Issues in DNA Replication and DNA Repair Mechanisms
1/15
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
16 Terms
3 Issues in DNA Replication
1) What happens when DNA is unwound?
2) What happens at the ends of eukaryotic linear chromosomes during replication?
3) How are mistakes found and corrected?
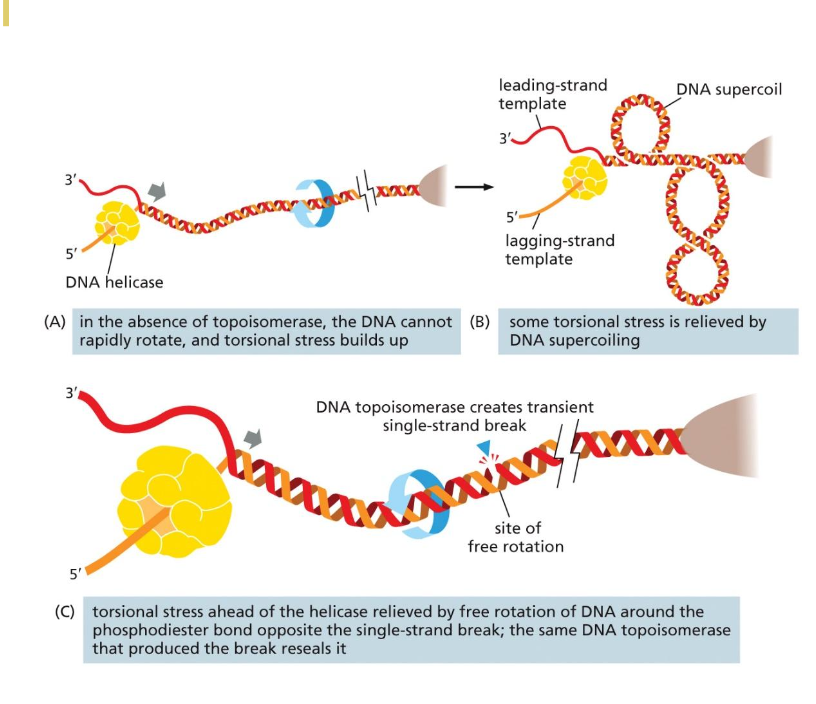
The unwinding problem
Why does it happen
Where is this problem more evident
What prevent tension? Does it work forever?
Main solution - what enzyme is involved
Supercoiling and torsional strain increases during unwinding process
Even though the DNA wants to unwind to relieve tension during unwinding, the DNA strand is very long and proteins involved that it is hard to unwind
Issue in circular chromosome and larger linear eukaryotic chromosomes
Solution can be supercoiling; however, there can only so much you can supercoil
Main solution
DNA topoisomerase: many types
Will cut the DNA, let the DNA unwind, and then reseals it
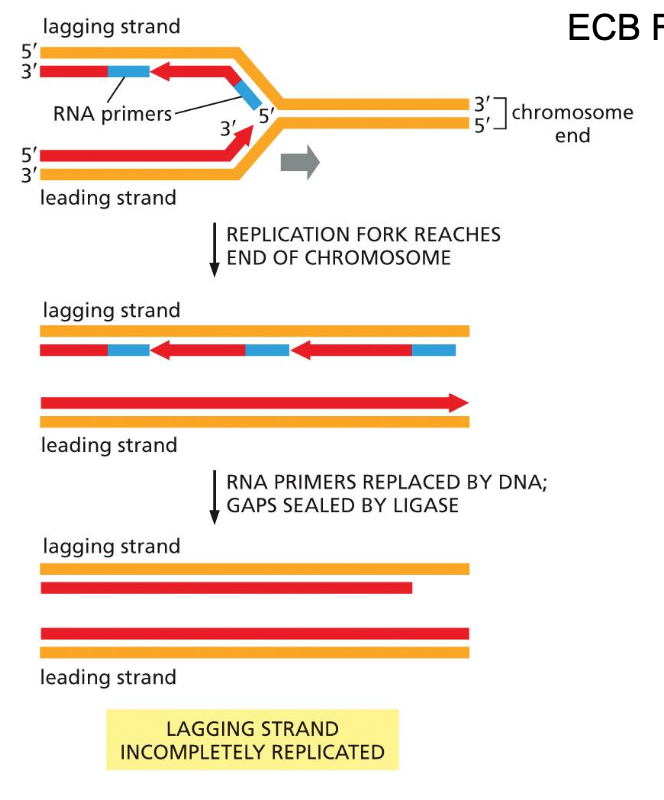
Issue at the end of linear chromosomes
which strand is this a major problem; why isn’t there an issue for the other leading strand?
Why is shortening of the end of the daughter DNA a problem?
Issue at the lagging strands
In the lagging strand
1) Primase is not very good at putting a primer at the very end
2) The RNA primer gets removed, now you have a 5’ end and you can add on to the 5’ end
Results: a shorter replicated strand
No issue for the leading strand
Prof notes
Look at the slide from 3.. What is the direction of DNA replication
The leading strand primer is removed to fill it in you add on to the 3’ end of the lagging strand of the other replication fork
Problem: loss of sequence information
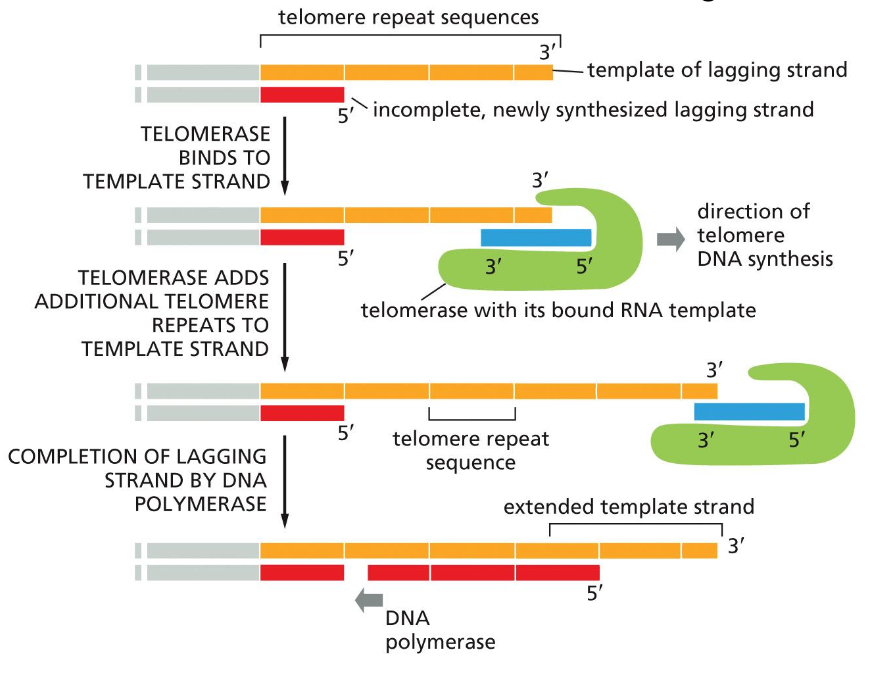
Solution to the issue regarding the end of the linear chromosome - Telomerase
The repetitive sequence that is added to the 3’ end of the parental strand (i.e. the lagging strand template) is determined by the RNA template to make a DNA complement in telomerase
RNA template —> DNA com
Telomere replication process
what does it not require
1) Telomerase has a bound RNA template that is repetitive for the lagging strand template
2) Resembles:
Reverse Transcriptase (reverse transcription)
3) Generates: G-rich ends (RNA template will have a lot of C’s)
4) telomere adds nucleotides to: 3’ ends of parental strand template during DNA synthesis
For the regular process:
the DNA synthesis for the lagging strand will
the removal of the primer results in the lose of repetitive DNA sequence from the RNA template of the telomerase during ligase process
Telomeres and Cancer
•Telomerase abundant in stem and germ-line cells, but not in somatic cells
•Loss of telomeres, which occurs normally during DNA replication limits the number of rounds of cell division
•Most cancer cells produce high levels of telomerase
How are mistakes found and corrected?
With a lot of DNA replicated, there will be a mistake eventually
Repair
If it doesn’t occur, then later on there is no way to detect there is an issue - a permanent DNA mutation
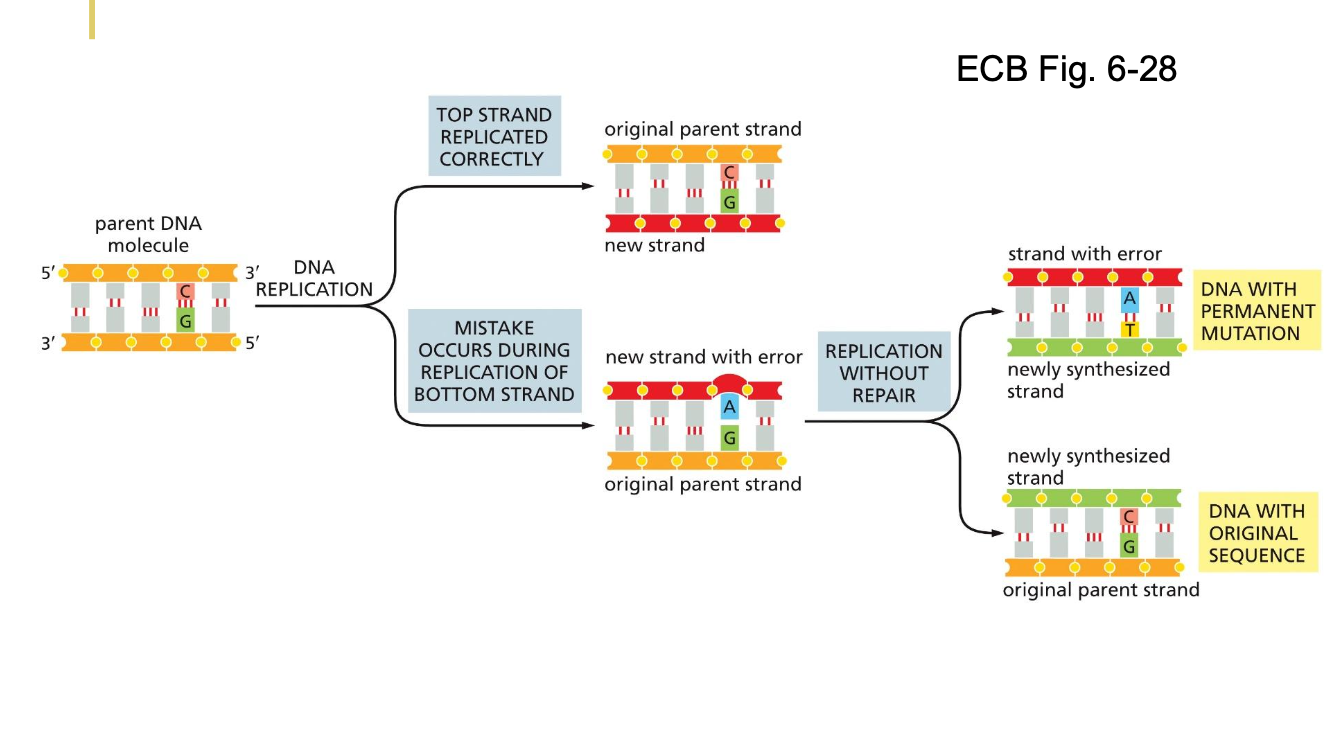
THE HIGH FIDELITY OF DNA REPLICATION
RNA polymerases have an error rate of 1 in 10^4
However, DNA polymerases have an error 1 in 10^9
For the genome, error is 3 nucleotides every time a cell divides
DNA proofreading and repair
2 mechanisms - list only no def
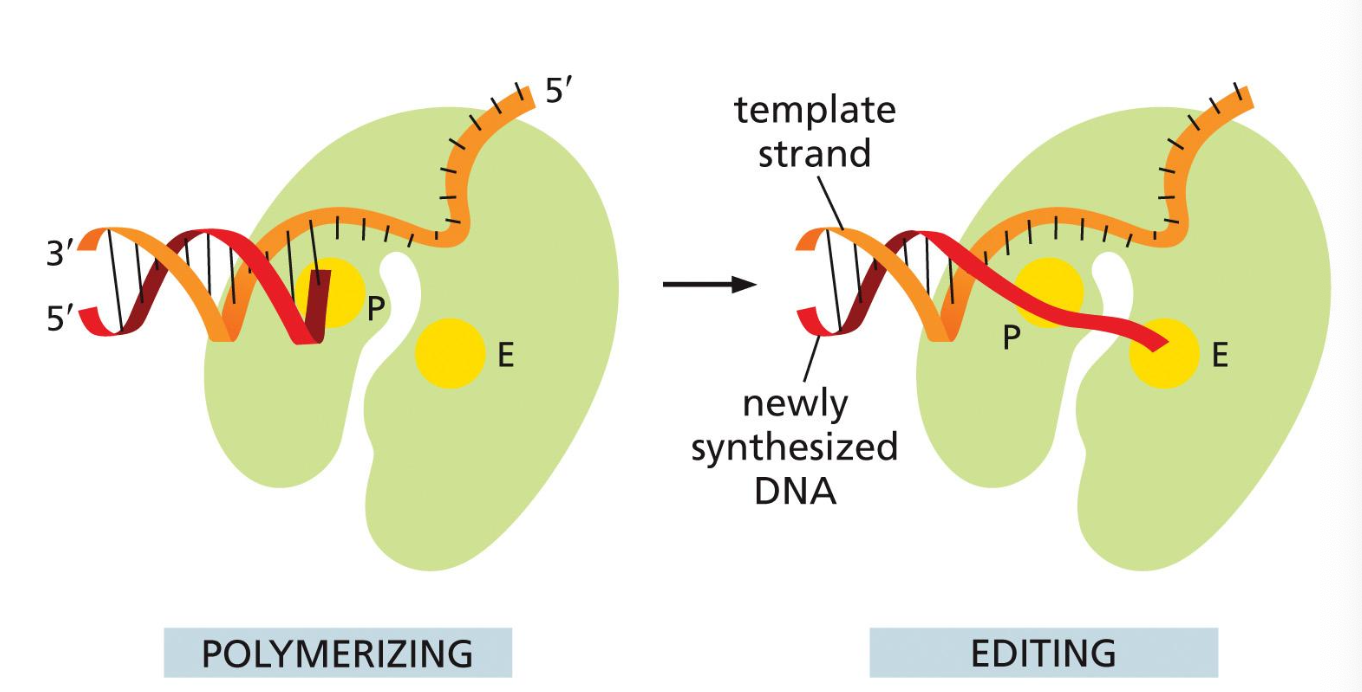
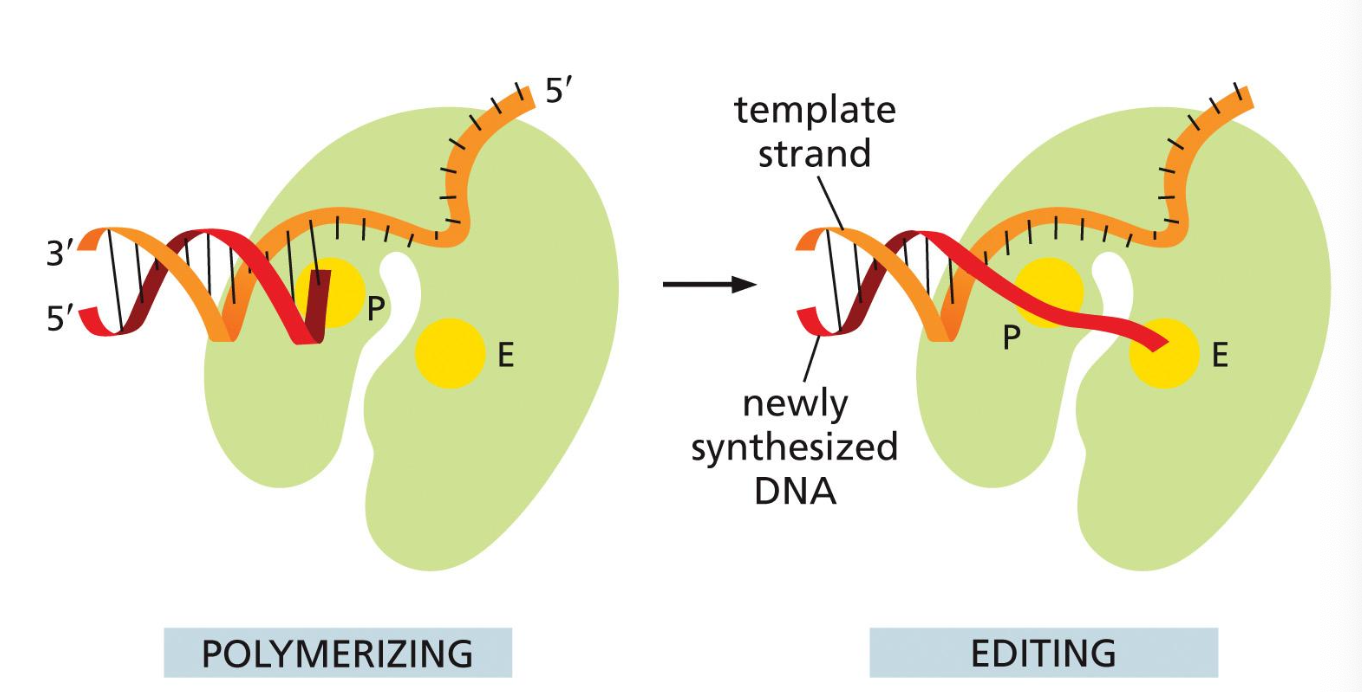
1) 3’ to 5’ exonuclease
It removes the misincorporated nucleotide
The DNA polymerase have exonuclease activity
“chop off a nucleotide at the end immediately“
3’ to 5’ backwards reading
“backspace button or 3’ to 5’“
DNA polymerase has two active sites
Polymerizing site
Editing/Exonuclease site
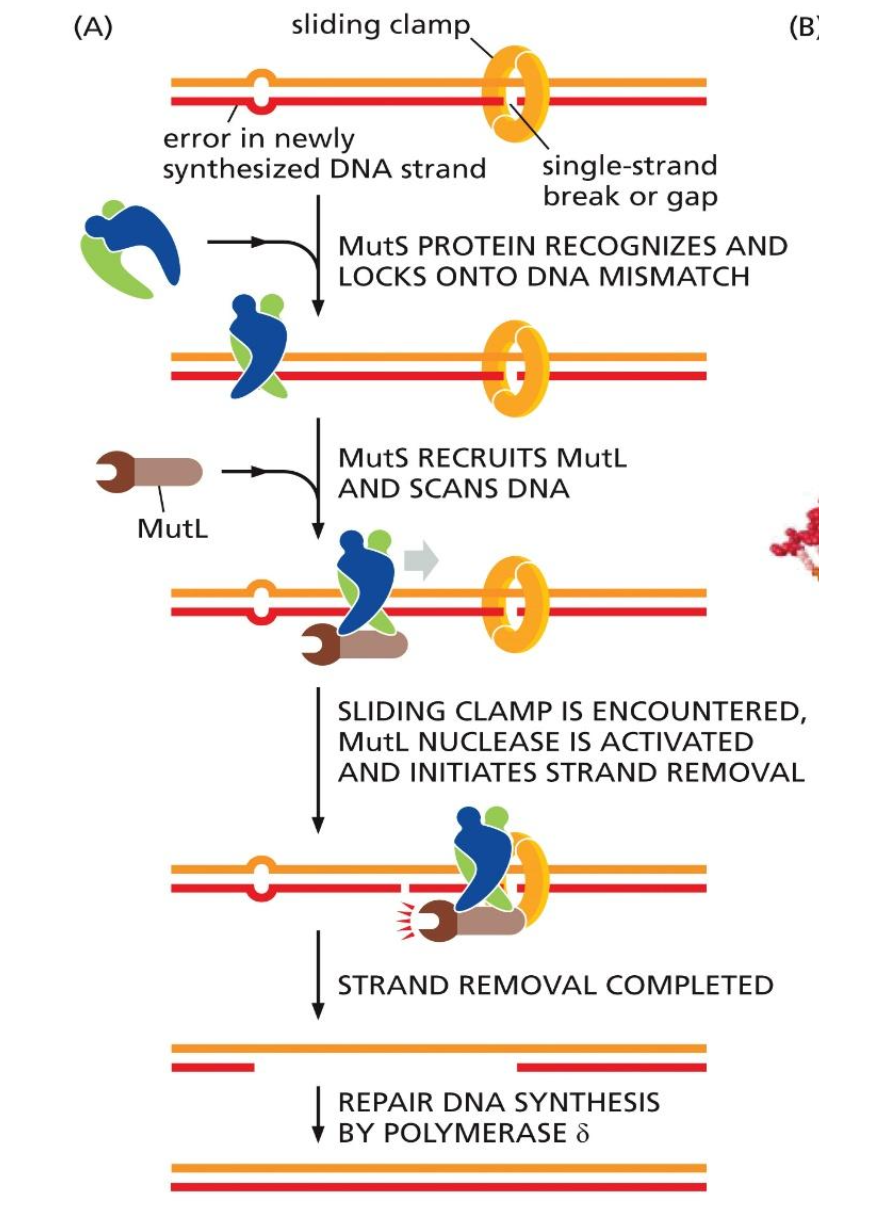
Strand-directed mismatch repair in eukaryotes
what about leading strand with no nicks?
2) Strand-direct mismatch repair
occurs when the proofreading fails “backspace fails”
MutS protein recognizes and sticks to the distortion in the geometry of the double helix
DNA ligase does not seal it, so MutL see the nick and knows its the synthesized strand
MutL removes the DNA strand
DNA synthesizes the gap during ligase process
Different between eukaryotes and prokaryotes
Difference is un-methylated adenines
Leading strand
Nick is on the other side of the replication fork
DNA Damage
why does it happen
What does it cause
why factor can damage DNA
Even after synthesis, DNA can get damaged and need repair
defects in repair mechanisms leads to human diseases
e.g. breat, colon, skin cancers
Factors
UV: causes pyrimidine dimers; two pryimidine dimers causes a covalent bonding when beside each other
Causes/Factors
**finish notes: Oxidation
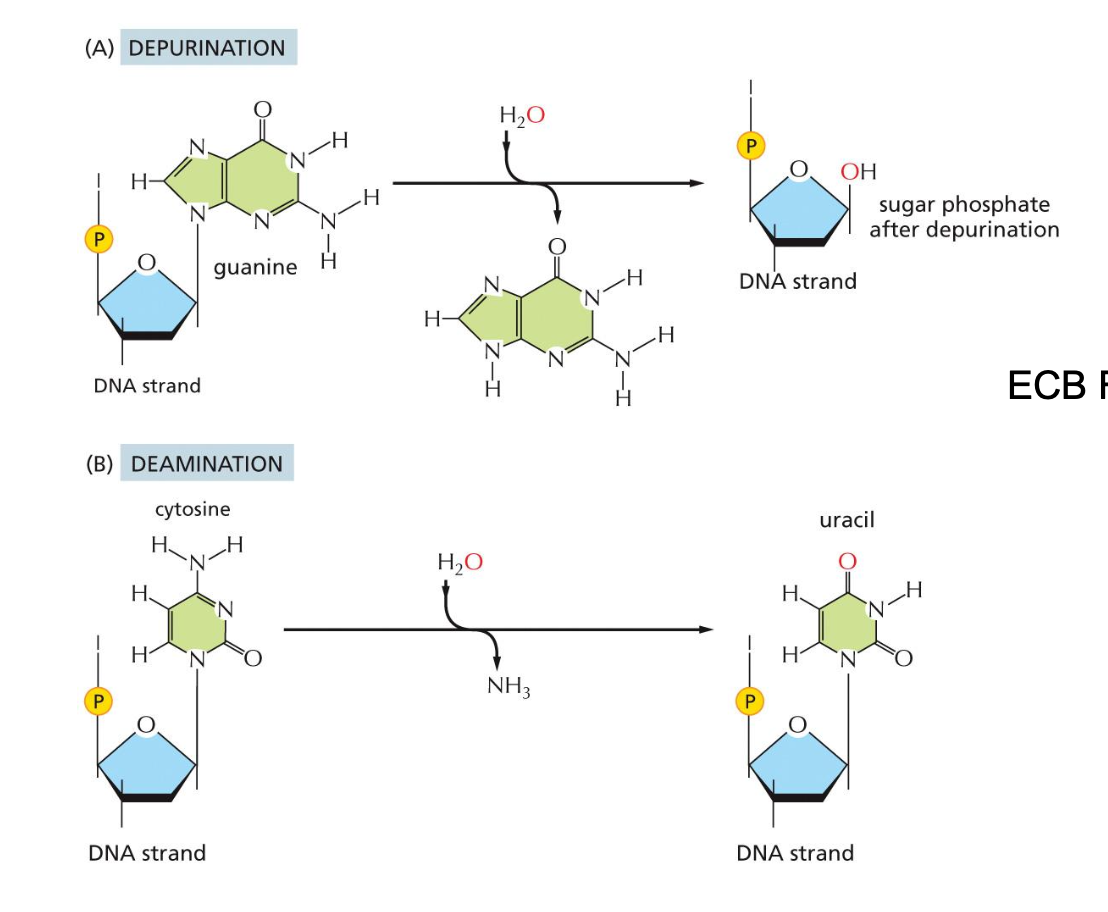
Spontaneous Damage to DNA can Also occur
provide example with water and guanine, another example with cytosine
2 Types of spontaneous damage
Depurination
loss of base, only sugar and phosphate remains
Could be adenine or guanine
Could be completely removed because they do not know what it is
Deanimation
Unique specifically for cytosine
amine lost results in the creation of uracil
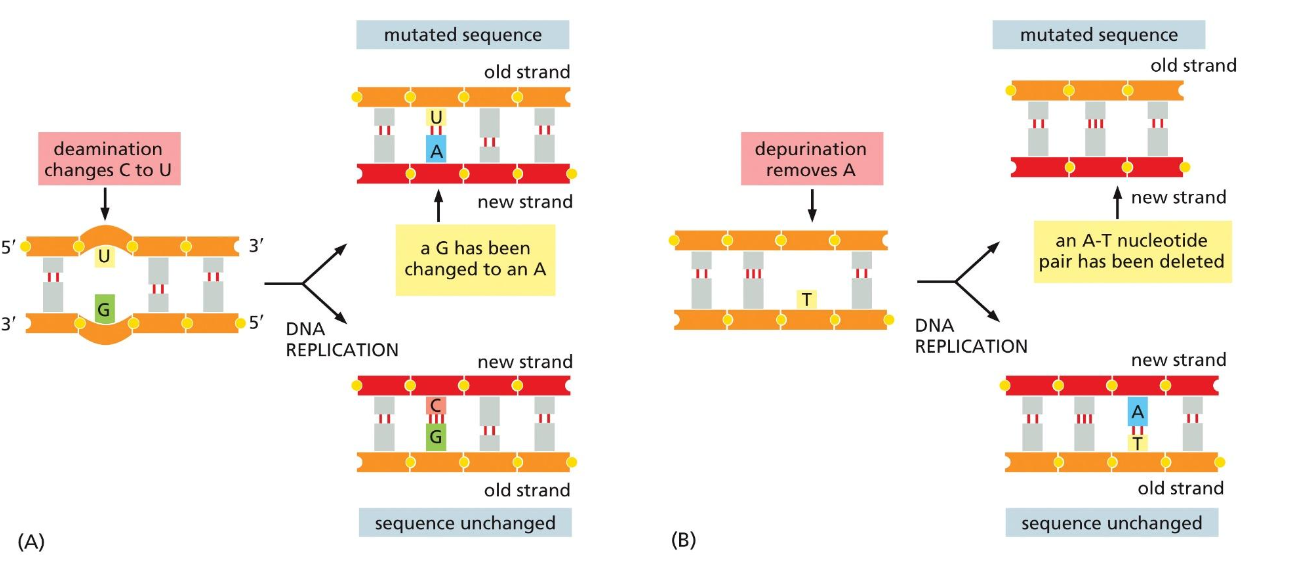
How mutations arise with spontaneous damage
if the deamination does not get fixed, it creates a permanent mutation
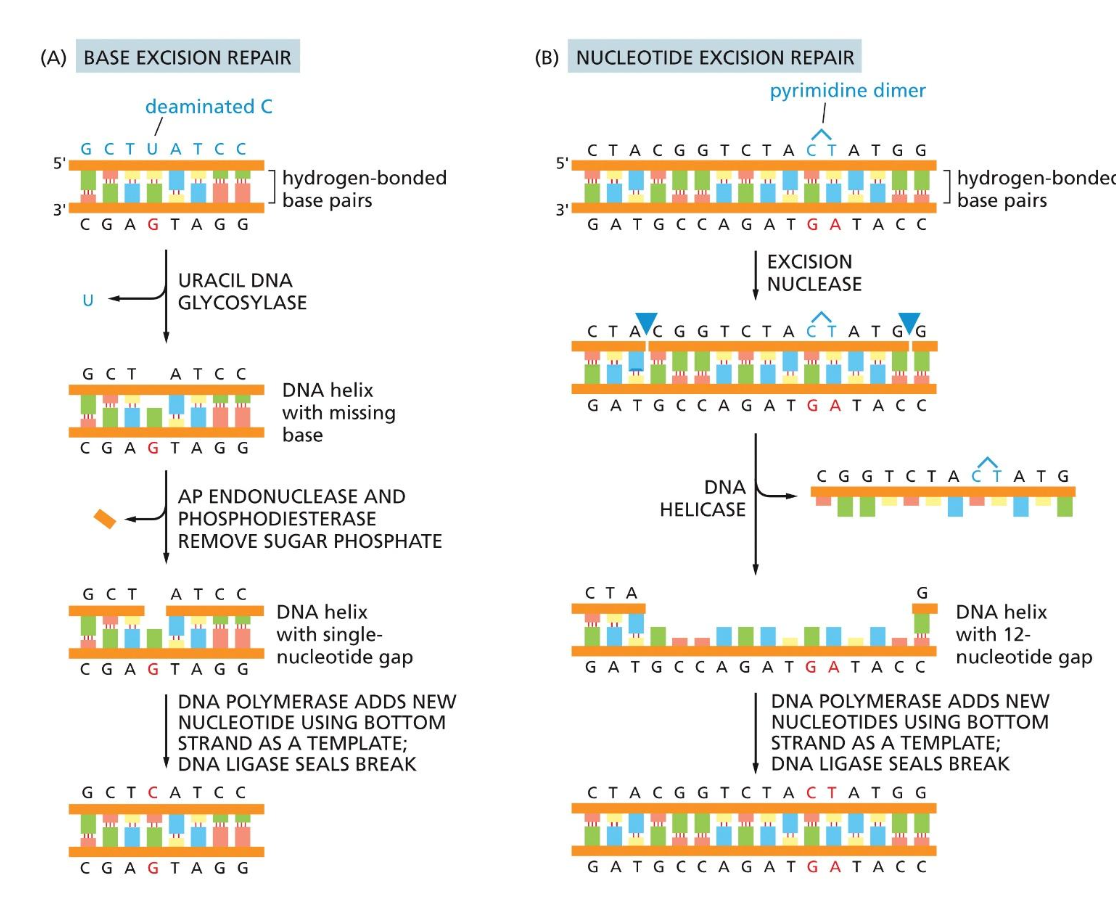
DNA repair
2 kinds of mechanisms
1) Base excision repair
Fixes at one nucleotide at a time
2) Nucleotide excision repair
Fixes a couple at a time
Perfect for pyrimidine dimers
Excision nuclease comes in and creates a large cut
DNA helicase comes in
DNA ligase
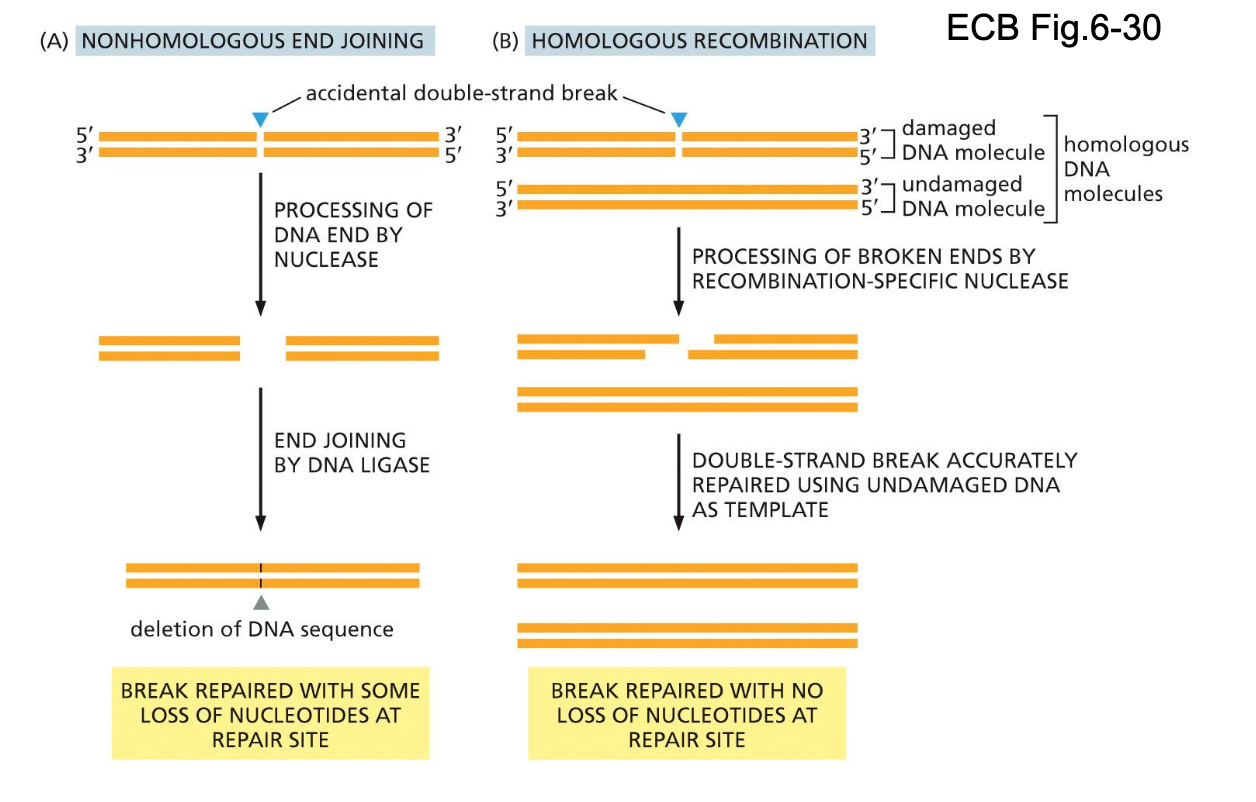
DNA repair of double-stranded breaks
1) Nonhomologous end joining
quickly seal together and get going
Quick + dirty
Prevent the wrong number chromosomes
2) Homologous recombination
using the information from the pair to correct
Slow but accurate