amino acids to proteins
1/227
Earn XP
Description and Tags
macromolecules pt. II
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
228 Terms
What are proteinogenic amino acids?
The 20 amino acids that create proteins in all organisms.
What are the three parts of proteinogenic amino acids?
An amino group, a carboxyl group, and a side chain (R group).
What are alpha-amino acids?
Amino acids with a central carbon directly adjacent to the carboxylic acid.

What is the stereochemistry designation of a-amino acids?
All except glycine are chiral (L) and all are S except cysteine which is designated as R due to its sulfur atom.
What are the classification categories for a-amino acids?
nonpolar: unable to hydrogen bond/dipole-dipole interact with water due to carbon and hydrogen having similar electronegativities
found in the hydrophobic environment
broken down into nonpolar aliphatic, branched alkyl chain, and nonpolar aromatic
polar: able to hydrogen bond with water due to oxygen and nitrogen (significant electronegativity difference)
found outside interacting with water
broken down into polar neutral, negatively charged (acidic) and positively charged (basic)
Name the nonpolar aliphatic amino acids.
Alanine, valine, isoleucine, leucine, methionine, glycine, proline.
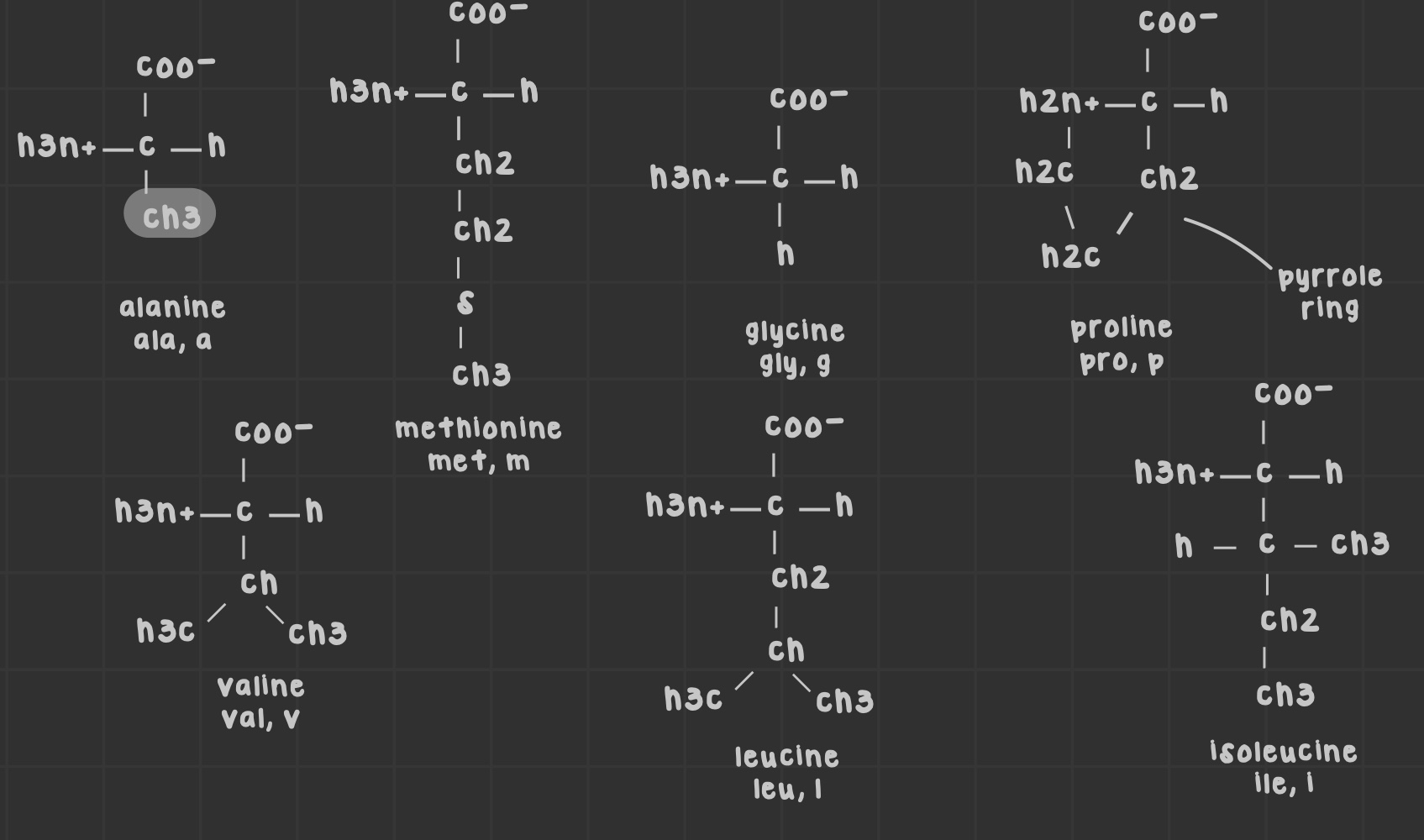
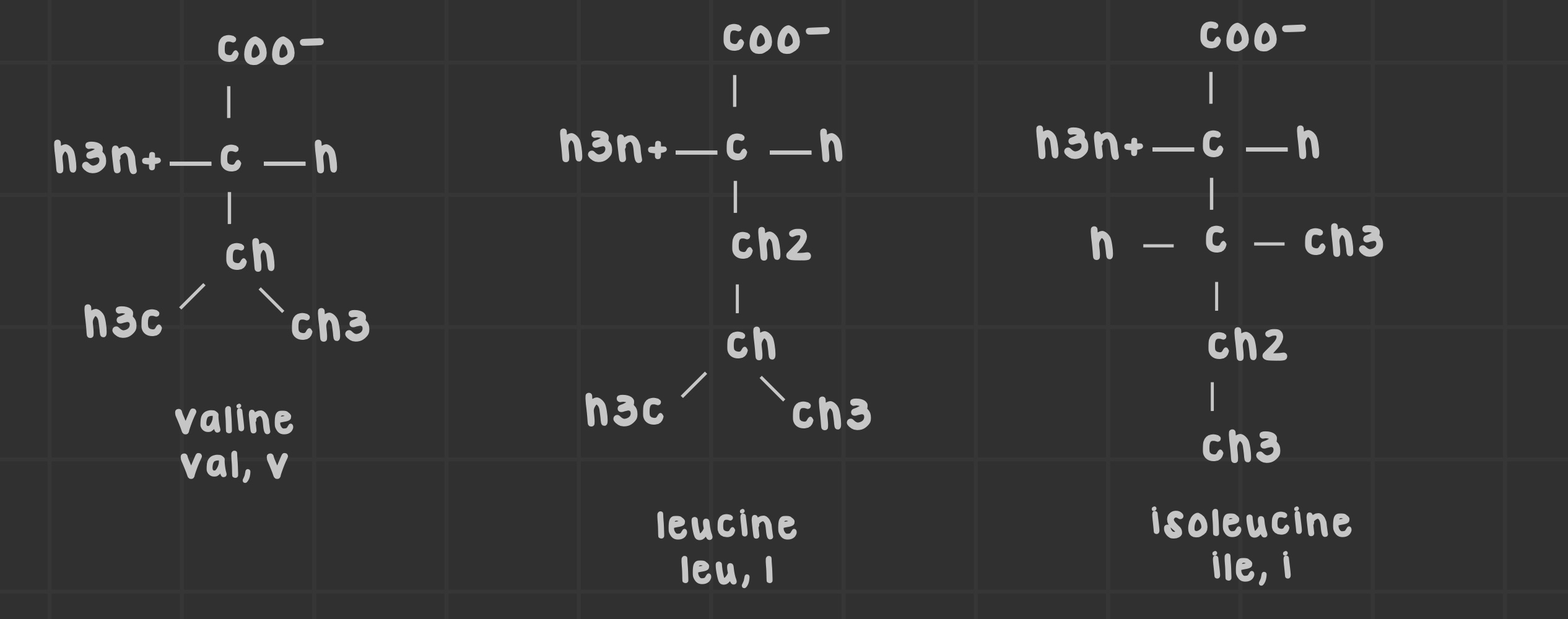
What are the branched alkyl chain amino acids?
Amino acids with branched side chains; leucine, isoleucine, and valine.
Why are tyrosine and tryptophan technically considered nonpolar?
Tyrosine’s large ring suppresses the polarity of the hydroxyl group; tryptophan’s indole ring suppresses nitrogen’s ability to act as a hydrogen bond donor
Name the polar uncharged amino acids.
Asparagine, glutamine, serine, threonine, cysteine.

Name the negatively charged (acidic) amino acids.
Glutamate and aspartate.

What is the difference between glutamic/aspartic acid and glutamate/aspartate?
Glutamic acid and aspartic acid are the non-ionized forms, while glutamate and aspartate are the ionized forms commonly found at physiological pH.
Name the positively charged (basic) amino acids.
Lysine, arginine, histidine.

Why are tyrosine, cysteine, and histidine able to be ionized?
Tyrosine’s hydroxyl is readily accessible to other aromatic amino acids, and resonance makes it deprotonate easier than other alcohols
Cysteine’s thiol is bent and the small hydrogen’s allow sulfur lone pairs to interact with water for stronger dipole-dipole interactions
Histidine’s side chain is mostly deprotonated and uncharged at normal pH with less than 10% protonated
What ionizable groups are present in all amino acids? What are their pKa values?
All amino acids possess an amino group (-NH2) and a carboxyl group (-COOH), which can ionize.
Amino pKa is 9.6, carboxyl pKa is 2.2
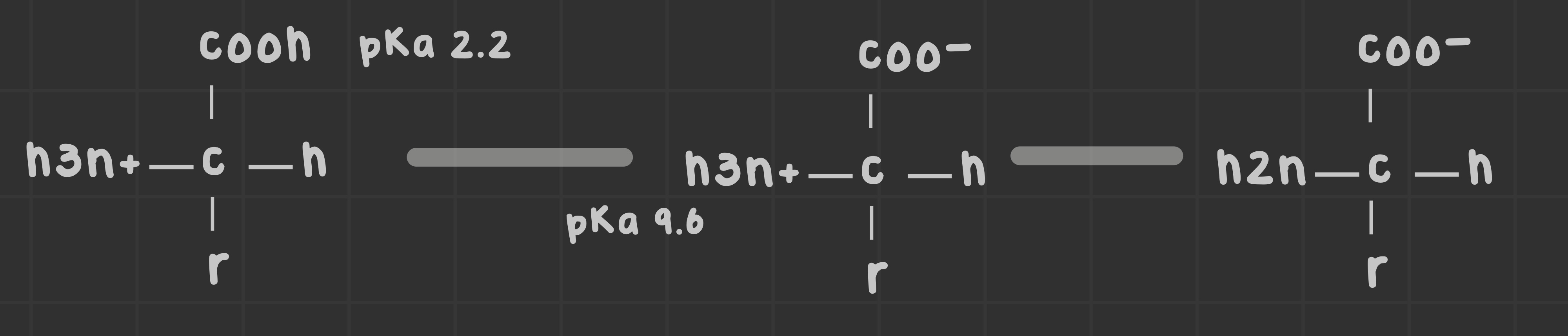
What are ionizable amino acids and their pkas?
Acids with three ionizable groups; R (12.5), K (10.5), H (6), E (4.3), D (3.7), C (8.0), Y (10.5)
What is a low pH environment for amino acids?
pH is below pKa values, resulting in protonation (more free protons) of amino groups and carboxyl groups, leading to a predominance of positively charged amino acids
What is a high pH environment for amino acids?
pH is above pKa values, resulting in deprotonation (less free protons) of amino groups and carboxyl groups, leading to a predominance of negatively charged amino acids.
What are nucleophilic amino acids?
Acids with functional groups that deprotonate to increase nucleophilicity; S, T, C, Y, K
What is pKa?
pH where 50% of the population is protonated and 50% is deprotonated.
If pH is less than pKa then,
the environment is acidic, leading to protonation of amino groups and carboxyl groups, resulting in a predominance of positively charged amino acids.
If pH is greater than pKa then,
the environment is basic, causing deprotonation of amino groups and carboxyl groups, leading to a predominance of negatively charged amino acids.
What is pI and what does it promote for amino acids?
pH where amino acids have a net charge of 0; promotes aggregation/decreased solubility as ionizable surface residues are neutralized
What are zwitterions?
Net neutral charge when the carboxyl group loses a proton and the amino group picks up a proton.
How does a titration work?
slow addition of strong base (NaOH) reacts with H3O+ to form water, and pH raises if solution pH is far from the pKa of the ionizable group.
What is the buffering region of a titration?
point where NaOH removes protons from the ionizable group (minimal pH change) and is 1 pH unit above/below pKa
What is the midpoint of a titration?
The point at which the amount of titrant added is equal to half the amount of the substance being titrated, resulting in the pH equal to the pKa of the ionizable group.
What is the central dogma?
DNA to mRNA to protein.
What is RNA and the three forms?
RNA: single stranded molecule that complementary base pairs with DNA (transcription)
mRNA: synthesized by RNA pol II from DNA to carry information to the ribosome for protein synthesis.
rRNA: transcribed by RNA pol I by ribosomal DNA in the nucleolus
tRNA: single-stranded RNA transcribed by RNA pol III that base pairs itself into a 3D L structure (translation)
What is a transcription unit?
Coding region where transcription begins and ends.
What is the role of the enzyme RNA replicase?
creates dsRNA from ssRNA
What is the hairpin loop formed by RNA?
RNA base pairs itself through close contact with complementary bases
What is transcription?
The process by which the genetic information in DNA is copied into messenger RNA (mRNA), which then serves as the template for protein synthesis.
What is a promoter?
Nucleotides in a unit that identify the start of transcription.
What is a response element?
DNA promoter region that binds specific transcription factors.
What is a terminator?
Nucleotides in a unit that identify the end of transcription.
What is upstream vs. downstream?
Upstream refers to sequences/promoters before transcription (negative sign)
Downstream refers to transcribed regions and terminators (positive sign)
What does RNA polymerase II do?
Reads 3’-5’ while assembling nucleotides in the 5' to 3' direction without a primer.
What is monocistronic?
1 mRNA creates 1 protein (eukaryotes)
What is polycistronic?
1 mRNA creates multiple proteins (prokaryotes)
What are the three steps of transcription?
Initiation, elongation, and termination.
What are the steps of initiation in transcription?
forming the transcription initiation complex where RNA pol II and general transcription factors bind the promoter
DNA helicase and topoisomerase unwind and relieve DNA supercoiling
RNA pol II attaches to gene of interest
transcription begins at the start site and forms the coding + noncoding strands
What is the TATA box?
A DNA sequence in the eukaryotic promoter region that helps initiate transcription by binding transcription factors and RNA polymerase II.
What is the Prinbow box?
A conserved DNA sequence found in bacterial promoters, recognized by RNA polymerase to initiate transcription.
What is the difference between the coding and noncoding strand of DNA?
The coding strand (sense strand) has the same sequence as the mRNA produced during transcription, and is in the 5’-3’ direction.
The noncoding strand (antisense strand) serves as the template for RNA synthesis, and is in the 3’-5’ direction.
What are the steps of elongation in transcription?
RNA pol II synthesizes 5’-3’ mRNA via complementary base pairing with the noncoding DNA strand
double helix reforms as RNA pol II progresses
What are the steps of termination in transcription?
Transcription complex reaches a terminator sequence in the DNA.
RNA polymerase detaches from the DNA, newly synthesized mRNA is released, and DNA strands re-anneal.
How do eukaryotes end transcription?
RNA pol II transcribes the polyadenylation signal sequence (3’=TTATT-5’) to separate the mRNA transcript and RNA pol II
Describe transcription factors and their components.
Transcription factors are proteins required for higher levels of gene expression that activate or inhibit gene expression.
DNA-binding domain: interacts with DNA binding sites in enhancer
activation domain: interacts with regulatory proteins/transcription machinery for transcription initiation
What are enhancers?
Regulatory DNA sequences in the proximal or distal end of the promoter that hold DNA binding site clusters for transcription factors
What are repressors?
Inhibition of initiation via competing with activators for enhancer binding sites or blocking activator binding
What is the mediator complex?
Carries messages from activators and repressors to RNA polymerase to activate or inhibit transcription.
What is gene duplication?
Copies a gene in series/parallel on the same chromosome to amplify expression.
What are ubiquitin proteins?
Proteins added to a target protein via ubiquitin ligase for proteasome recognition. The protein is hydrolyzed into small oligopeptides for further cytosol degradation.
What is ncRNA?
RNA that does not code for mRNA, tRNA, or rRNA, and forms ncRNA-protein complexes that interact with mRNA for mRNA degradation or to block translation.
What is translation?
mRNA transformed into a protein nucleotide sequence.
What are codons?
Three nucleotides of an amino acid that reads 5' to 3'.
What is the start codon?
AUG/methionine; initiates translation
What are the stop codons?
UGA, UAG, UAA
What is miRNA?
Less specific ssRNA that regulates multiple mRNA targets
somewhat complementary to the mRNA transcript, translation is blocked
perfectly complementary to the mRNA transcript, mRNA degrades
What is siRNA?
Highly specific dsRNA
perfectly complementary to the mRNA transcript: mRNA degradation.
What are cis regulators?
Elements that regulate DNA locally - promoter and enhancer regions.
What are trans regulators?
Elements that regulate DNA distantly - transcription factors.
What is the amber codon?
TAG/UAG
What is the opal codon?
TGA/UGA
What is the ochre codon?
TAA/UAA
What is an anticodon?
Complementary three nucleotide sequence to a codon (read 3'-5') that is used by tRNA to match the correct amino acid with its specific codon.
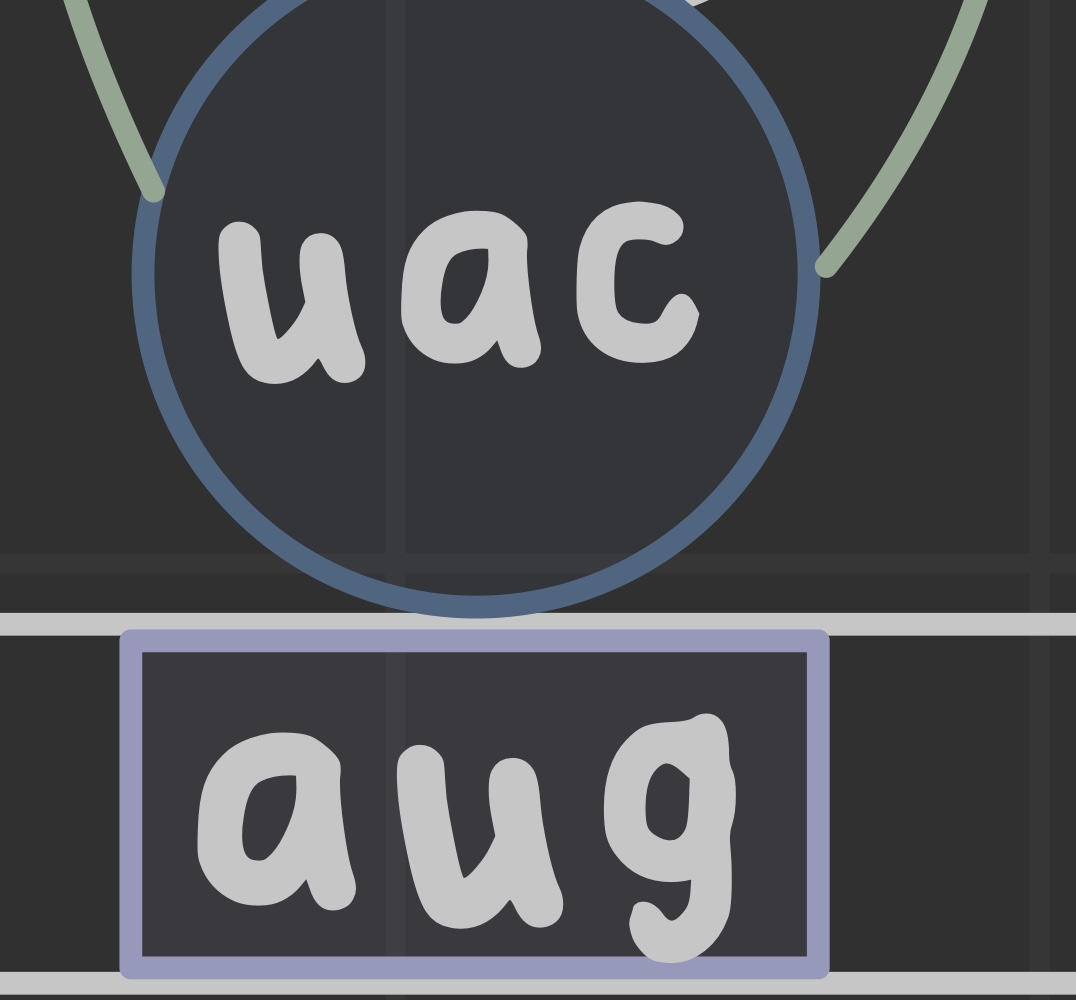
What is wobble pairing?
Third nucleotide varies from watson-crick base pairing.
What does aminoacyl-tRNA synthetase do?
Pairs amino acids to the correct tRNA molecule via ester linkages through ATP hydrolysis
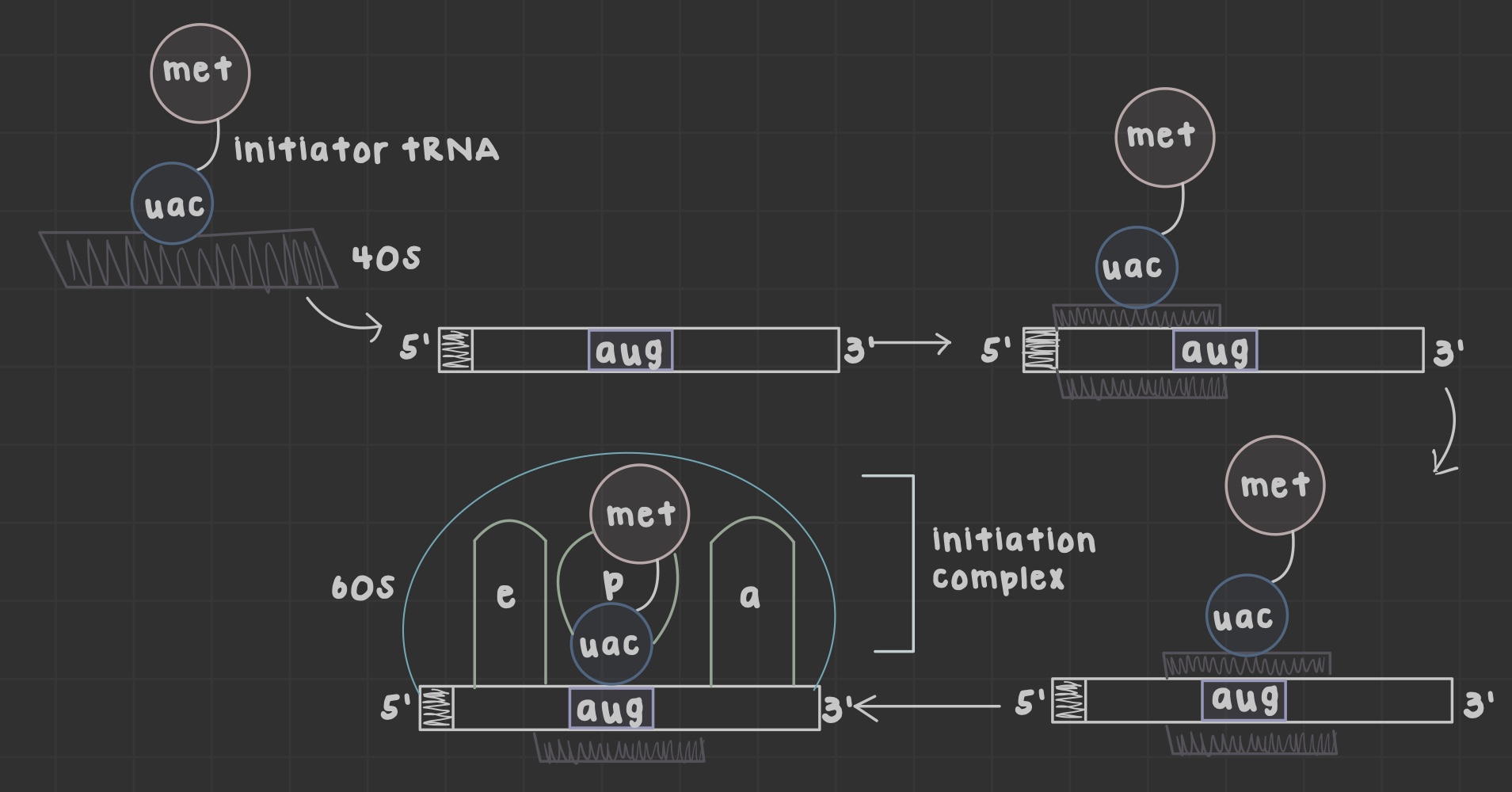
What are ribosomes?
Site of translation that allow mRNA and tRNA to interact, as well as catalyze peptide bond formation.
What does the peptidyl (P) site do?
Holds tRNA carrying the growing chain
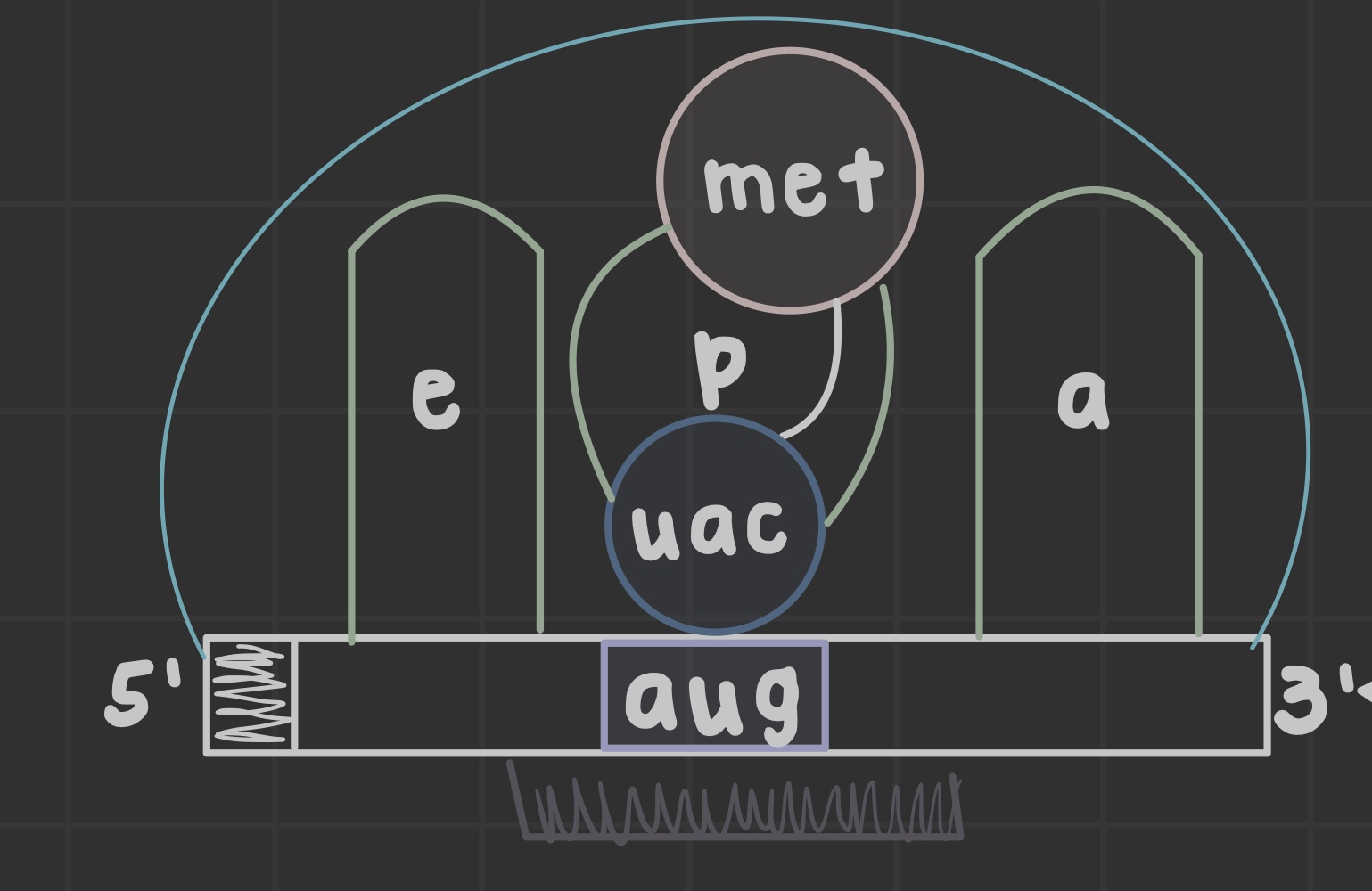
What does the aminoacyl (A) site do?
Holds tRNA carrying the next amino acid
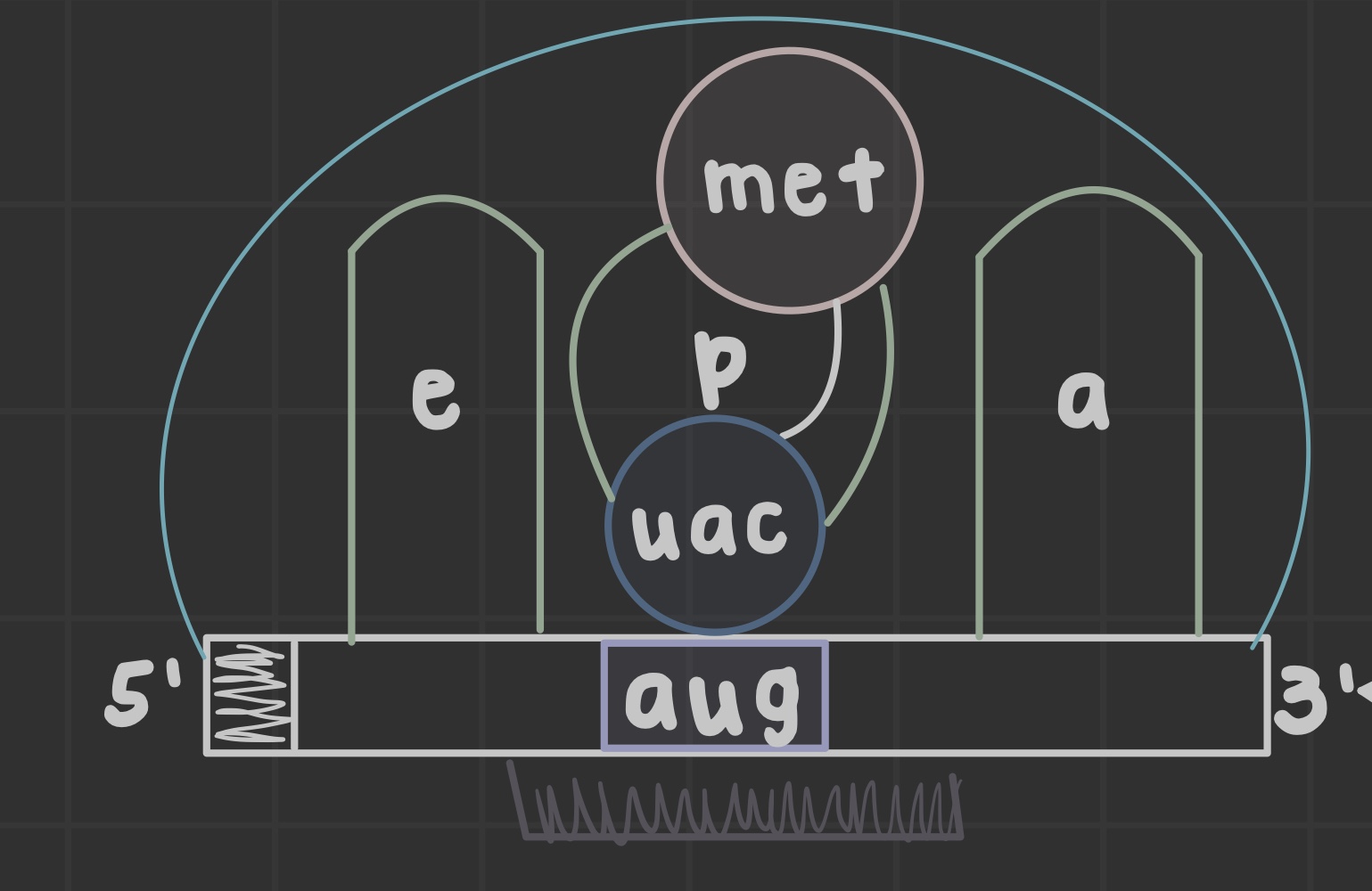
What is the exit (E) site?
Discharged tRNA leaves the ribosome after amino acid transfer
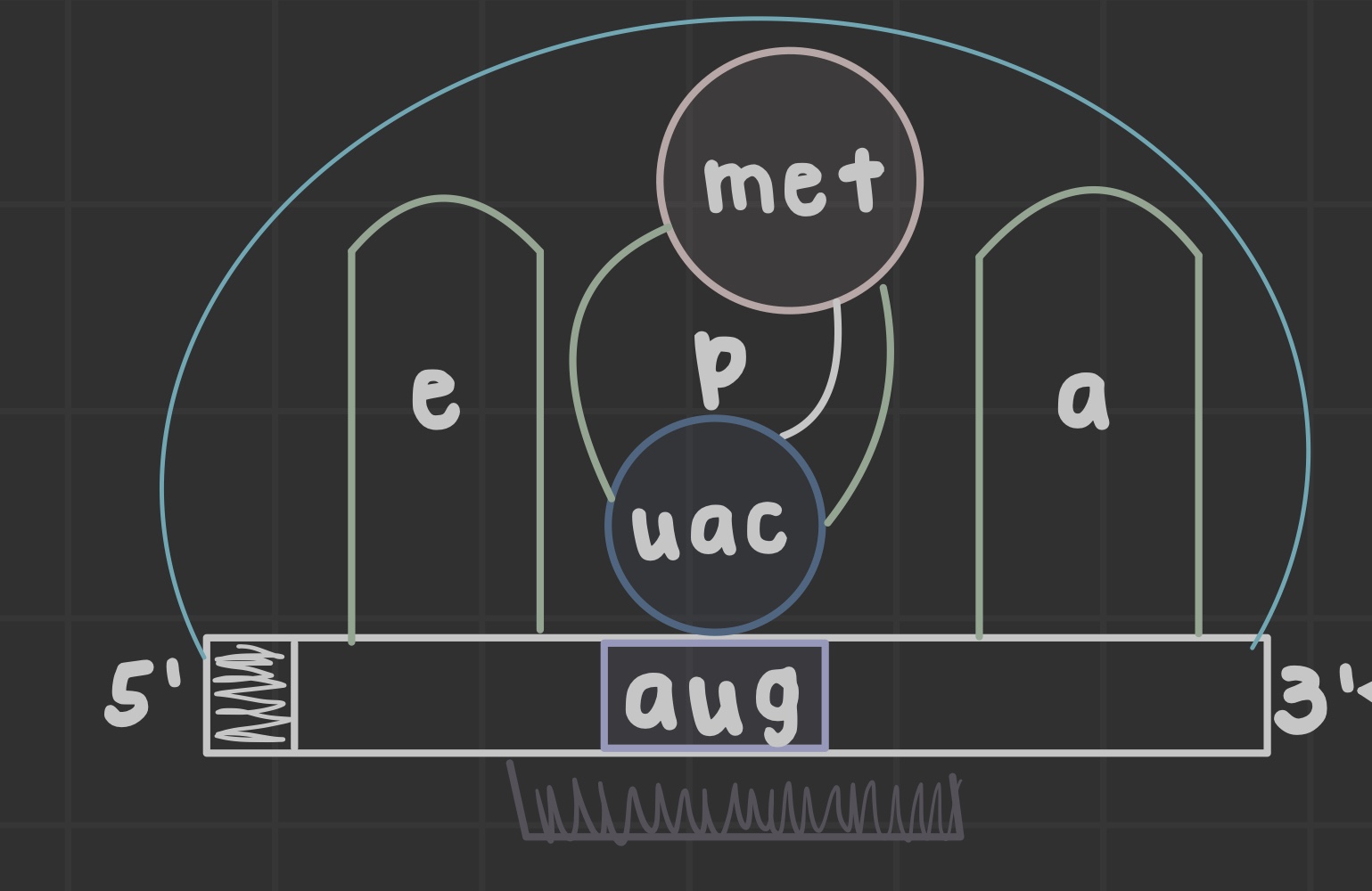
What is an open reading frame?
the mRNA portion translated by ribosomes formed after initiator tRNA anticodon base pairs the AUG codon
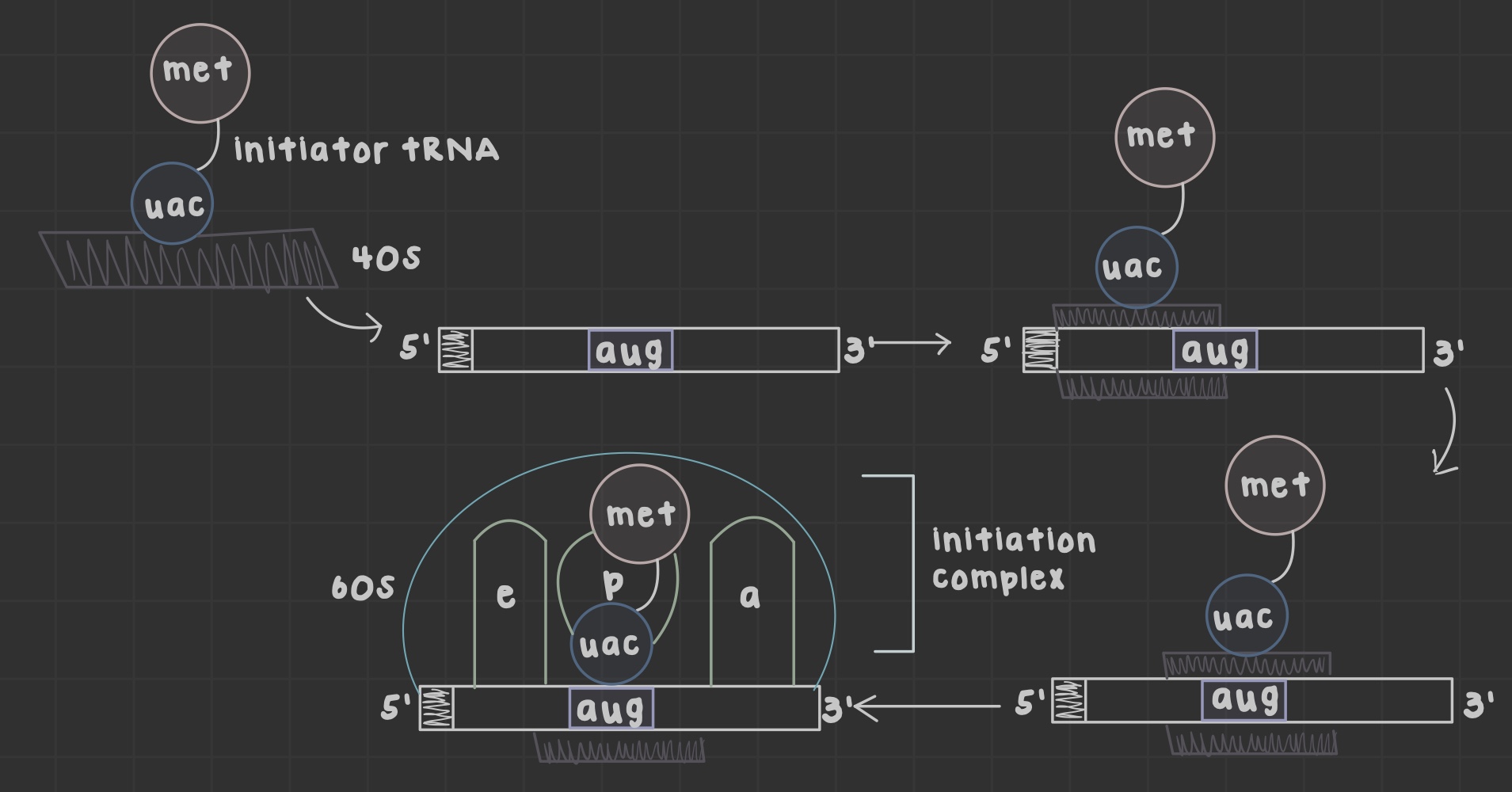
What does the peptidyl transferase center do?
Catalyzes peptide bond formation in the 60S subunit
What do elongation factors do?
Assist tRNAs by recruiting their correct aminoacyl-tRNAs throughs exchanging GDP for GTP
What does the release factor do?
Promotes ester linkage hydrolysis between the protein and tRNA in the P site
What is a signal sequence?
20 amino acids added to the N-terminus that designates correct organelle destination after translation and are cleaved off once the protein arrives.
What occurs during initiation in prokaryotic translation?
30S subunit binds the shine-dalgarno sequence in the 5'-UTR and searches for fmet-tRNA
What occurs during elongation in prokaryotic translation?
Ribosome reads mRNA 5'-3' and synthesize proteins from N- to C-terminus.
What occurs during termination in prokaryotic translation?
Release factor binds stop codon and no amino acid-tRNA exists for stop codon.
What occurs during n-linked glycosylation?
-NH2 groups of N side chains are glycosylated
What occurs during o-linked glycosylation?
-OH groups of S or T side chains are glycosylated
What are heat shock proteins?
Chaperones that prevent cellular damage from high temperatures.
What are cofactors?
Non-amino groups required for protein factors (single metal ions or organic molecules).
What are signal peptides?
Proteins that direct ribosomes to a translocation complex on the ER membrane surface where protein synthesis continues and are cleaved at the ER lumen
What do chaperones do?
Assist in protein folding and other modifications to make proteins functional.
phosphorylation: uses kinases to transfer a y-phosphate from ATP to a S, T, or Y sidechain
glycosylation: glycosyltransferases add a carbohydrate group in the lumen of the ER and GA
ubiquitination: targets the N-terminus/side chain (often of K) and nucleophilically attacks the C-terminus of ubiquitin to direct proteins to a proteasome for recycling
acetylation: K residues have an acetylene group added (i.e. histones)
prenylation: isoprene groups added to membrane-bound vesicles via C residues
lipidation: a nucleophilic amino acid (K, C, or S) attacks a carbonyl linked to a hydrocarbon
proteolysis: removes protein groups when a protease hydrolyzes a peptide bond for protein activation
What are housekeeping genes?
Genes that are always expressed/needed.
What does acetylation do to histone proteins?
Upregulates transcription and forms euchromatin.
What does deacetylation do to histone proteins?
Downregulates transcription and forms heterochromatin.
What are transposons?
Transposable elements created by transposase that move around the genome and insert in random locations after transfer via horizontal gene transfer.
What is an operon and its components?
Expresses all prokaryotic genes necessary for the same process/function and utilizes the Jacob-Monod model.
regulatory region: promoter and operator; encodes repressor protein that binds operator site
operator site: repressor protein binds
structural gene: cluster downstream the regulatory region that codes for enzymes and proteins needed under specific conditions
regulatory genes: constitutively expressed and code for a repressor that binds the operator
promoter: binds RNA pol
What are inducible operons?
Repressed until conditions are favorable (negative feedback)
What are repressible operons?
Always expressed until conditions that inhibit gene expression occur.
What are point mutations?
Replace one nucleotide and its complementary partner with another nucleotide pair.
What is a missense mutation?
Open reading frame substitution that causes a different amino acid to be placed into the proteins during translation.
What is a nonsense mutation?
Open reading frame substitution that causes a premature stop codon.
What is a silent mutation?
Open reading frame substitution that does not change the amino acid.
What is a synonymous mutation?
DNA sequence is not changed due to wobble effect.
What is a frameshift mutation?
Deletion/insertion of not 3 nucleotides within the open reading frame that alters tRNA codon recognition and is nonfunctional.
What is an inframe mutation?
Deletion/insertion of three nucleotides and is functional.