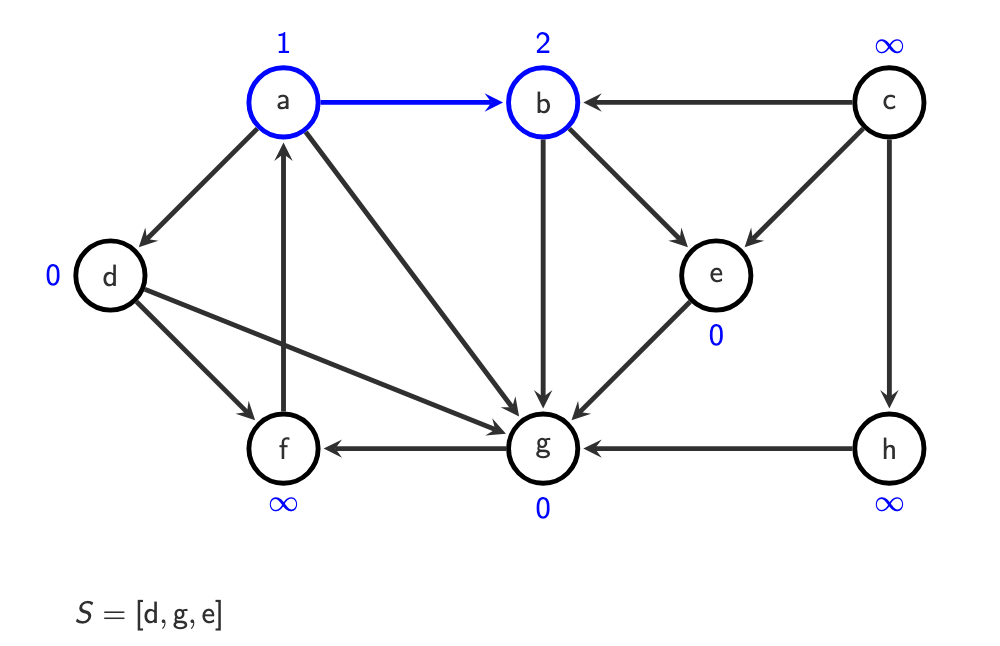
g is visited, because it’s 0 we do with g afterwards:
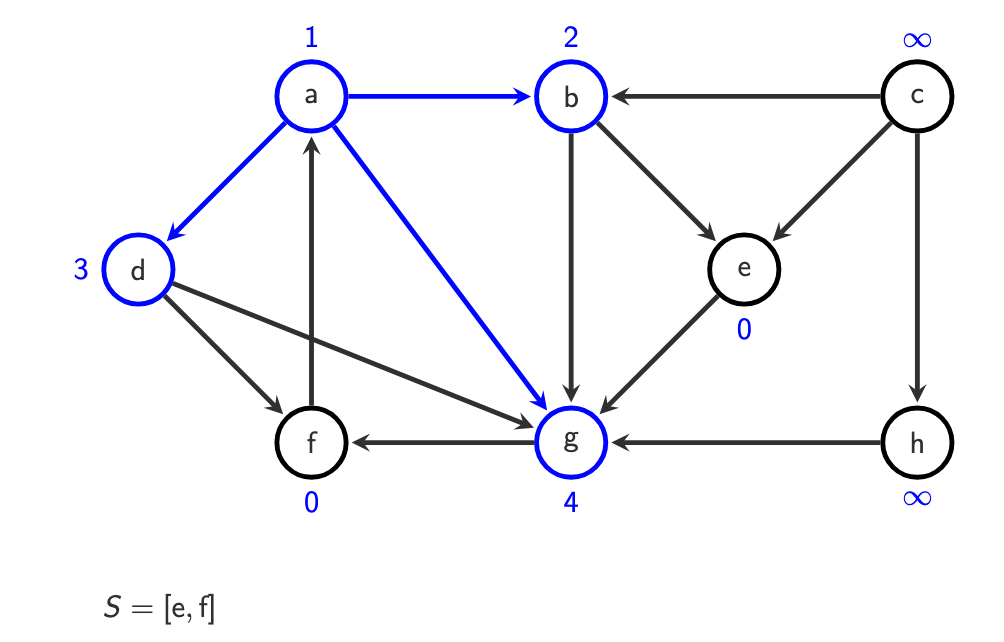
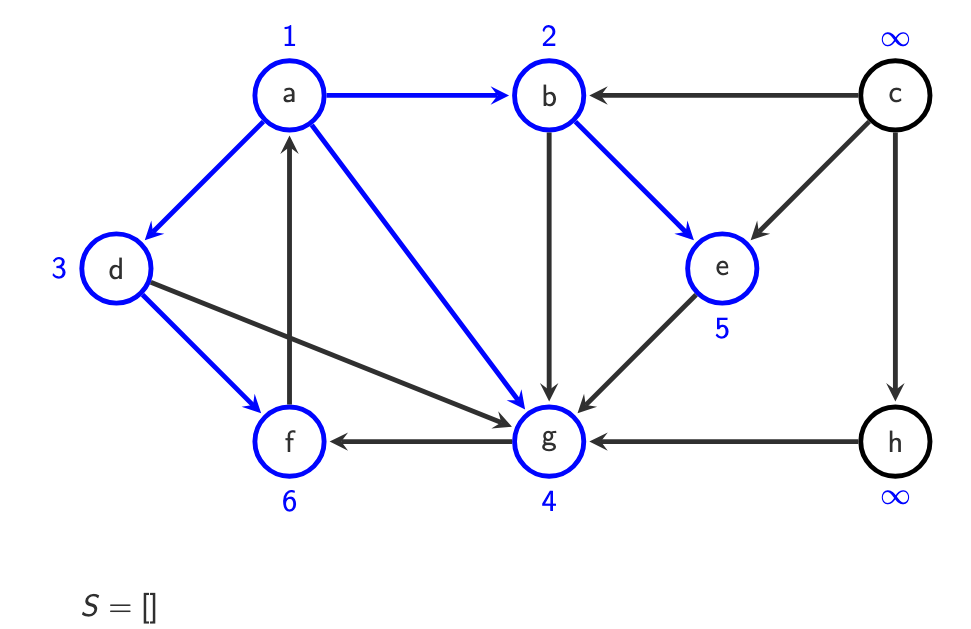
do with c afterwards, it has infinity. Its neighbour is h, we put it in.
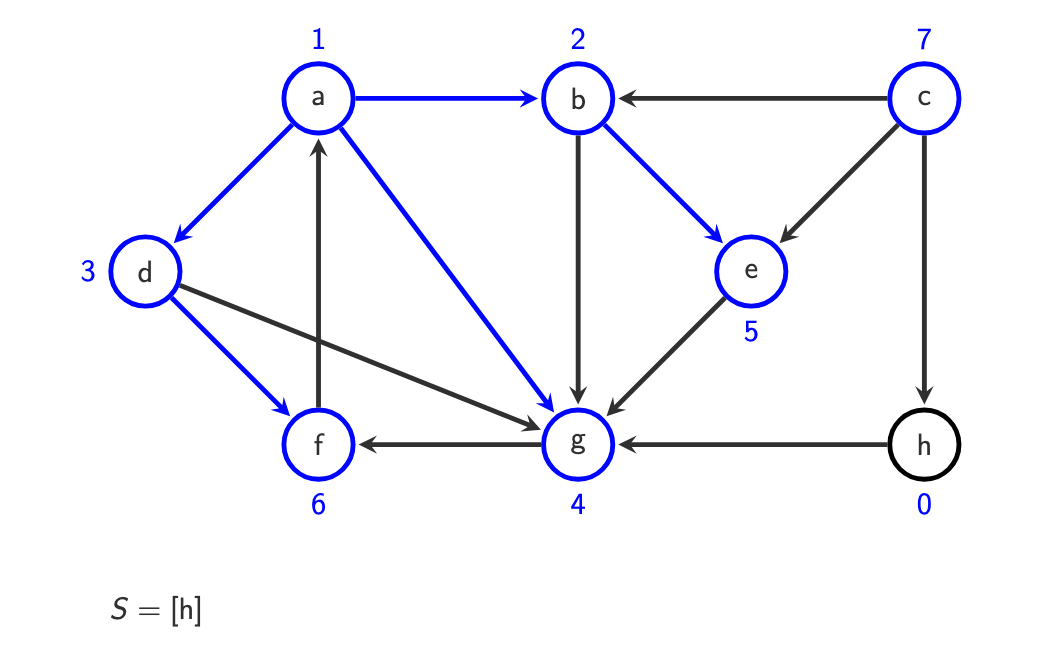
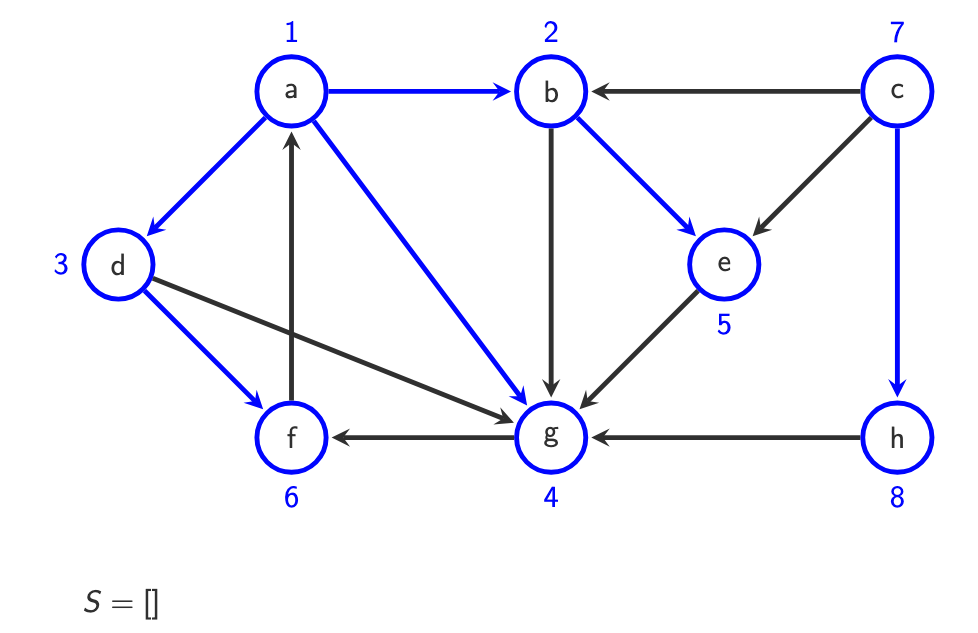
Why is BFS suitable for finding shortest paths in unweighted graphs? BFS limitiation
Key observation:
The queue in BFS is automatically ordered by distance from the source.
Because:
vertices at distance
kare discovered beforevertices at distance
k+1
reconstruct shortest paths by storing predecessors
BFS works for:
unweighted graphs
or graphs where all edges have equal weight
For weighted graphs with non-negative weights:
BFS generalizes to Dijkstra’s algorithm
which uses a priority queue instead of a FIFO queue
What is a topological order, and when does it exist?
A topological order is an assignment of numbers to vertices:
o : V -> N
such that for every directed edge: (u, v) in E
we have:
o(u) < o(v)
Meaning:
umust come beforevin the order
A topological order exists if and only if the graph is a DAG (directed acyclic Graph)
Important notes
A topological order is not unique
Multiple valid orders may exist
What are typical applications of topological sorting?
Topological sorting is used when there are dependencies.
Typical examples:
task scheduling
instruction ordering
resolving dependencies (e.g. build systems)
Detecting cycles.
Find shortest paths from a source in some weighted DAG in linear time.
Explain the idea of topological sort using predecessor counts.
For every vertex v, compute:
pre(v) = number of incoming edgesVertices with:
pre(v) = 0
Step 2 — Find vertices with zero predecessors
have no dependencies
can be placed first in the order
Put them into a queue or stack.
Step 3 — Remove them
Repeatedly:
Take a vertex
vwithpre(v) = 0Output
vin the topological order
For every edge v -> u
decrease:
pre(u) = pre(u) - 1
If
pre(u)becomes0, adduto the queue
Step 4 — Stop condition
If all vertices are output → success
If vertices remain with
pre(v) > 0→ cycle exists
Small example (intuitive)
Edges:
a -> b
a -> c
a -> d
b -> d
c -> b
Initial predecessor counts:
pre(a)=0
pre(b)=2
pre(c)=1
pre(d)=2
Start with: queue = [a]
[a]
pre(b)= 2 - 1 = 1
pre(c)= 1 - 1 = 0
pre(d)= 2 - 1 = 1add c to queue: [a, c]. continue with c. for c→b, pre(b) becomes 1 less
pre(b)= 1 - 1 = 0
pre(d)= 1continue with b. Add to queue: [a, c, b]. For b→d, pre(d) = 1-1= 0. add to queue [a, c, b, d]
Describe the topological sort algorithm and its running time.
Initialization
For all vertices:
o(v) = ∞
pre(v) = number of predecessors
Create:
S = empty queue
i = 1
Main loop
for each v:
if pre(v) = 0:
push v into S
Processing
while S not empty:
v = pop S
o(v) = i
i = i + 1
for each u in vE:
pre(u) = pre(u) - 1
if pre(u) = 0:
push u into S
Cycle detection
If after the algorithm:
exists v with o(v) = ∞
then:
graph has a cycle
Running time
O(|V| + |E|)How can topological sort also be implemented using DFS?
Alternative idea:
run DFS
record finishing times
Key rule:
A vertex is finished after all its successors are finished.
If we sort vertices by decreasing finishing time, we get: a topological order.
What is the SCC discovery problem, and what are the main ideas to solve it?
The SCC discovery problem is to find all strongly connected components of a directed graph.
It can be solved in linear time, for example using Tarjan’s algorithm or by running DFS twice: first to compute finishing times, then on the reversed graph in decreasing finishing time order, where each DFS tree forms one SCC.
What is a spanning tree and spanning subgraph and a minimum spanning tree (MST)
We start with an undirected graph:
G = (V, E)
Spanning subgraph
A subgraph is spanning if:
it contains all vertices
but possibly fewer edges
Spanning tree
A spanning tree is a spanning subgraph that:
is connected
has no cycles
So it connects all vertices using the minimum number of edges.
An MST is a spanning tree with minimum total weight among all spanning trees.
What are important properties of spanning trees and minimum spanning trees?
1⃣ Existence
A spanning tree exists only if the graph is connected
If not connected → each connected component has its own spanning tree
2⃣ Number of edges
All spanning trees of a graph have the same number of edges: |V| - 1
3⃣ Negative weights
Negative edge weights are not a problem:
you can add a constant to all weights
the MST does not change
4⃣ Maximum spanning tree
A maximum spanning tree can be found by:
w'(e) = -w(e) and then computing an MST with w'
Explain Kruskal’s algorithm for computing a minimum spanning tree.
Kruskal’s algorithm builds the MST by adding edges in increasing weight order, while avoiding cycles.
Step 1 — Sort edges
Sort all edges by increasing weight:
L = edges sorted by w(e)
Step 2 — Initialize
Start with empty edge set:
S = {}
Initialize a Union-Find structure over vertices:
U
Union-Find keeps track of connected components.
Step 3 — Process edges
For each edge (u, v) in sorted order:
Check components:
U.find(u) != U.find(v)
If true:
add edge to MST:
S = S ∪ {(u, v)}merge components:
U.union(u, v)If false:
skip edge (it would create a cycle)
Step 4 — End condition
If all vertices are connected → MST found
Otherwise → graph was not connected (you get a minimum spanning forest)
Kruskal’s algorithm sorts edges by weight and repeatedly adds the cheapest edge that connects two different components
What is the running time of Kruskal’s algorithm and why?
1⃣ Sorting edges
Sorting |E| edges:
O(|E| log |E|)2⃣ Union-Find operations
Each edge causes at most:
2
findoperationssome
unionoperations
With efficient Union-Find:
O(|E| α(|V|)) where:
αis the inverse Ackermann functiongrows extremely slowly (almost constant)
Final running time
Dominated by sorting:
O(|E| log |E|)What is the basic idea of Prim’s algorithm for computing a minimum spanning tree?
Prim’s algorithm builds the MST by:
starting from one vertex and repeatedly adding the cheapest edge that connects the current tree to a new vertex.
So unlike Kruskal:
Kruskal grows many components and merges them
Prim grows one tree, step by step
Key intuition
Start with a single vertex
Always extend the tree using the minimum-weight outgoing edge
Never create cycles, because you only connect to new vertices
Prim’s algorithm how it works? Running time? How different from kruskal?
Prim’s algorithm computes a minimum spanning tree by growing one tree starting from an arbitrary vertex.
For each vertex v, it stores the cheapest edge weight c(v) that connects v to the current tree, and a predecessor pre(v).
A priority queue is used to always select the vertex with minimum c(v).
When a vertex is chosen, it is added to the tree, and the costs of its neighbors are updated if a cheaper connecting edge is found.
This process continues until all vertices are included. O(E + VLogV)
Prim looks asymptotically better
• However, in practice, usually Kruskal is faster, unless the graph is
large and dense