L4 Post-Translational Modifications in Biomolecules
1/65
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
66 Terms
Post-Translational Modifications
PTMs are chemical changes made to a protein after it has been synthesised by ribosomes.
Locations of PTMs
Usually occur in the endoplasmic reticulum, Golgi apparatus, or cytoplasm.
Post-Translational Modifications (PTMs)
Chemical modifications that fine-tune a protein's properties, activity, location, and stability.
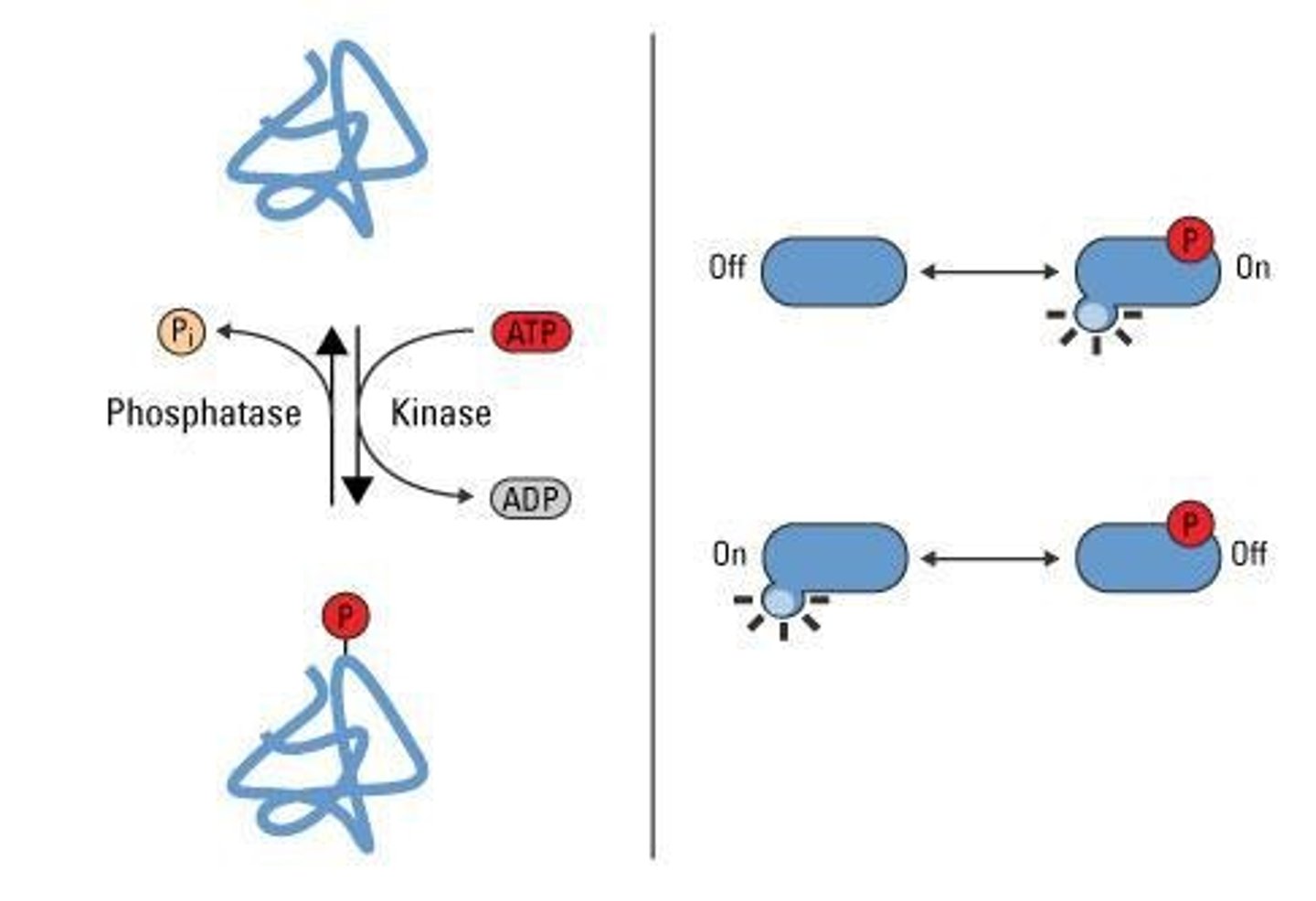
Phosphorylation
- Addition of a phosphate group (PO4^3) to a protein after it is made; one of the most common and important PTMs in cells.
- Protein-OH + ATP → Protein-O-PO3; -Kinase transfers the phosphate from ATP to the hydroxyl group on the protein.
Glycosylation
Attachment of sugar chains to proteins.
Ubiquitination
Tags proteins for degradation.
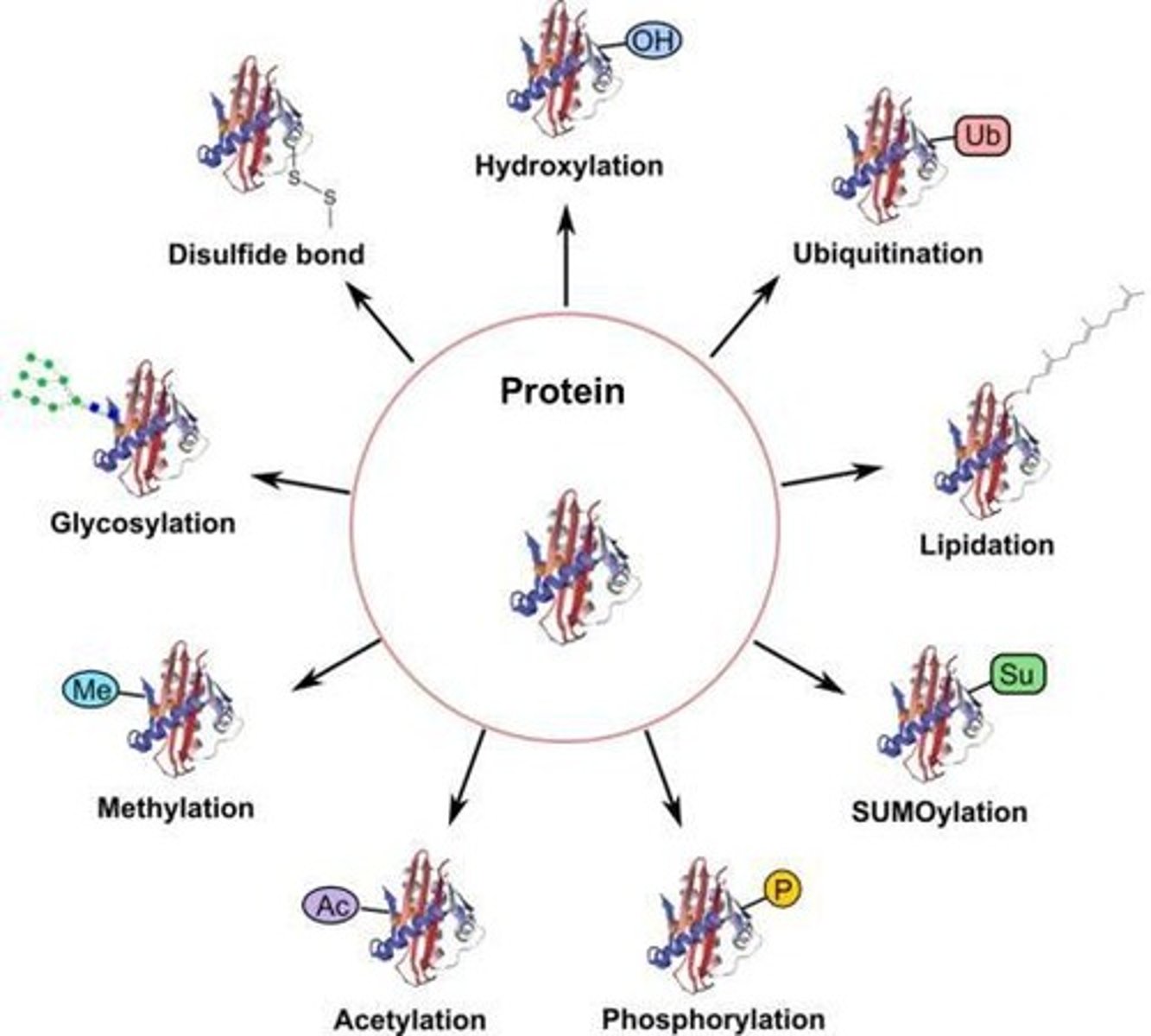
Acetylation
Addition of an acetyl group to proteins.
Methylation
Addition of a methyl group to proteins.
Prenylation
Attachment of a lipid group to proteins.
Myristoylation
Attachment of myristoyl fatty acid to proteins.
Palmitoylation
Attachment of palmitoyl fatty acid to proteins.
Phosphate donor
ATP (adenosine triphosphate) is the phosphate donor in phosphorylation.
Serine (Ser)
Most common phosphorylation site; small side chain, abundant in proteins.
Threonine (Thr)
Similar chemistry to serine but bulkier and less common.
Tyrosine (Tyr)
Less abundant but critical in signalling; aromatic ring affects local structure.
Receptor Kinases
Membrane-bound receptors that detect extracellular signals and convert them into intracellular phosphorylation events.
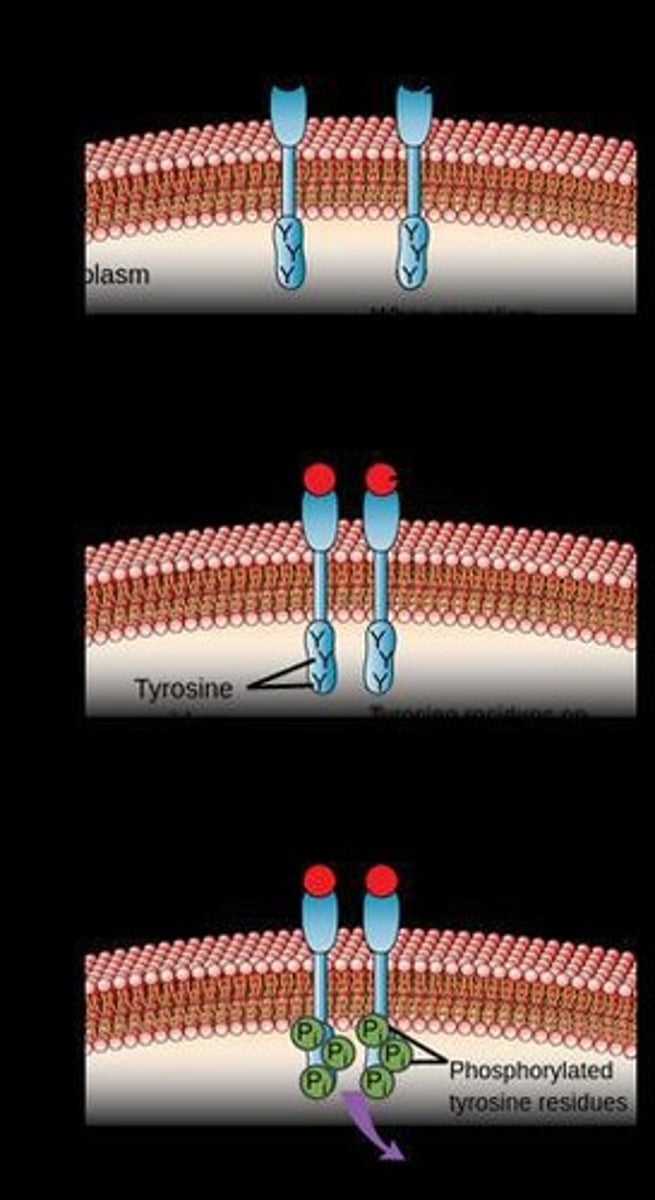
Ligand Binding
Triggers autophosphorylation of the receptor, activating downstream signalling proteins.
Epidermal Growth Factor (EGF)
Binding to EGFR causes a conformational change in the dimerised receptor, activating signalling pathways.
Receptor Tyrosine Kinases (RTKs)
Most receptor kinases; involved in cell growth, differentiation, and metabolism.
Downregulation of Receptor Kinases
Occurs through endocytosis and degradation, phosphatases removing phosphate groups.
Autophosphorylation
Phosphorylation of a receptor by itself after ligand binding.
Signalling Cascade
A series of biochemical events triggered by receptor activation that leads to multiple signalling pathways being activated.
Phosphorylation Motifs
Short amino acid sequences around the phosphorylation site that kinases recognise.
Function of Phosphorylation Motifs
It acts like a 'postal address' that kinases use to recognise where to dock and transfer the phosphate.
Phosphorylated Residue Location
The phosphorylated residue is usually in the centre of the motif.
Kinase Specificity
Kinases can't just phosphorylate any Ser/Thr/Tyr - they have specificity based on shape and charge.
Example of Phosphorylation Motif
[R/K]-X-[S/T], where R/K arginine/lysine, X = any amino acid, S/T = serine/threonine.
Another Example of Phosphorylation Motif
[S/T]-X-X-[E/D].
Phosphorylation Cascades
Sequential activation of multiple kinases in a chain where one kinase phosphorylates the next, amplifying the signal.
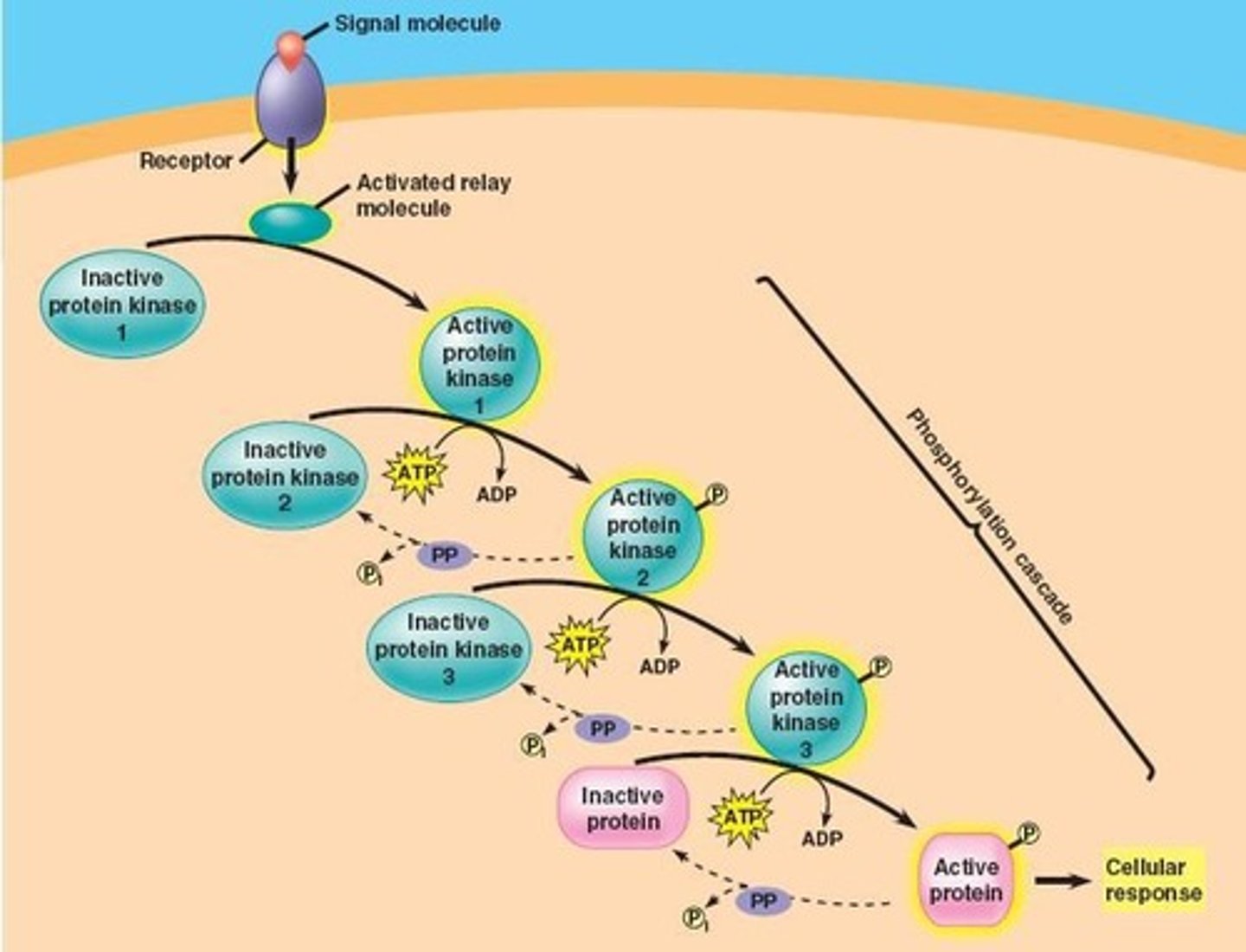
Example of Phosphorylation Cascade
Ras → Raf → MEK → ERK.
What Can Be Phosphorylated?
Enzymes, receptors, transcription factors, structural proteins, transport proteins, and scaffold proteins.
Prevalence of Glycosylation
Affects >50% of all proteins in humans.
Functions of Glycosylation
Aids protein folding and stability
Mediates cell-cell recognition
Signalling
Modulates immune recognition.
Catalysts of Glycosylation
Catalysed by glycosyltransferases, reversed partially by glycosidases.
General Reaction of Glycosylation
Protein + Sugar-Nucleotide → Glycoprotein + Nucleotide.
Where is found Mannose and what are its functions?
Often found in the core structure of N-linked glycans and essential for forming the initial core sugar structure.
Role of Mannose
Important for protein quality control in the ER, signaling whether a glycoprotein is correctly folded.
Sialic Acid
Typical terminal sugar in glycans that adds a negative charge to the polypeptide.
Influence of Sialic Acid
Influences circulation half-life of glycoproteins in blood and protects glycoproteins from clearance.
Where is N-acetylglucosamine found?
Found in core linkages of N-linked glycans and O-linked glycosylation initiation.
O-GlcNAcylation
A dynamic PTM performed inside the cytoplasm and nucleus that regulates several cellular processes.
O-linked Glycosylation
Attachment of a sugar(s) to the hydroxyl oxygen of serine.
N-linked Glycosylation
Glycan is added to asparagine residues.
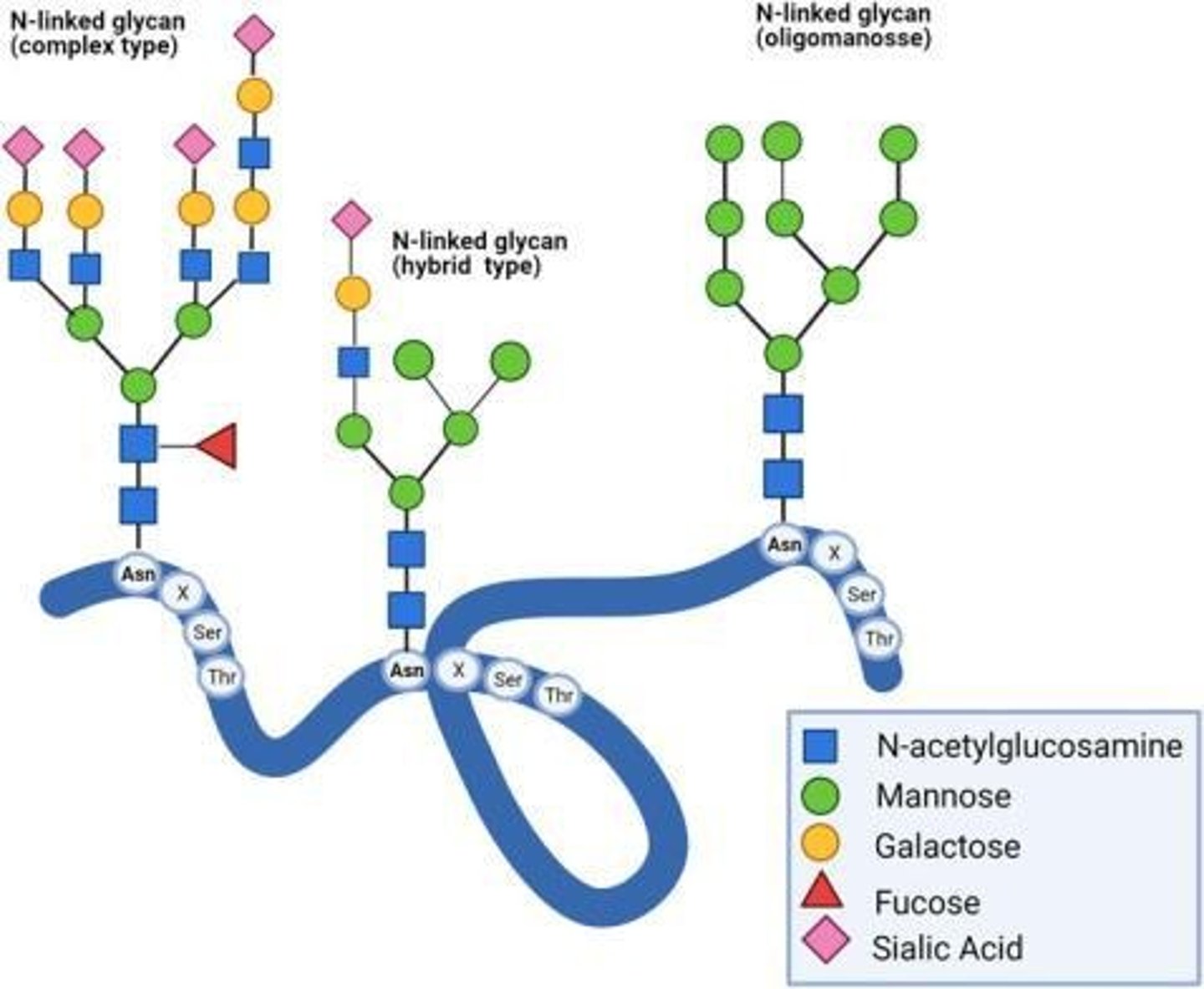
Characteristics of O-glycans
Generally smaller but more diverse than N-glycans.
What Can Be Glycosylated?
Proteins like secreted proteins, membrane-bound receptors, extracellular matrix proteins, and lipids such as glycolipids.
Function of Ubiquitination
Regulates protein fate through degradation by the proteosome and influences protein trafficking, DNA repair, and signalling.
Ubiquitination Reaction
Involves an enzyme cascade that attaches ubiquitin to lysine residues on substrate proteins.
Activation
Ubiquitin is activated by an E1 enzyme (ubiquitin-activating enzymes) using ATP.
Conjugation
Activated ubiquitin is transferred to an E2 enzyme (ubiquitin-conjugating enzyme).
Ligation
An E3 ligase facilitates transfer of ubiquitin from E2 to the substrate protein.
E1 (Ubiquitin-activating enzyme)
Activates ubiquitin by forming a high energy bond with its C-terminal glycine.
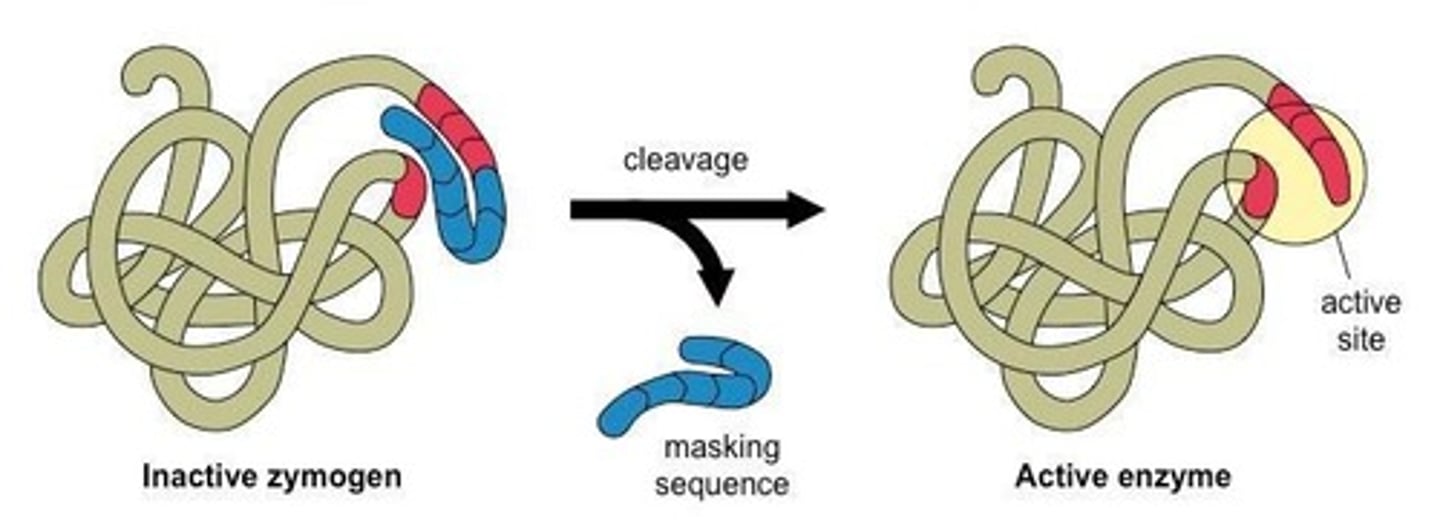
E2 (Ubiquitin-conjugating enzyme)
Receives activated ubiquitin from E1 and carries ubiquitin to the substrate.
E3 (Ubiquitin ligase)
Recognises specific substrate proteins and catalyses the transfer of ubiquitin from E2 to substrate lysines.
What Can Be Ubiquitinated
Proteins with exposed lysine residues can be ubiquitinated.
Substrates for Ubiquitination
Includes misfolded/damaged proteins for degradation, regulatory proteins controlling cell cycle, DNA repair, signal transduction, and histones involved in chromatin remodelling.
Where Does Ubiquitination Take Place
Ubiquitination occurs primarily in the cytoplasm and nucleus.
Target Proteins
Can be soluble, membrane-bound, or nuclear in nature.
What is the 26S Proteosome?
The 26S Proteosome is a large protein complex responsible for degrading ubiquitinated proteins.
Recognition by Proteosome
Recognises proteins tagged with polyubiquitin chains, especially those linked via lysine.
Deubiquitinating Enzymes (DUBs)
Remove attached ubiquitin molecules before degradation, freeing ubiquitin for reuse.
Protein Unfolding
Removal of the ubiquitin allows the proteins to be unfolded and fed into the proteosome core.
Proteolytic Sites
Cleaves the protein into small peptides (~3-25 amino acids) within the proteosome core.
Protease Activities
The proteosome has multiple protease activities (chymotrypsin-like, trypsin-like, etc.) for broad substrate cleavage.
Why are Release of Small Peptides
The small peptides are released into the cytoplasm or nucleus for further degradation by peptidases.
Immune System Priming
Small peptides may be up-taken by immune cells and used to prime the immune system to recognise unknown antigens.
Informing the Immunoproteosome
Viral/bacterial proteins, abnormal tumour proteins, or normal turnover proteins can inform the immunoproteosome to protect the cell/host.