Biochemistry I Lesson 6: DNA Regulation
1/76
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
77 Terms
A skin cell and an eye cell have which of the following in common?
(A) They have the exact same proteins.
(B) They have the exact same mRNA.
(C) They have the exact same DNA.
(D) None of the above.
(C) They have the exact same DNA.
A skin cell, an eye cell, and every other cell in your body all have the exact same DNA. What makes cells different from one another is the different expression of that DNA. Skin cells will express the DNA needed to make skin proteins, and eye cells will express the protein needed to make eye proteins.
Gene expression is regulated at the level of:
(A) Replication
(B) Transcription
(C) Translation
(D) Post-translational Modification
(B) Transcription
Gene expression is regulated at the level of transcription, meaning that at certain times only certain DNA will be expressed and transcribed into RNA. This way, we only produce the proteins we need at a given time.
Why wouldn't it be a great idea for gene expression to be regulated at the protein or mRNA level?
It would not be a great idea for gene expression to be regulated at the mRNA or protein level because it would be a complete waste of energy! It takes a lot of ATP to put together proteins and mRNA, so it would be inefficient to make a bunch of proteins or mRNA that the cell will never use.
CRB Which of the following are laboratory techniques used to quantify the amount of mRNA (or RNA in general) to study Gene Expression?
I. Microarrays
II. RNAseq
III. Histone Methylation
(A) III only
(B) I and II only
(C) II and III only
(D) I, II and III
(B) I and II only
Microarrays and RNAseq are often used to quantify the amount of mRNA (or RNA in general) to study Gene Expression.
An operon is a group of ___________ under the control of ___________.
(A) exons, several promotors
(B) exons, a single promotor
(C) genes, several promotors
(D) genes, a single promotor
(D) genes, a single promotor
An operon is a group of genes under the control of a single promotor.
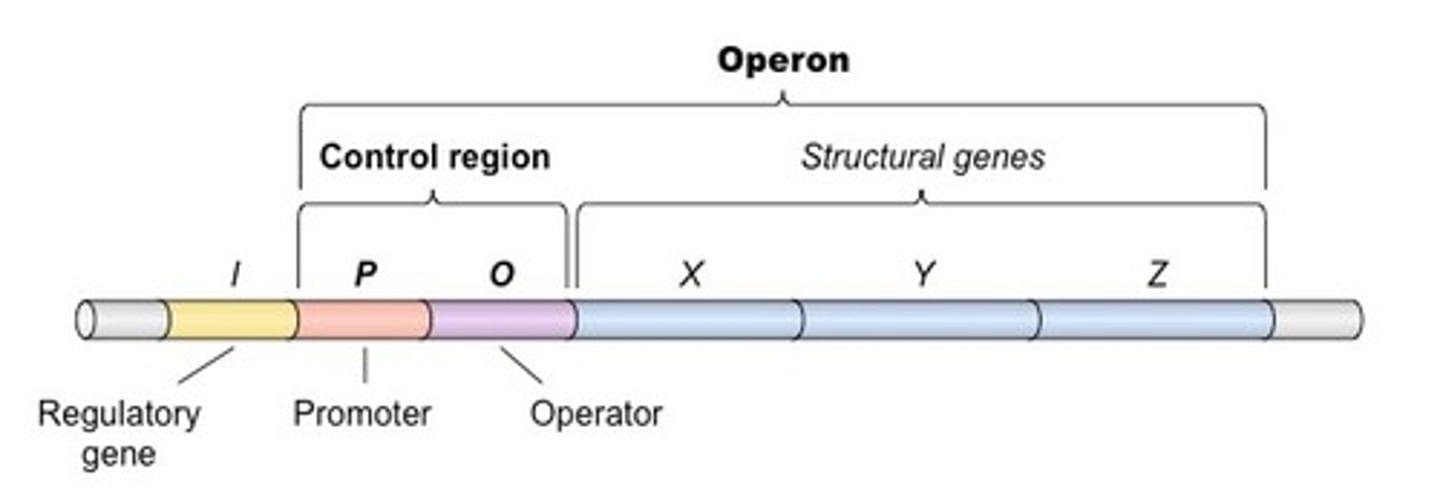
The Lac operon is a group of genes that code for the enzymes that help e. Coli break down:
(A) a polypeptide
(B) a disaccharide
(C) a phospholipid
(D) a hormone
(B) a disaccharide
The Lac operon is a group of genes that code for the enzymes that help e. Coli break down lactose, the disaccharide found in milk.
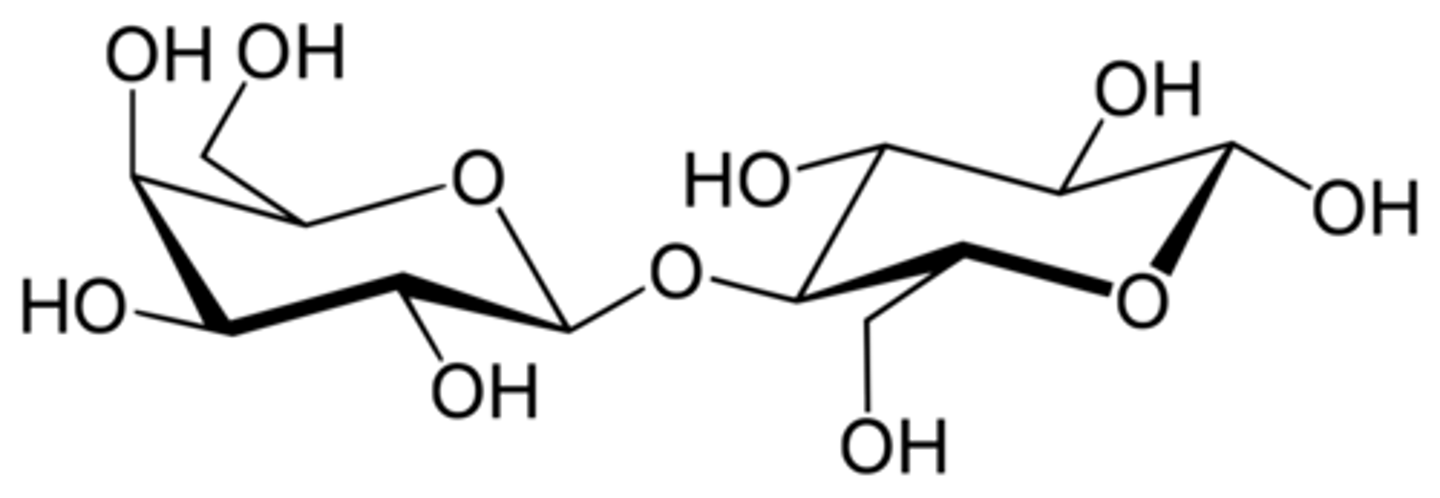
Lactose is composed of which two sugar monomers?
(A) glucose and mannose
(B) galactose and fructose
(C) glucose and galactose
(D) fructose and glucose
(C) glucose and galactose
Lactose is composed of glucose and galactose.
Why is regulating the lac operon important for e. Coli?
e. Coli does not need any enzymes to breakdown lactose when there is no lactose present in the first place. Using regulation, e. Coli is able to be more efficient in which proteins it produces.
What does the lac Z gene code for? What role does that protein have?
(A) Thiogalactoside transacetylase
(B) Acyl Galactase
(C) Lactose permease
(D) Beta-galactosidase
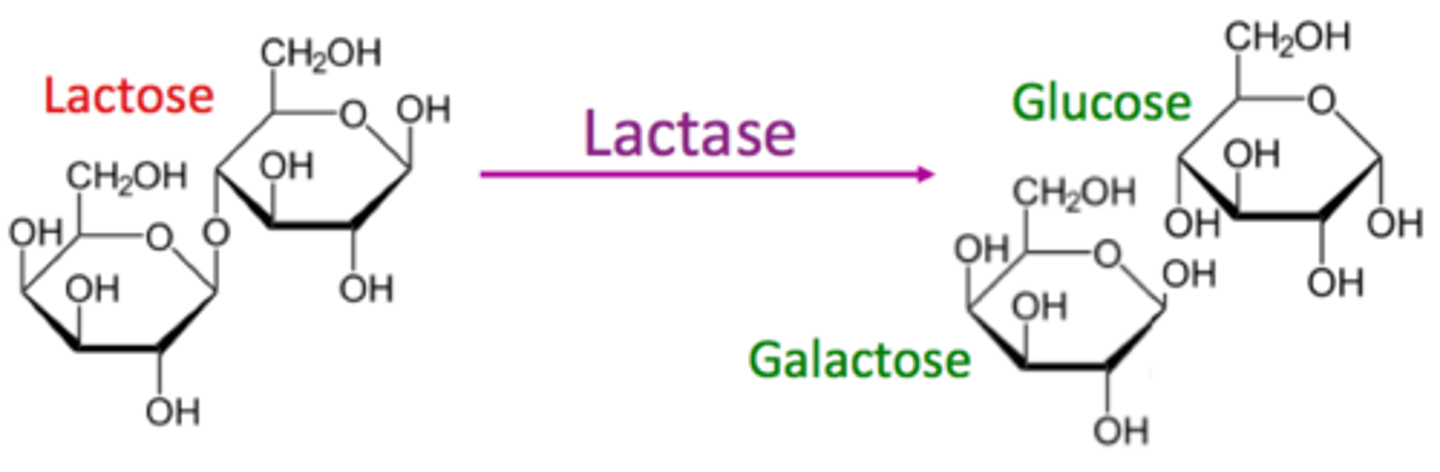
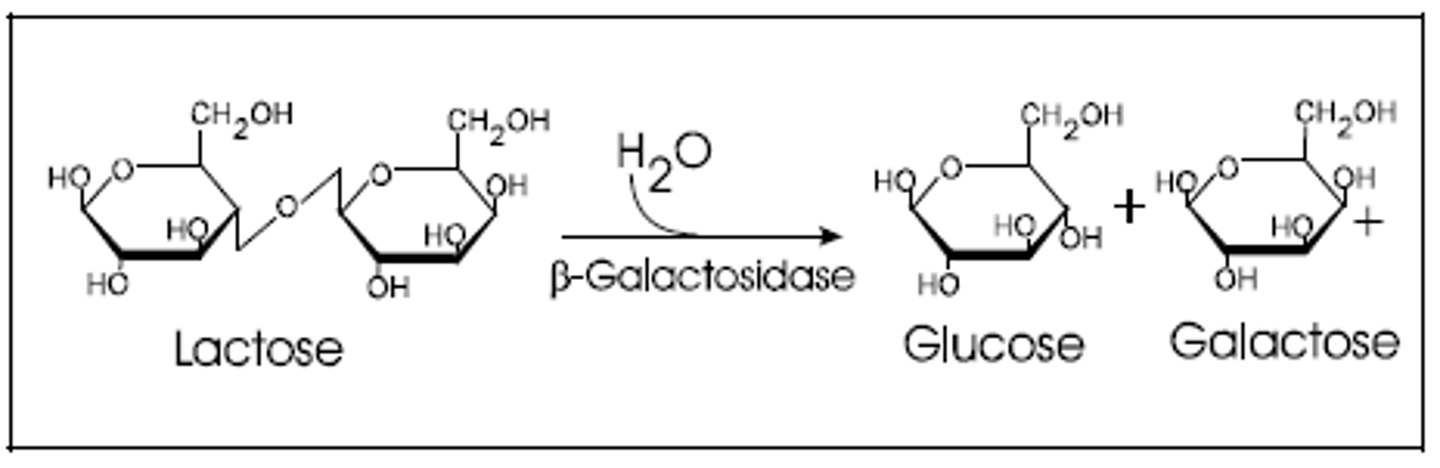
(D) Beta-galactosidase
The lac Z gene codes for the enzyme beta-galactosidase (aka Lactase). Beta-galactosidase cleaves lactose into glucose and galactose.
What does the lac Y gene code for? What role does that protein have?
(A) Thiogalactoside transacetylase
(B) Acyl Galactase
(C) Lactose permease
(D) Beta-galactosidase
(C) Lactose permease
The lac Y gene codes for the enzyme lactose permease. Lactose permease is a cytoplasmic membrane protein that transports lactose into the cell.
What does the lac A gene code for?
(A) Thiogalactoside transacetylase
(B) Acyl Galactase
(C) Lactose permease
(D) Beta-galactosidase
(A) Thiogalactoside transacetylase
The lac A gene codes for thiogalactoside transacetylase. Its function isn't worth memorizing.
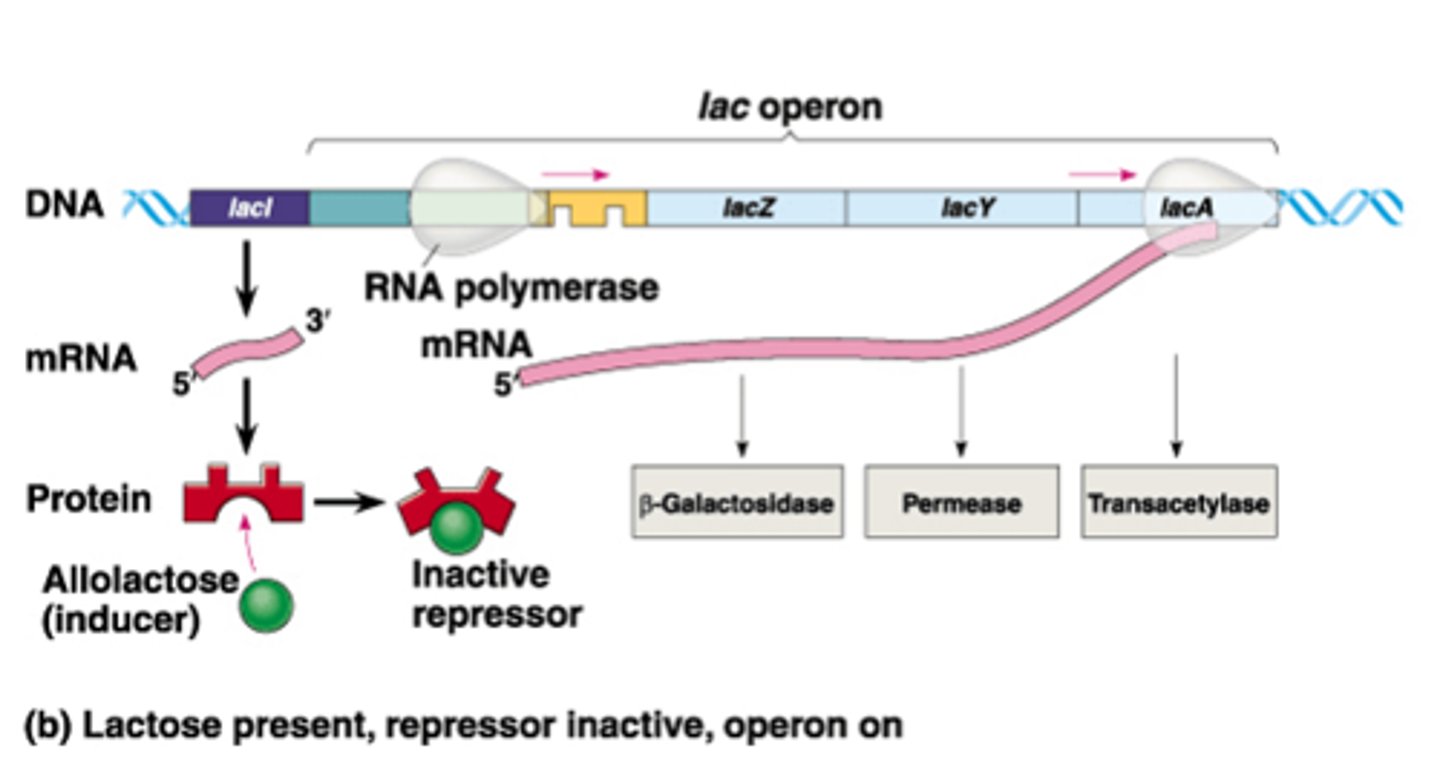
What is the difference between structural genes and the regulatory genes in terms of the lac operon?
The structural genes are the genes that code for the enzymes that are directly involved in lactose metabolism such as the Lac Z, Y, and A genes. The regulatory genes lie upstream of the lac operon and regulate whether or not the structural genes get transcribed.
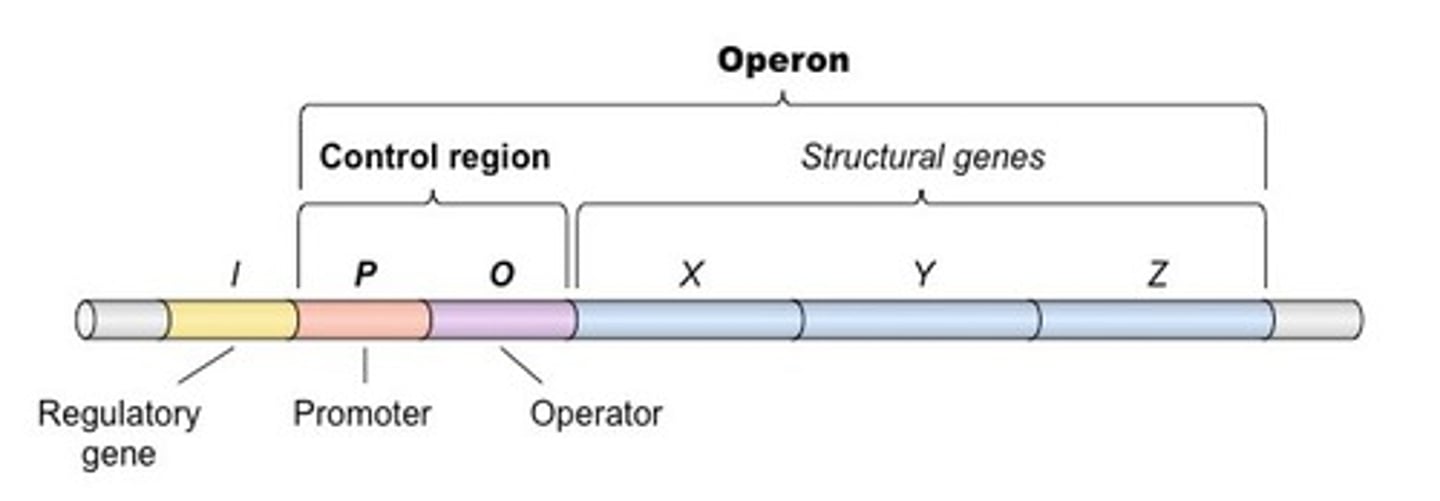
What binds to the promotor versus the operator?
RNA Polymerase will bind to the promotor. The repressor protein will bind to the operator.
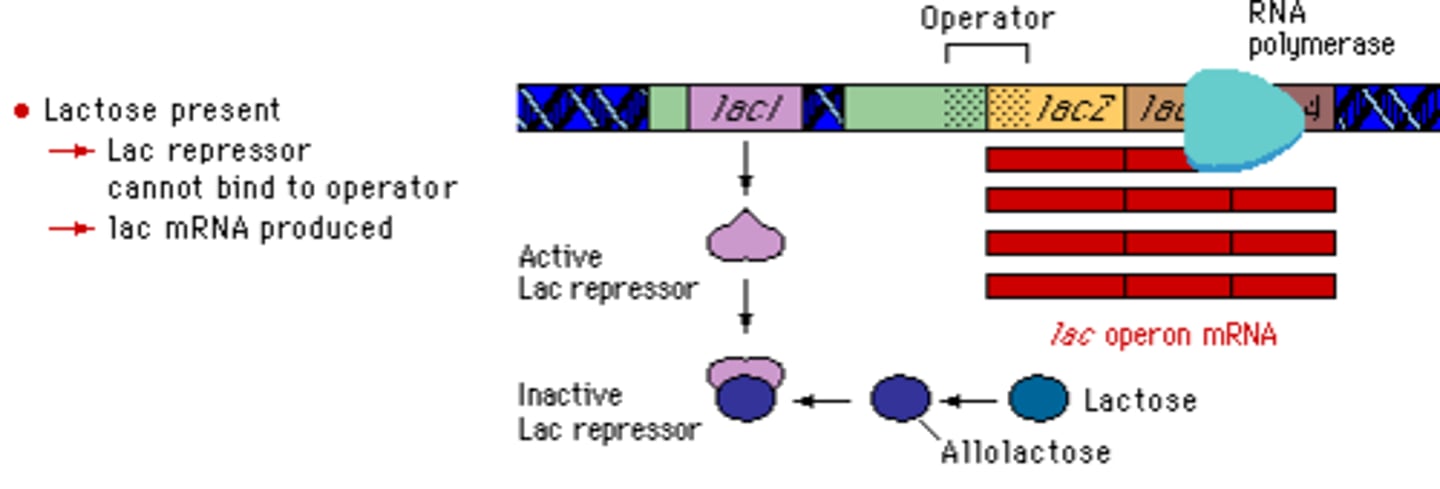
What causes the repressor protein to change its conformation and fall off the operator gene, allowing RNA polymerase to transcribe the lac operon genes (lac Z, lac Y, lac A)?
A metabolite of lactose ("allolactose") binds to the repressor protein, causing it to fall off the operator gene. This allows the RNA polymerase to transcribe the lac operon genes.
CRB Fill in the blanks: The Lac-Operon is a famous example of a(n) ____________. The trp (Tryptophan-producing) Operon is a famous example of a(n) __________________.
(A) Repressible System, Repressible System.
(B) Inducible System, Repressible System
(C) Repressible System, Inducible System
(D) Inducible System, Inducible System.
(B) Inducible System, Repressible System
The Lac-Operon is a famous example of an Inducible System. The trp (Tryptophan-producing) Operon is a famous example of a Repressible System.
What is the difference between a repressible and inducible operon?
A repressible operon is on until it is turned off ("repressed").
An inducible operon is off until it is turned on ("induced").
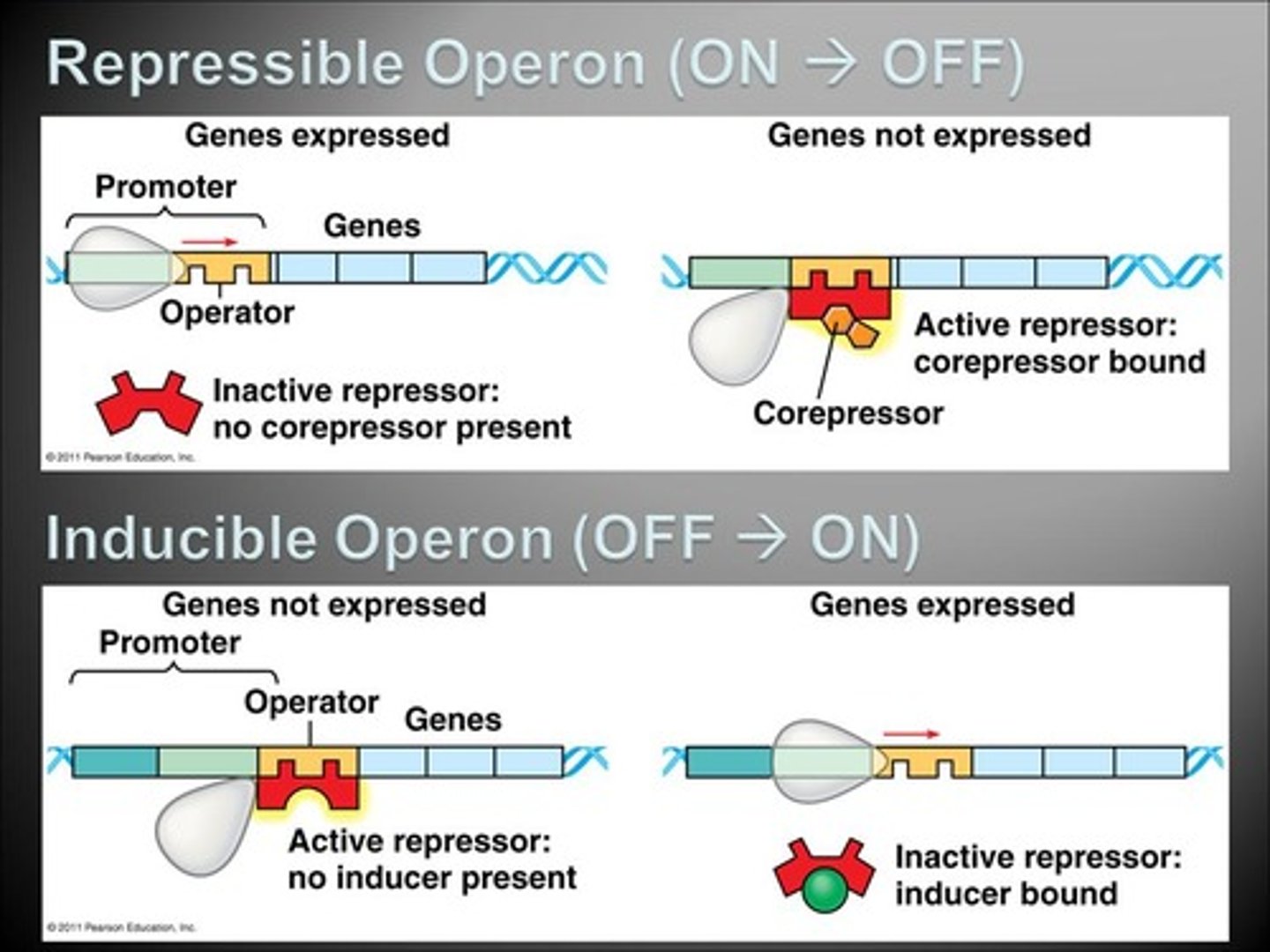
CRB Fill in the blanks: There are many inducible ______________ enzymes and repressible ______________ enzymes. This helps to maintain a stable amount of products in the prokaryote.
(A) Catabolic, Metabolic
(B) Metabolic, Anabolic
(C) Anabolic, Catabolic
(D) Catabolic, Anabolic
(D) Catabolic, Anabolic
There are many inducible Catabolic enzymes and repressible Anabolic enzymes. This helps to maintain a stable amount of products in the prokaryote.
What is the inducer of the lac operon? Why is it considered an inducer?
The inducer of the lac operon is allolactose because its presence induces transcription of the lac operon by causing the repressor protein to detach itself from the operator gene.
The Lac Operon is an example of a repressible or inducible operon?
The Lac Operon is an example of an inducible operon.
What does it mean to say that the repressor protein is "constitutively expressed"?
To say that something is "constitutively expressed" means that its gene is always turned on; thus, in this case, the repressor protein's mRNA is continuously being produced.
Describe how the repressor protein regulates gene expression of the lac operon when lactose concentration is high? low?
When high concentrations of lactose are present in e. Coli, allolactose binds to the repressor, thus removing the repressor and allowing RNA polymerase to transcribe the lac operon genes in order to make enzymes to break down the lactose.
If there are low concentrations of lactose, the repressor would block the RNA polymerase from transcribing the lac operon.
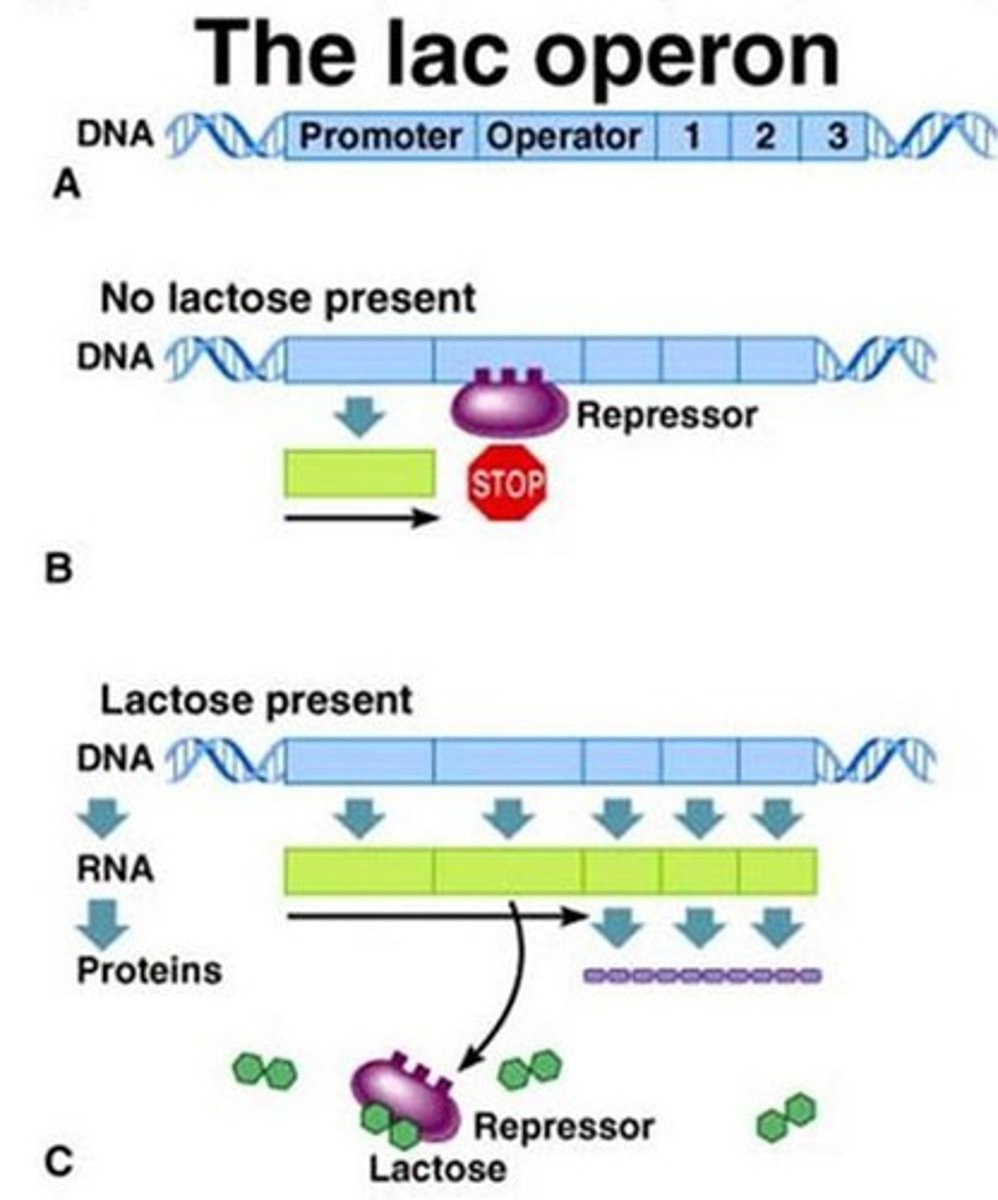
What happens to the transcription of the lac operon when glucose and lactose are both present? How is this an example of the inducer exclusion effect?
When there are equal amounts of glucose and lactose, the lac operon will not be transcribed because glucose that is transported into the cell signals an intermediate protein to bind to lactose permease and prevent it from transporting lactose into the cell. Now, because there is no lactose in the cell, its metabolite, allolactose, will not bind to and inhibit the repressor protein. For this reason, RNA polymerase is unable to transcribe the lac operon genes.
The inducer exclusion effect is when a molecule (the intermediate protein that binds to lactose permease) stops the transcriptional inducer (allolactose) from entering the cell/nucleus, preventing it from binding to its target repressor protein.
CRB There are also other methods of Transcriptional Regulation in Eukaryotes. Which of the following is NOT one of them?
(A) mRNA Surveillance, so that defective transcripts and stalled transcripts (stuck in the ribosome in translation) are degraded.
(B) RNA Interference, where miRNA and siRNA can complex with mRNA and prevent translation and/or cause degradation.
(C) RNA Translocation, where mRNA may be trafficked to specific parts of the cell, and would only be translated at those specific places.
(D) All three of these are valid Eukaryotic Transcriptional Regulation mechanisms.
(D) All three of these are valid Eukaryotic Transcriptional Regulation mechanisms.
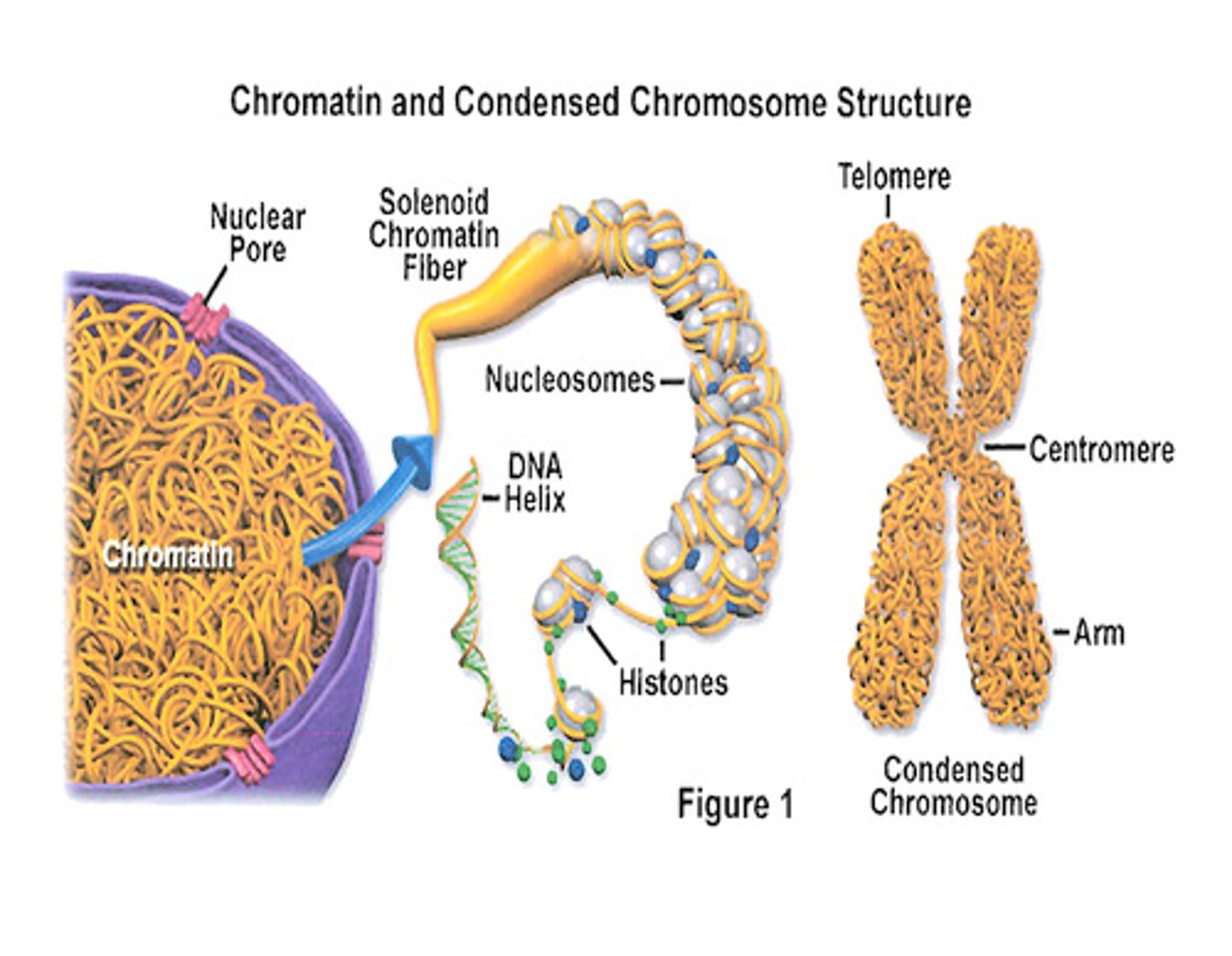
What is the difference between chromatin and chromosomes?
Chromatin is composed of DNA that is wrapped around histone proteins and can be described as "bead-like" structures in the nucleus.
A chromosome (supercoiled DNA) is what chromatin is condensed into in preparation for cell division.
Which of the following is not
a major component of chromatin?
(A) DNA
(B) Proteins
(C) RNA
(D) Histones
(C) RNA
Chromatin is made up of DNA, histones, and non-histone proteins.
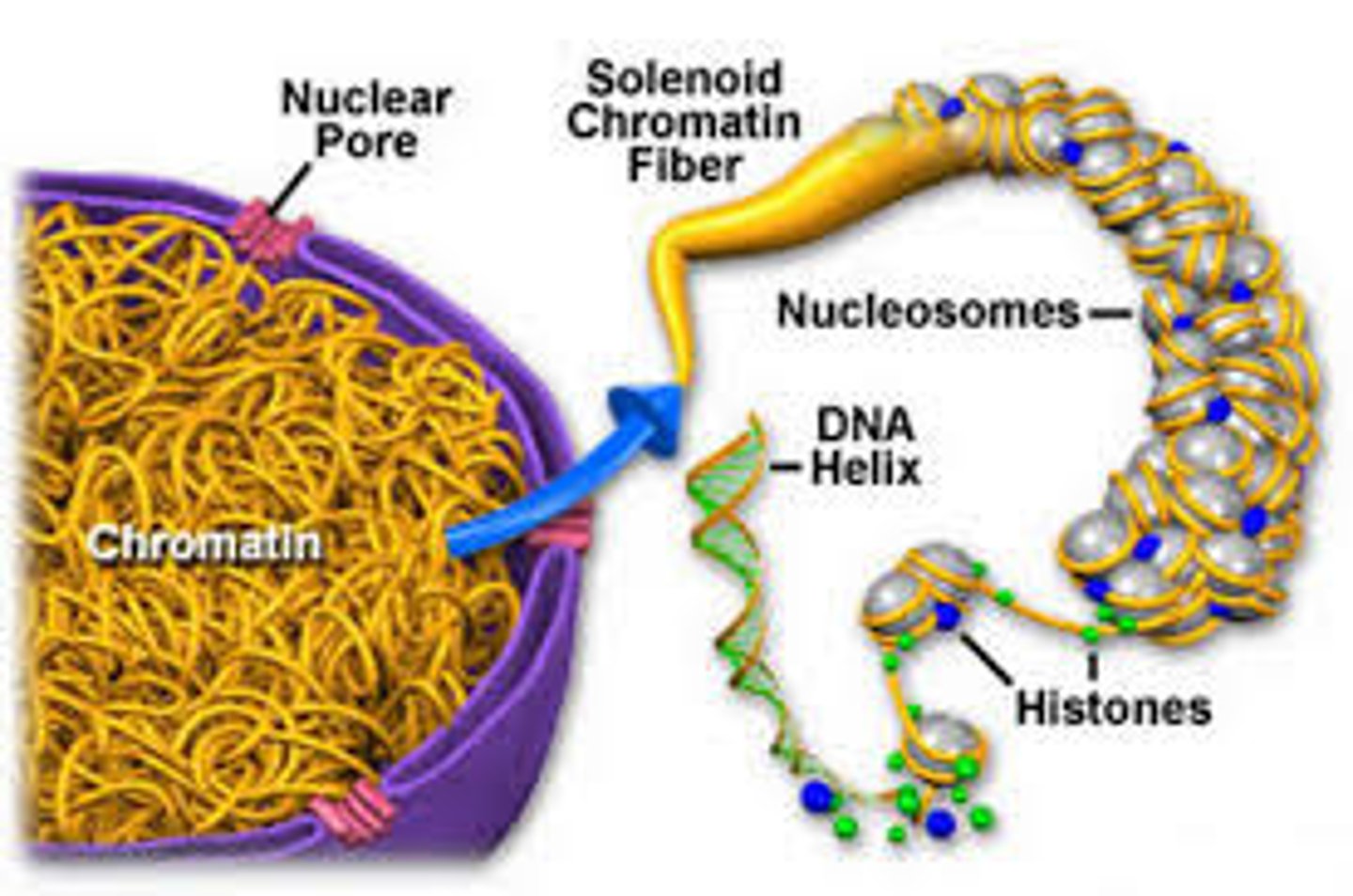
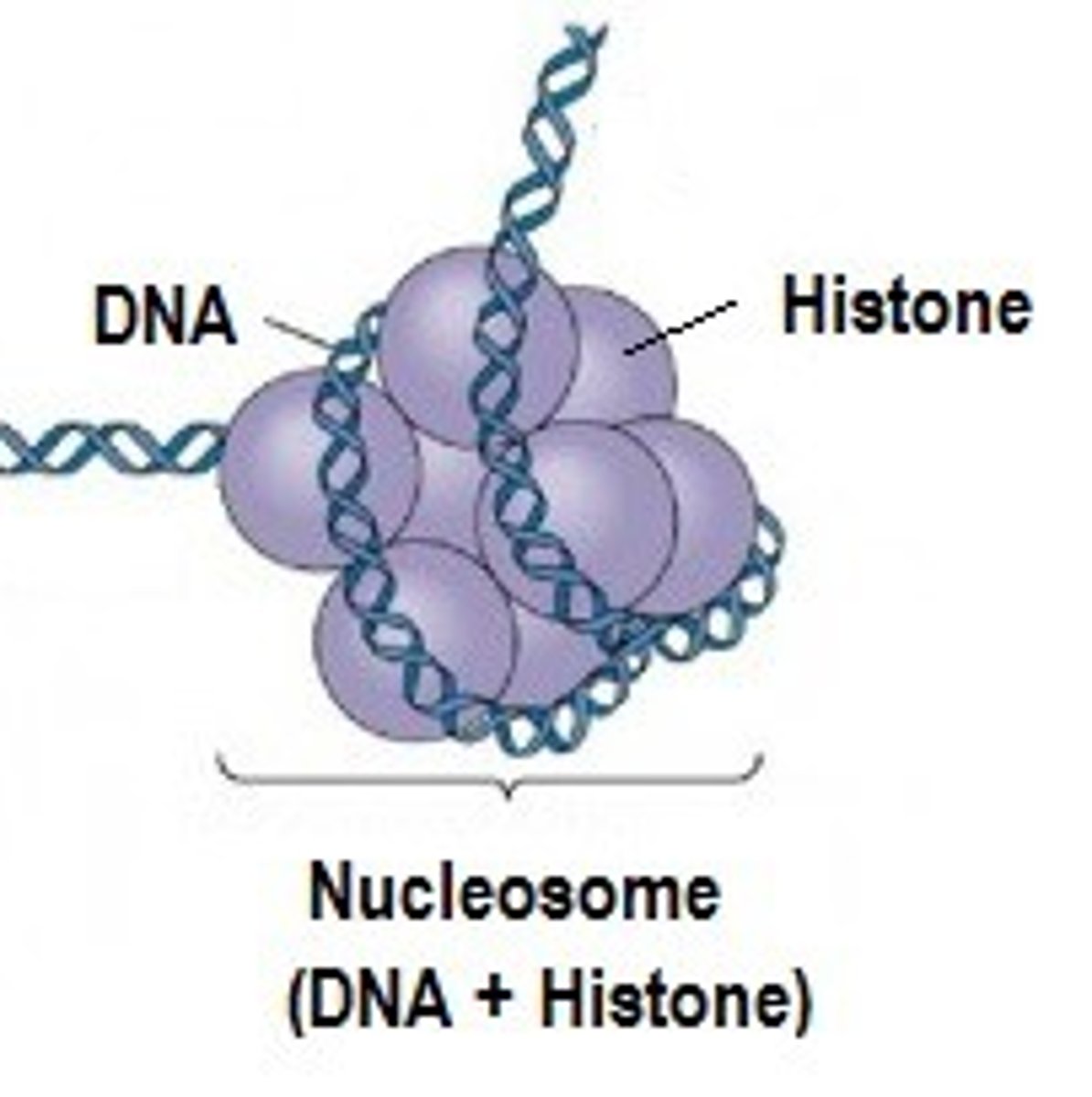
How do nucleosomes relate to chromatin?
Nucleosomes are the repeating unit of chromatin. They are composed of a DNA helix wrapped around a core of 8 histone proteins.
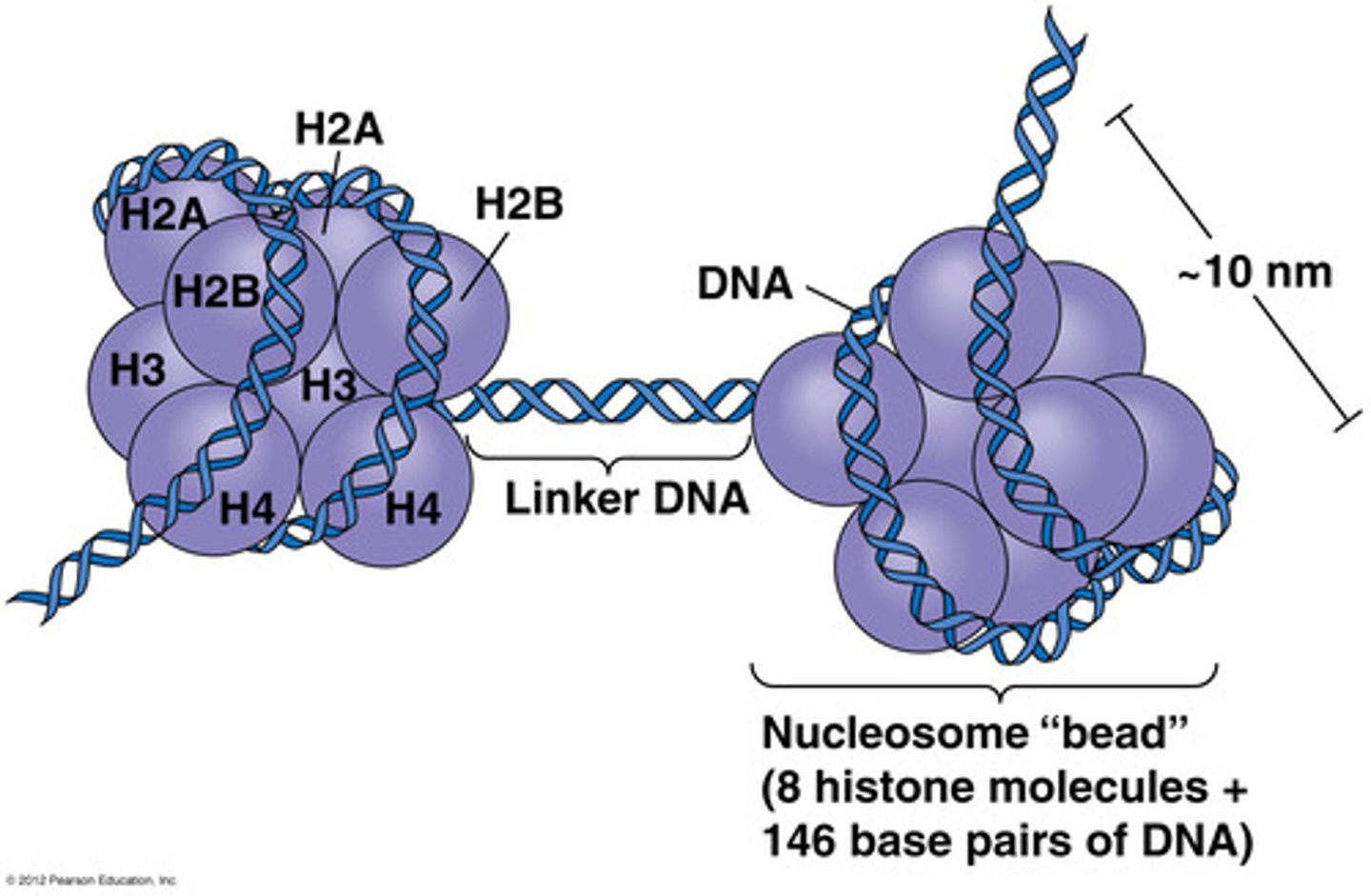
What 4 histone proteins are part of the histone core of a nucleosome?
H2A, H2B, H3, and H4 are the histone proteins that are part of the histone core of a nucleosome.
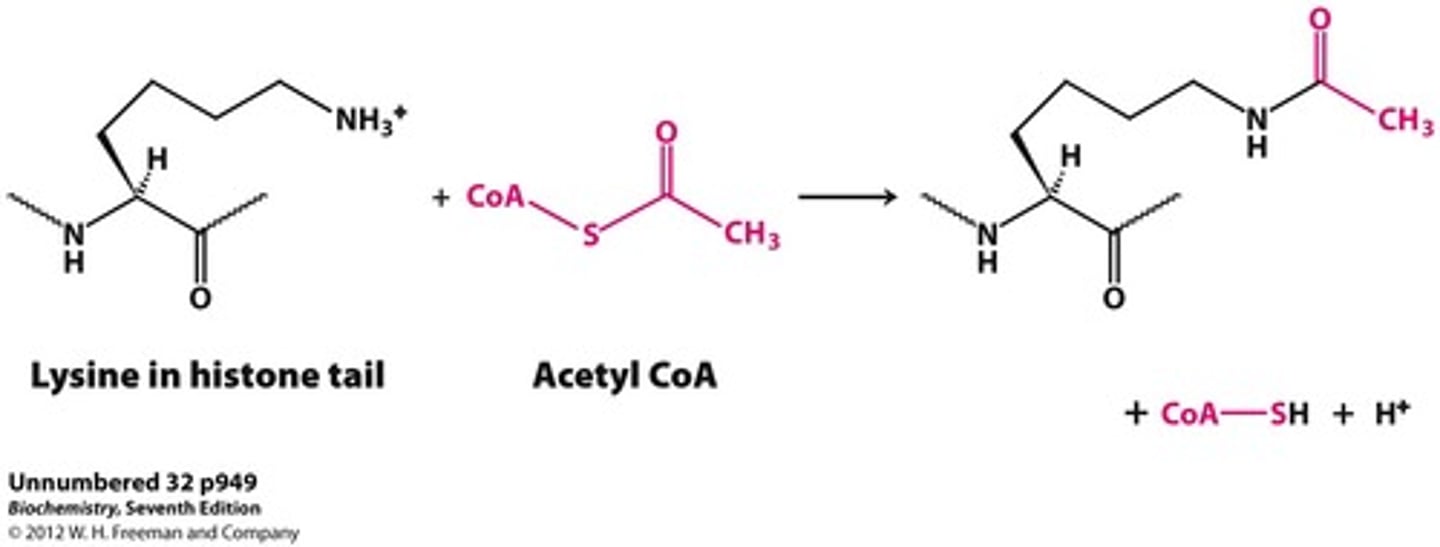
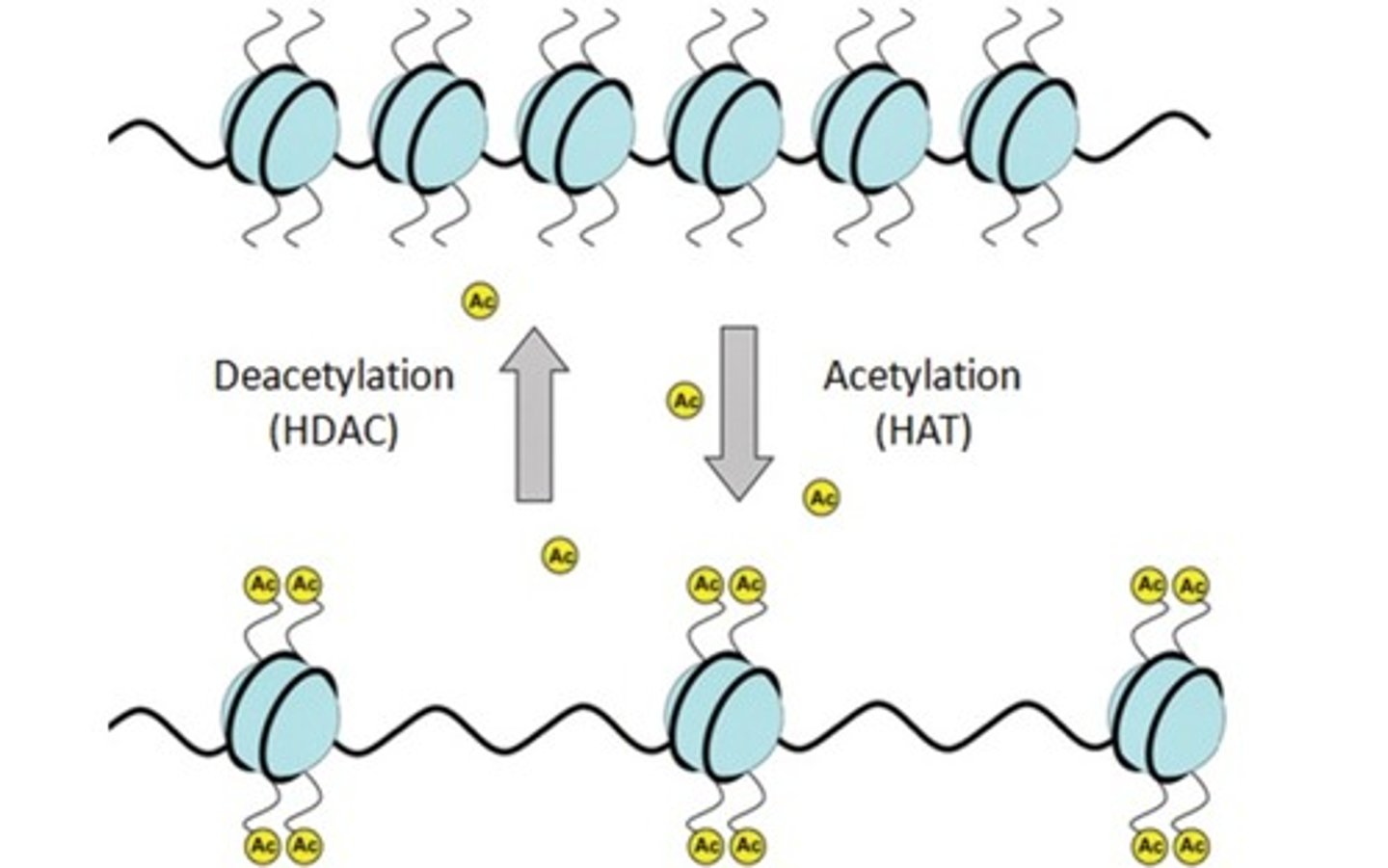
What does histone acetyltransferase (HAT) do? What does histone deacetylase (HDAC) do?
Histone acetyltransferase (HAT) is the enzyme responsible for adding acetyl groups to the Lysine residues of histone proteins.
Histone deacetylase (HDAC) is the enzyme responsible for removing acetyl groups from the Lysine residues of histone proteins.
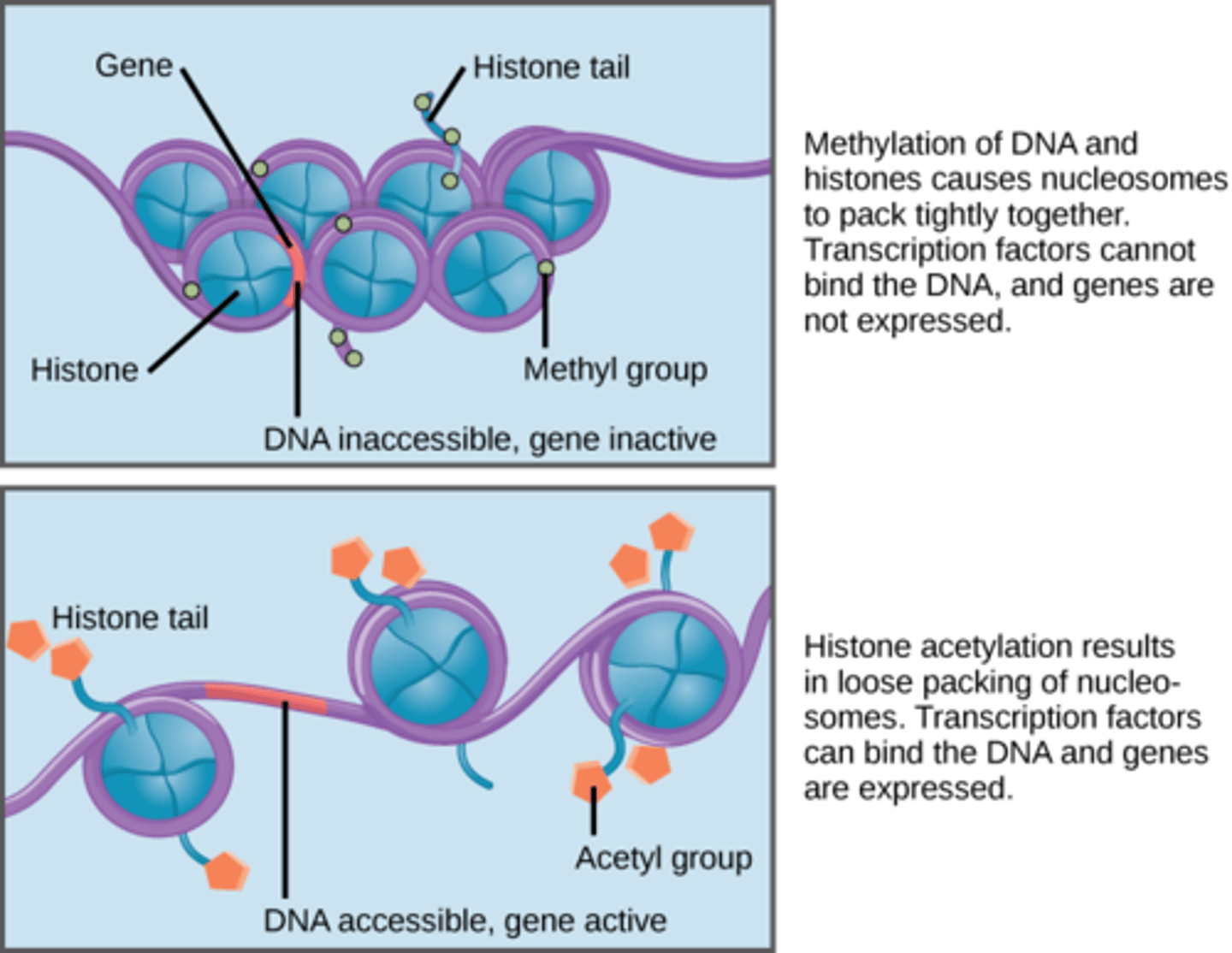
What happens to nucleosomes when histone proteins are acetylated? How does this effect gene expression?
What happens to nucleosomes when histone proteins are deacetylated? How does this effect gene expression?
Acetylation leads to gene expression. When histone proteins are acetylated, the histone proteins uncoil, thus allowing the transcriptional machinery to gain access to the DNA and express those genes.
Deacetylation prevents gene expression. When histone proteins are deacetylated histone proteins coil into a more compact structure, preventing the transcriptional machinery from accessing the DNA and expressing those genes.
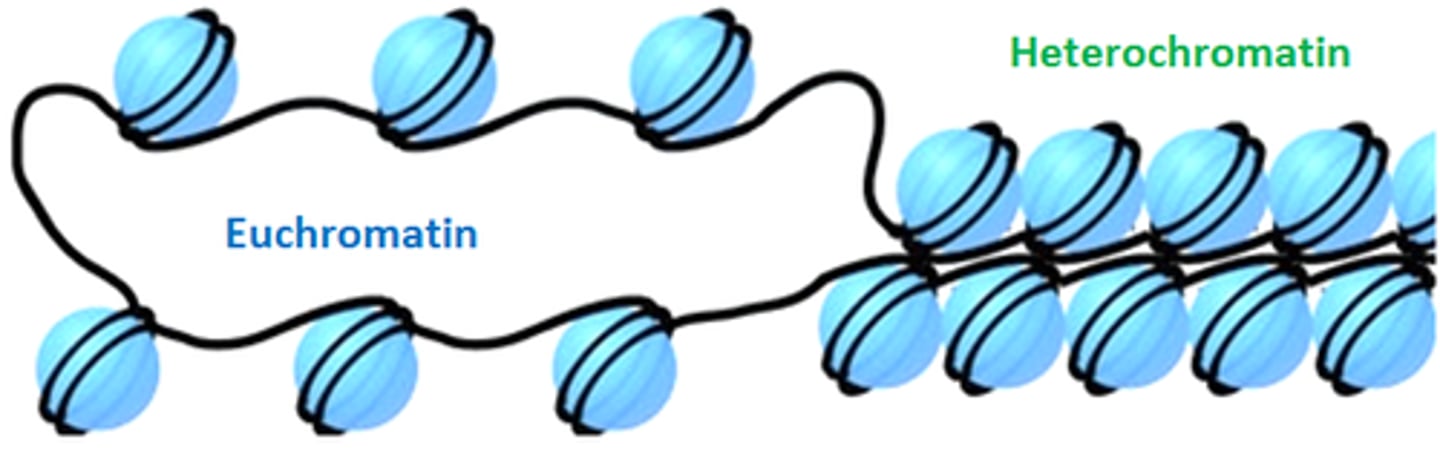
What is the difference between heterochromatin and euchromatin?
Heterochromatin is densely packed transcriptionally inactive DNA.
Euchromatin is less dense transcriptionally active DNA.
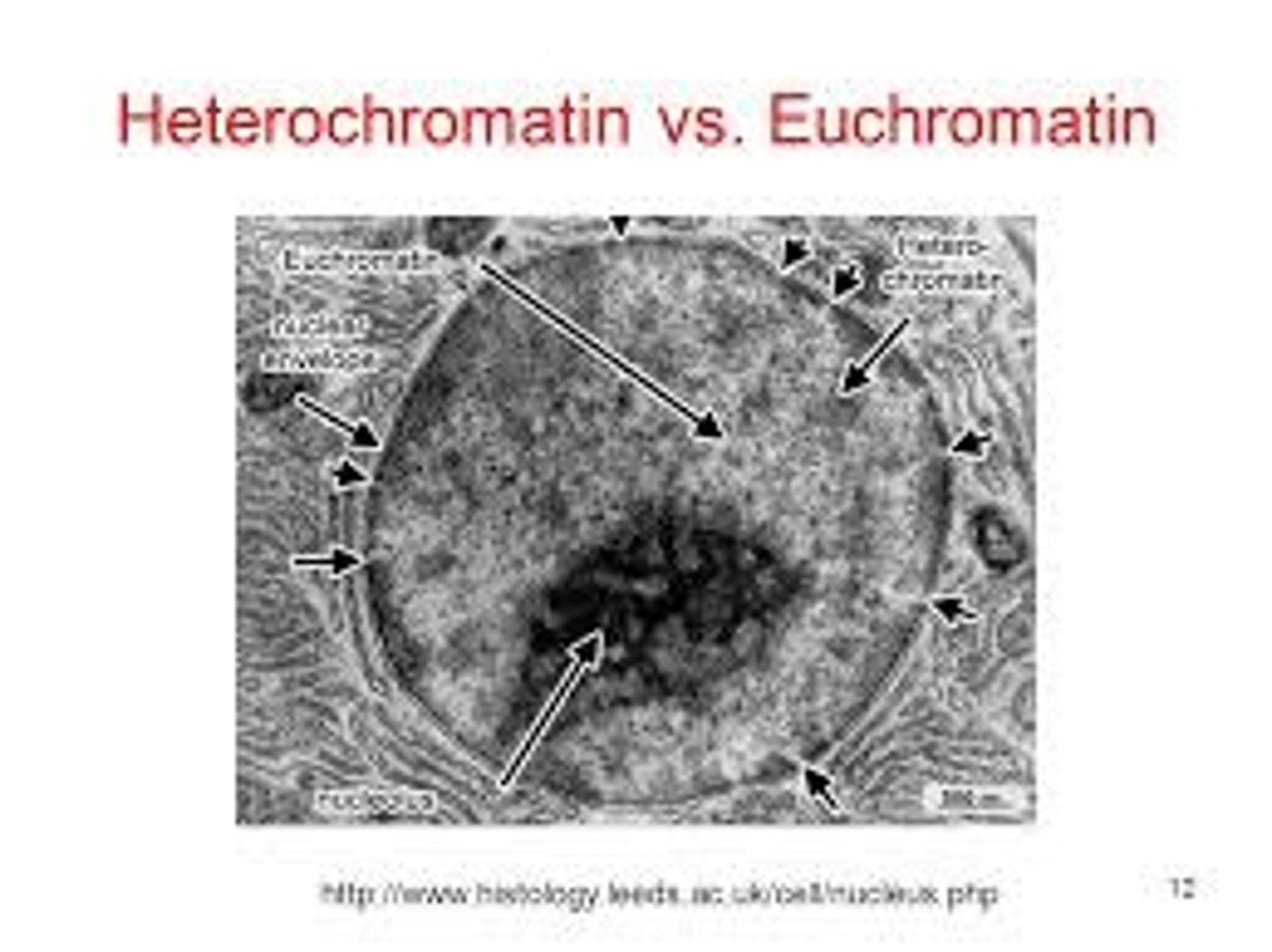
You look through a microscope at DNA and see dark spots and light spots. Which is heterochromatin and which is euchromatin?
The dark spots are heterochromatin because they are more dense. The light spots are euchromatin becuase they are less dense.
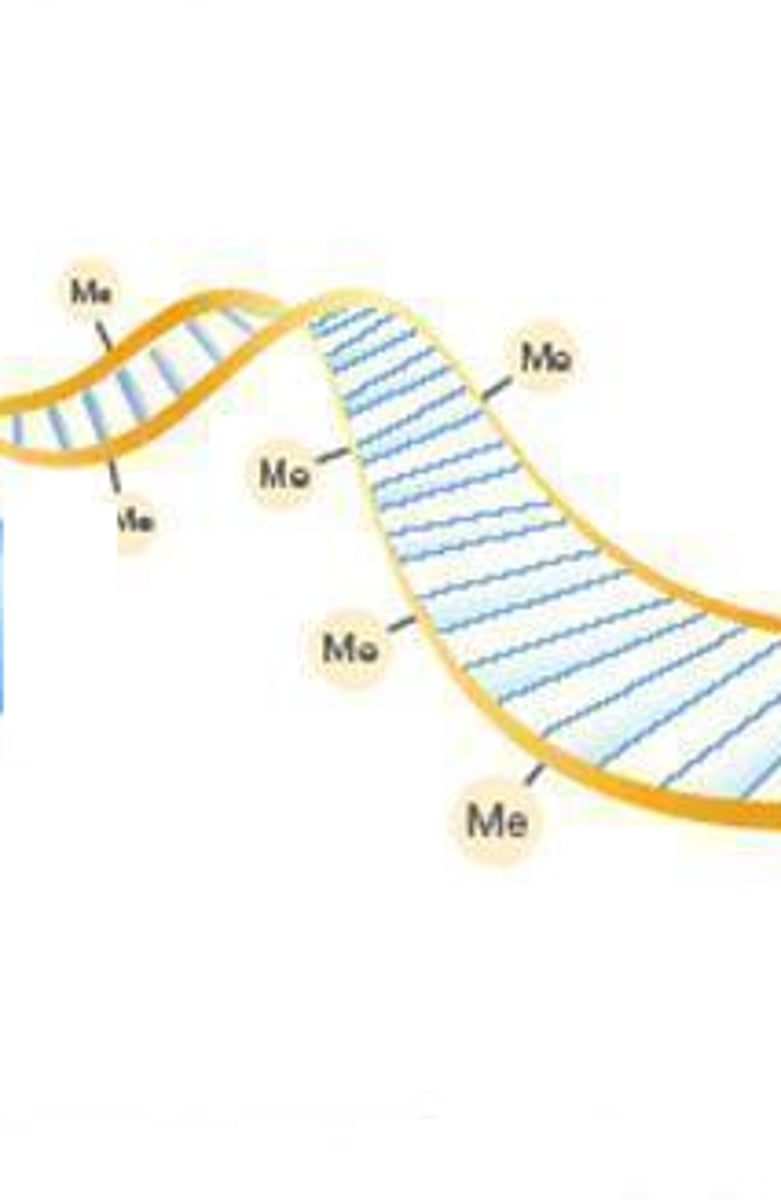
What does methyltransferase do? How does this affect gene expression?
Methyltransferase is the enzyme responsible for adding methyl groups to DNA. This prevents the expression of those genes.
What is the similarity and difference between deacetylation and methylation?
Both deacetylation and methylation inactivate the expression of DNA.
The difference is that methylation is a permanent method of down-regulating genes while deacetylation is a reversible mechanism.
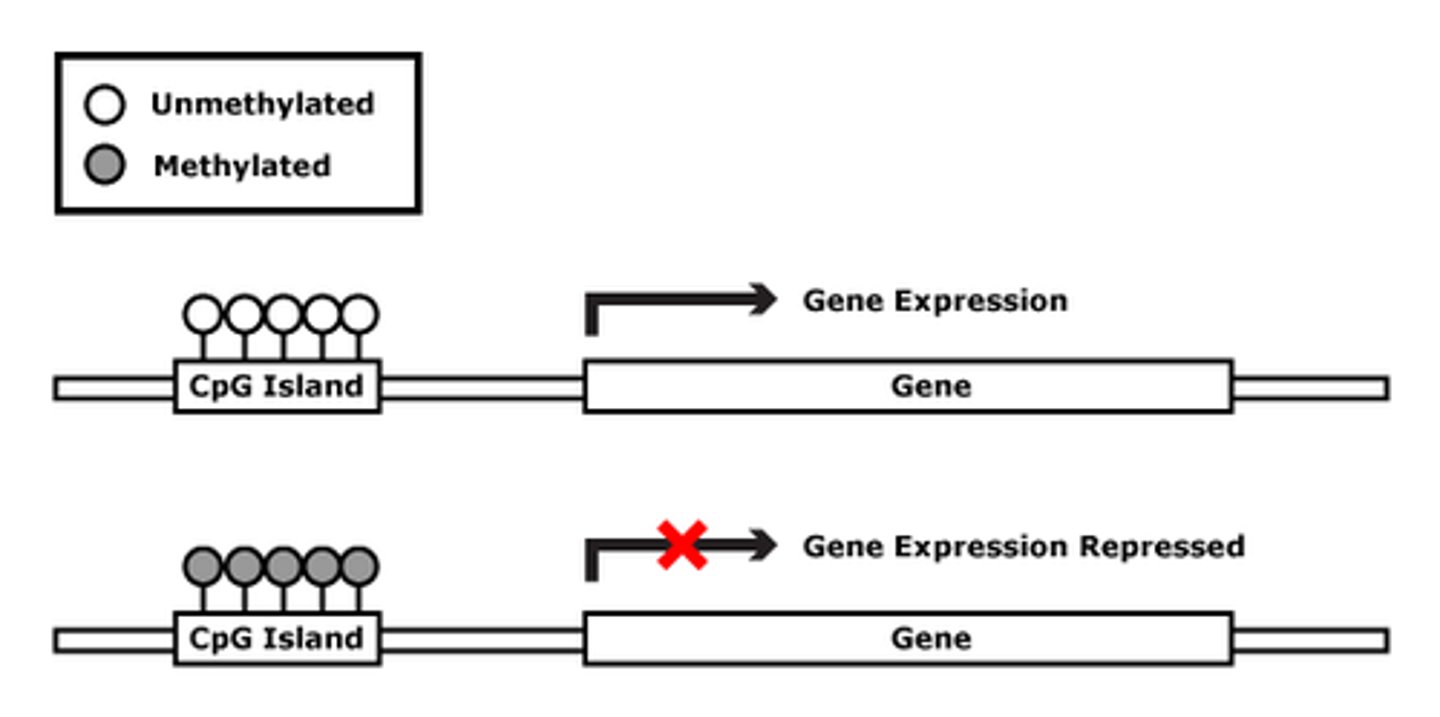
Methylation typically occurs in the DNA regions rich in which nucleotide?
(A) A
(B) U
(C) T
(D) C
(D) C
Methylation is most common in cytosine and guanine rich sequences called CpG Islands.
As cells divide and differentiate from stem cells into specific tissues which gene regulatory mechanism is commonly involved? Why?
(A) Acetylation
(B) Deacetylation
(C) Methylation
(D) Both A and B
(C) Methylation
Methylation is involved as cells divide and differentiate from stem cells into specific tissues because methylation alters gene expression in a stable (more permanent) manner.
CRB True or false? DNA Methylation has been seen in every vertebrate genome studied to date.
True. DNA Methylation has been seen in every vertebrate genome studied to date.
Which DNA regulatory mechanism is associated with genomic imprinting and x-chromosome inactivation?
(A) Acetylation
(B) Deacetylation
(C) Methylation
(B) Both A and B
(C) Methylation
Methylation is associated with genomic imprinting and x-chromosome inactivation.
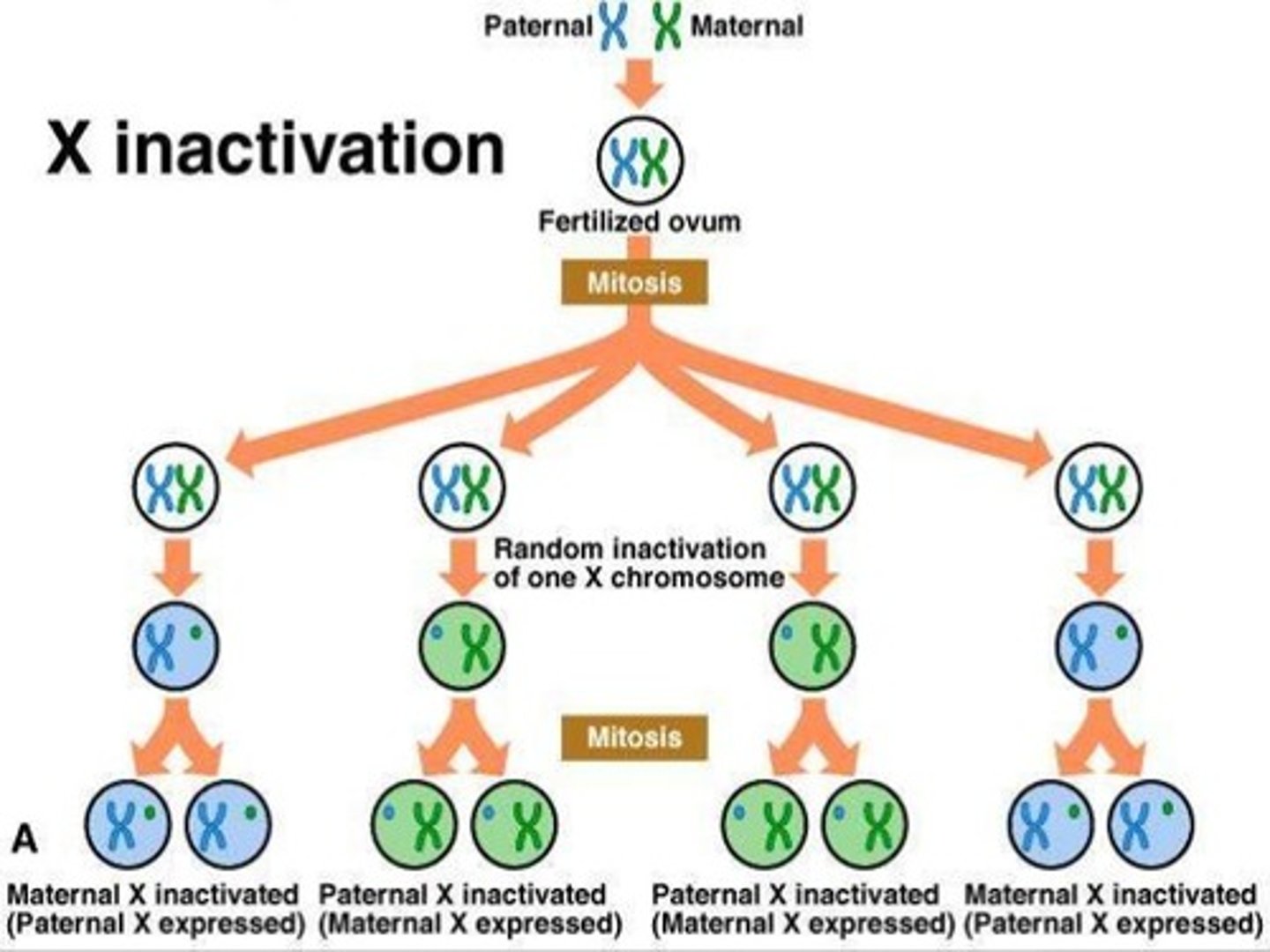
CRB Which of the following statements about X-inactivation is FALSE?
(A) X-inactivation mechanisms vary across different animals.
(B) Some marsupials always have the paternal-X chromosome inactivated.
(C) An inactivated chromosome is expected to be very condensed and methylated.
(D) None of these statements are false.
(D) None of these statements are false.
CRB True or false? Recently, non-coding RNA passed in the gametes has also been shown to affect genomic imprinting.
True. Recently, non-coding RNA passed in the gametes has also been shown to affect genomic imprinting.
CRB Which of the following statements about Genomic Imprinting is true?
(A) Genomic Imprinting is stable from generation to generation.
(B) Genomic imprinting is typically free of Epigenetic Marks.
(C) Genes that undergo Genomic Imprinting tend to be clustered together in the genome.
(D) More than one of these statements are true.
(C) Genes that undergo Genomic Imprinting tend to be clustered together in the genome.
CRB Which of the following terms best describes the focus on gene expression levels that can be heritable (but not caused by changing DNA sequences) or have long-term effects?
(A) Imprinting
(B) Inactivation
(C) Epigenetics
(D) Mendelian Inheritance
(C) Epigenetics
Epigenetics refers to the changes in gene expression that are heritable (but not caused by changing DNA sequences) or have long-term effects.
True or False? Abnormal DNA methylation can lead to carcinogenesis.
True. Abnormal DNA methylation can lead to carcinogenesis (cancer).
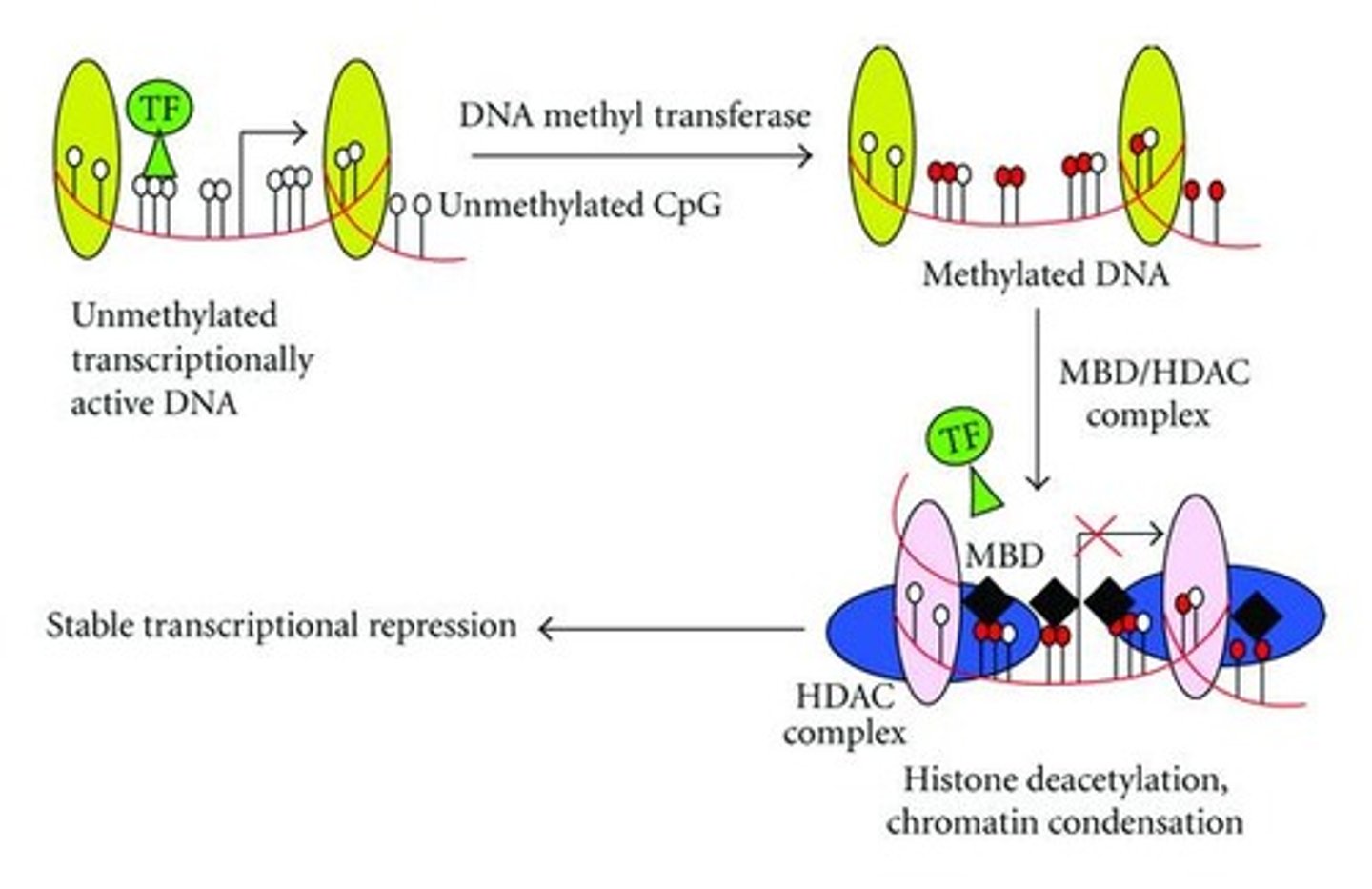
How does methylation affect DNA transcription directly? Indirectly?
Methylation affects DNA transcription directly by impeding the binding of transcriptional proteins on the genes.
Methylation can affect DNA transcription indirectly by attracting proteins called methyl cpg-binding domain proteins (MBDs), which recruit additional chromatin remodeling proteins such as histone deacetylases (HDACs) to the gene site, resulting in the condensation of the chromatin into heterochromatin.
Fill in the blanks: Multiple General Transcription Factors (also called Basal Transcription Factors) are needed for transcription to occur in ____________, whereas only one General Transcription Factor, called Sigma Factor, is needed for Transcription in ____________.
(A) Eukaryotes, Prokaryotes
(B) Eukaryotes and Prokaryotes, Archaea
(C) Eukaryotes, Prokaryotes and Archaea
(D) Eukaryotes and Archaea, Prokaryotes
(D) Eukaryotes and Archaea, Prokaryotes.
Multiple General Transcription Factors (also called Basal Transcription Factors) are needed for transcription to occur in Eukaryotes and Archaea, whereas only one General Transcription Factor, called Sigma Factor, is needed for Transcription in Prokaryotes.
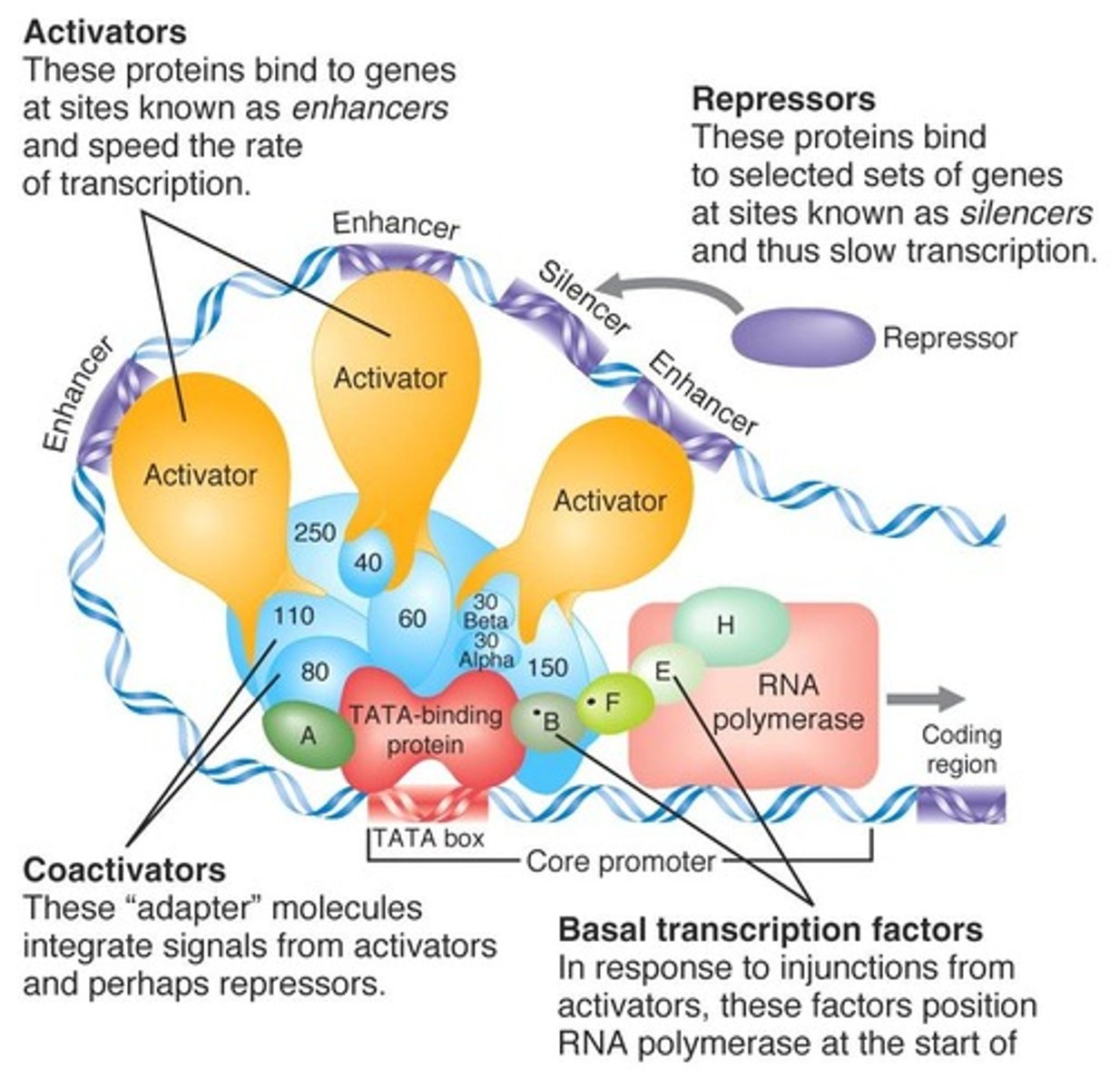
Which of the following is NOT a component of the basic transcription apparatus?
(A) General transcription factors
(B) Enhancers
(C) RNA polymerase
(D) mediator multiple protein complex
(B) Enhancers
General transcription factors (GTFs), RNA polymerase, and mediator multiple protein complex all constitute the basic transcription apparatus found in prokaryotic cells.
Enhancers are DNA sequences, not proteins!

The basic transcription apparatus is found in prokaryotic or eukaryotic cells?
The basic transcription apparatus is found in prokaryotic cells.
CRB True or false? Some prokaryote promoters are more effective than others, and this helps regulate transcription levels.
True. Some prokaryote promoters are more effective than others, and this helps regulate transcription levels.
CRB Explain how having promoter strengths as the main mode of regulating transcription could cause issues.
If transcription is only regulated by the strength of the promoter, then there is no way to modify transcription in response to environmental stressors. This could be very dangerous!
Because of this, most prokaryotes have other transcription mechanisms too, like the Lac-Operon.
What is the role of an activator protein?
The DNA binding protein activator is responsible for enhancing the interaction of RNA polymerase and a particular promoter thus encouraging the expression of a gene.
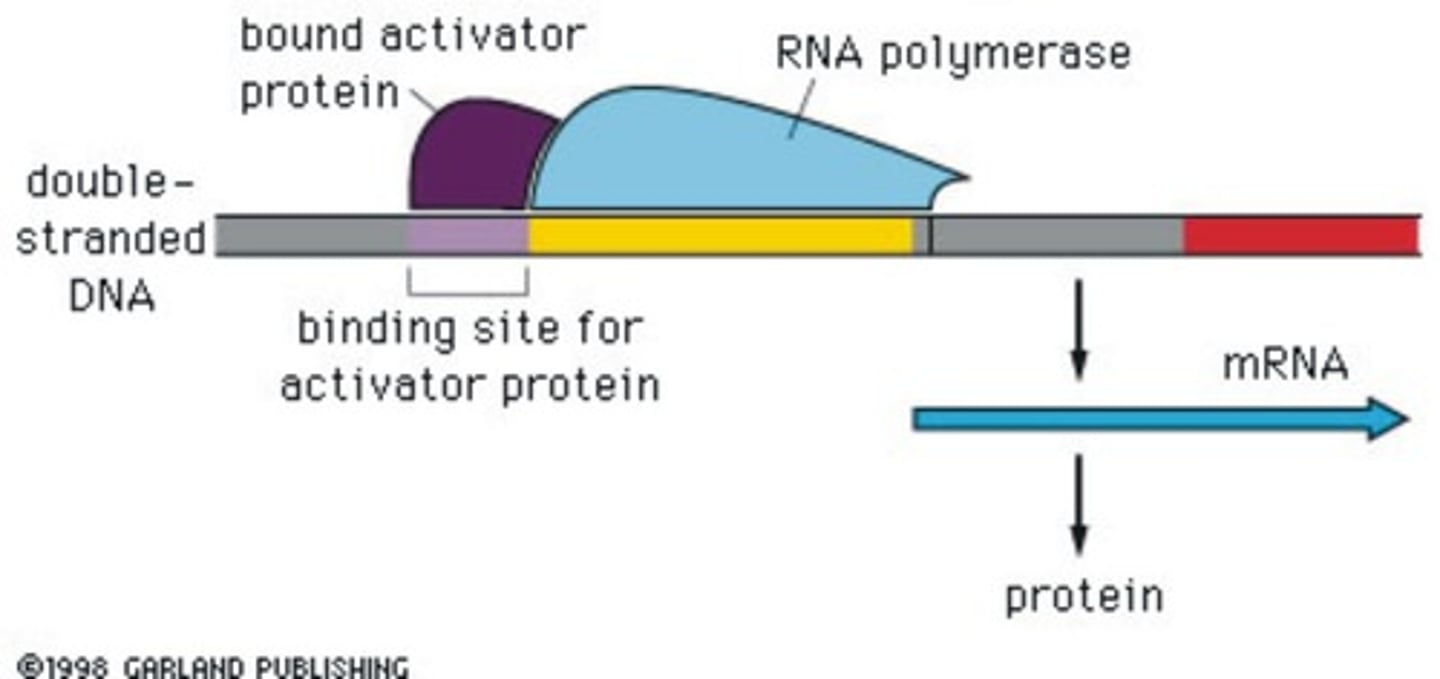
CAP is an example of an activator protein. Describe the role and mechanism of action of Catabolite Activator Protein (CAP) in e. Coli.
In E. Coli, cyclic adenosine monophosphate (cAMP) is produced during glucose starvation. Consequently, cAMP binds to a protein called CAP which causes a conformational change that allows the CAP protein to bind to a DNA site adjacent to the promoter. Then the CAP protein recruits RNA polymerase to the promoter..
It is an example of an activator protein because it interacts with the RNA polymerase to help it attach to the promoter sequence thus encouraging the expression of a gene.
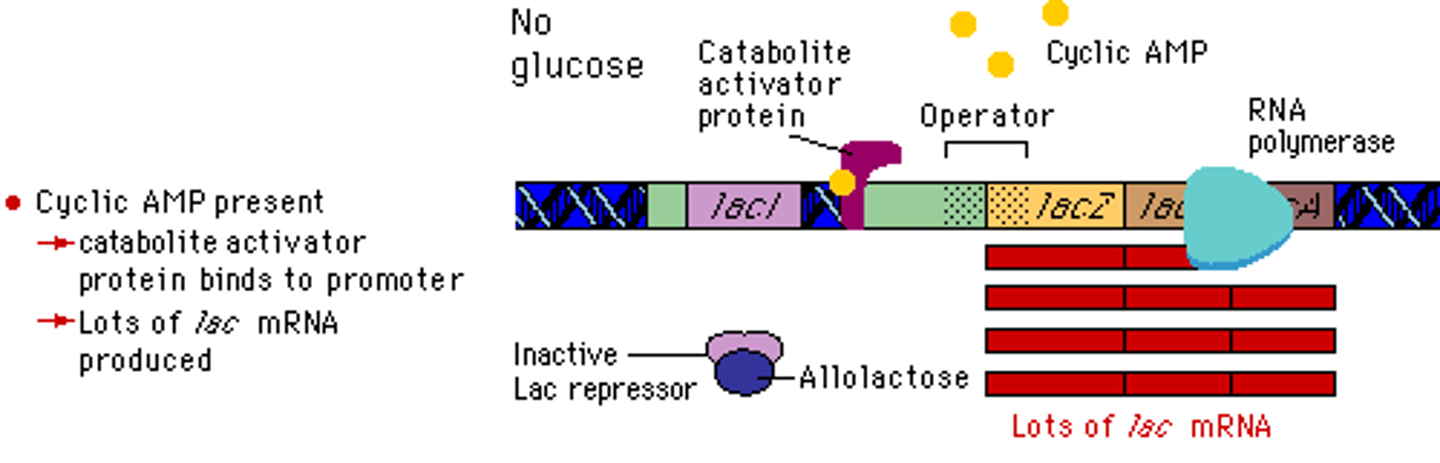
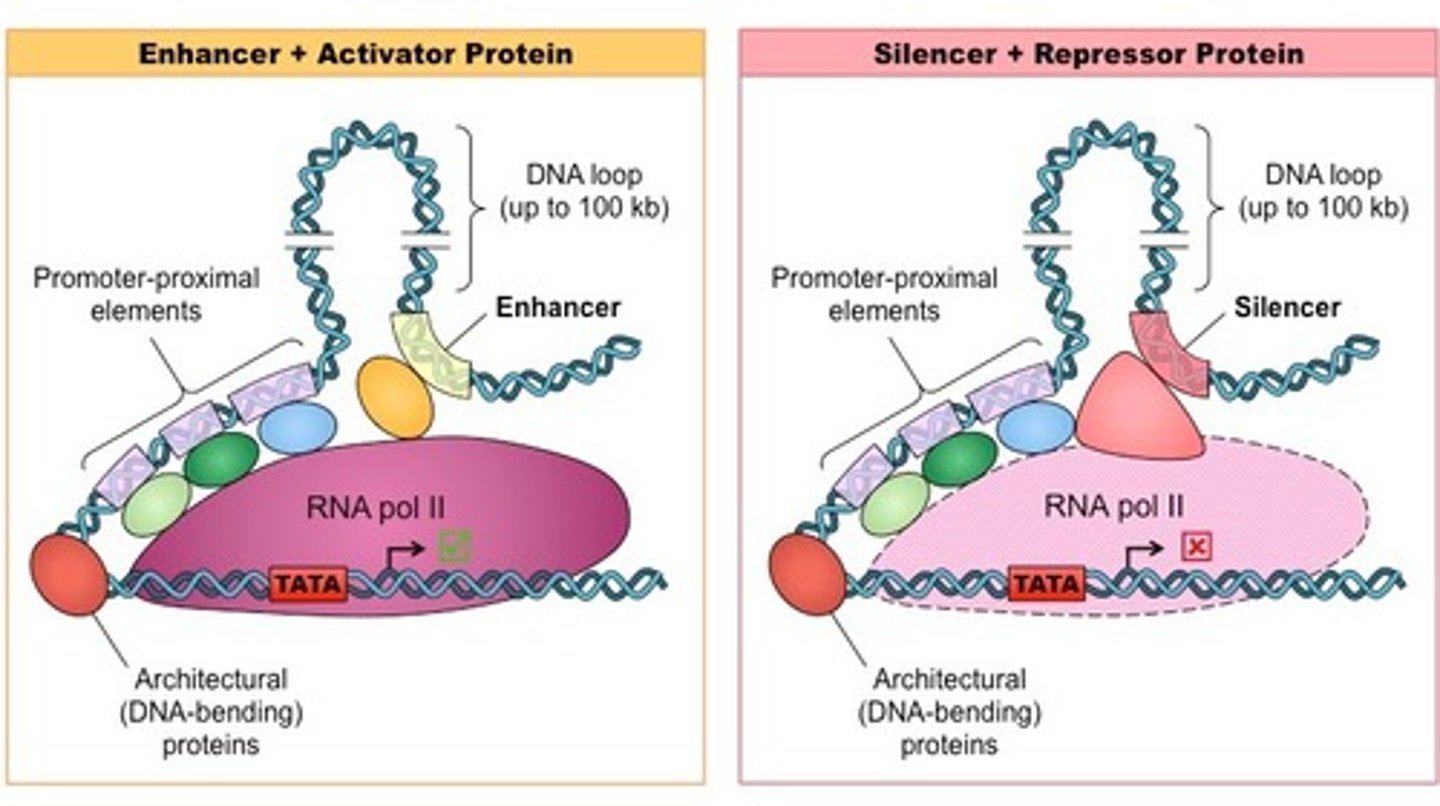
Compare enhancers and promoters.
Enhancers sequences of DNA that are distant from the gene(s) they regulate. They are bound by activators which loop the DNA in a certain way, bringing other transcription factors and mediator proteins to the promotor, enhancing the expression of that DNA.
Promotors, on the other hand, are sequences of DNA located immediately upstream from the gene(s) that they regulate. RNA Polymerase and activator proteins may bind here.
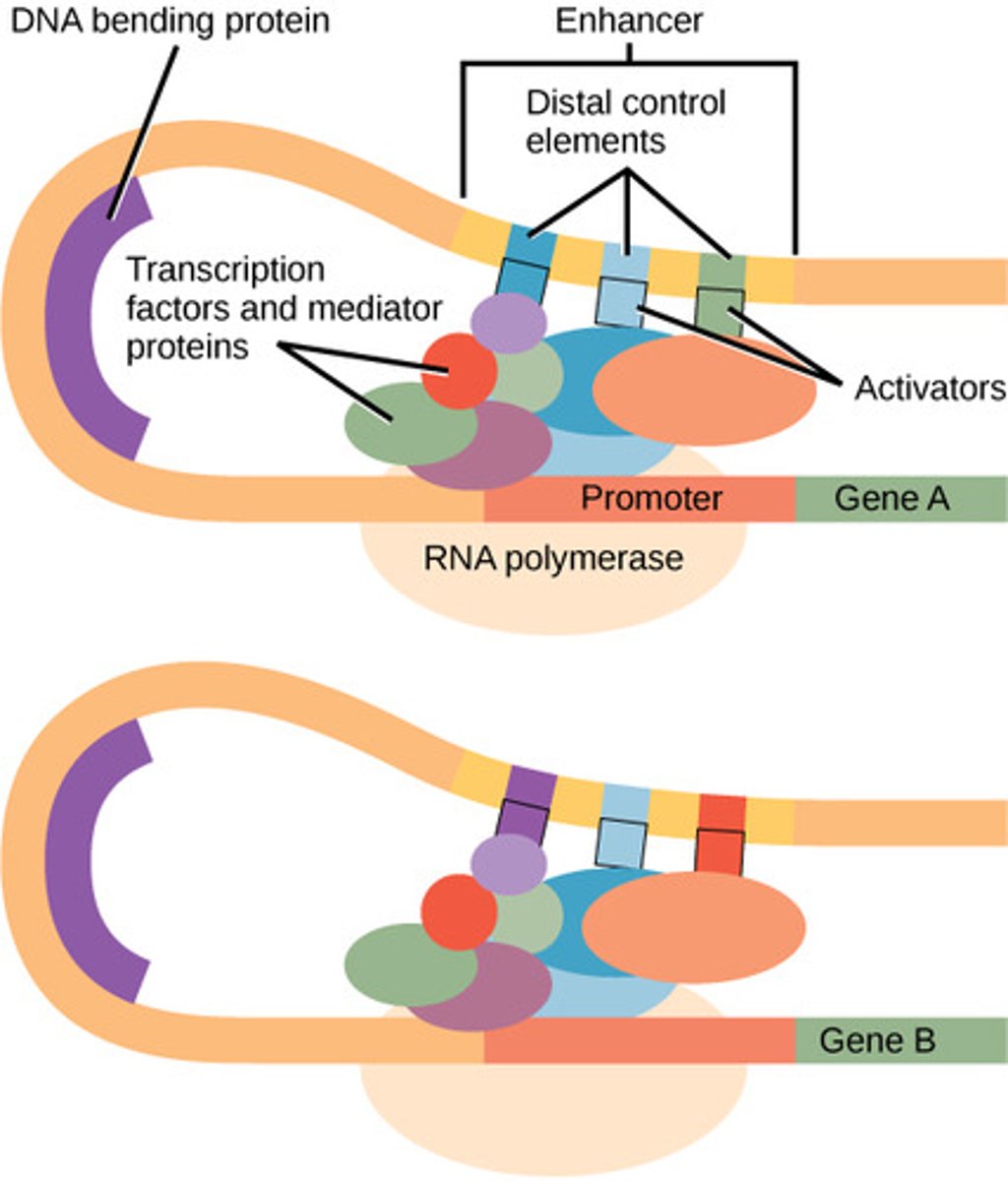
True or False? Enhancers do not need to be cis-acting (on the same chromosome as the promoter it will work on).
True. Enhancers do not need to be cis-acting (on the same chromosome as the promoter it will work on). Enhancers can affect the expression of distant genes.
Compare silencers and operators.
Silencers are regions of DNA that are distant from the gene(s) they regulate. When a repressor protein binds to these sequences, RNA Polymerase is prevented from binding to the promoter.
Operators are regions of DNA that are located directly upstream from the gene(s) they regulate. When a repressor protein binds to these sequences, RNA Polymerase is unable to transcribe the genes.
Which of the following is not a possible way to increase/enhance transcription?
(A) Increase activators
(B) Increase transcription factors
(C) Increase DNA enhancers
(D) Increase histone acetylation
(C) Increase DNA enhancers
DNA is not increased or decreased; thus, you cannot increase the amount of enhancers, promoters, silencers, or operators. Only the expression of DNA can be modified, which is done via increasing/decreasing the concentration of regulatory proteins or using post-translational modifications of those proteins.
True or False? DNA transcription is not regulated in prokaryotes.
False. In prokaryotes, the regulation of transcription is needed for the cell to be able to quickly adapt to the changing environment; thus, a combination of activators, repressors, and on rare occassion enhancers determine whether genes are transcribed.
How is transcription regulated in eukaryotes as compared to prokaryotes?
In eukaryotes, transcriptional regulation involves a combination of interactions between several transcription factors which allows for a more sophisticated response. In addition, eukaryotes have a spatial and temporal control of gene expression due to the nuclear envelope.
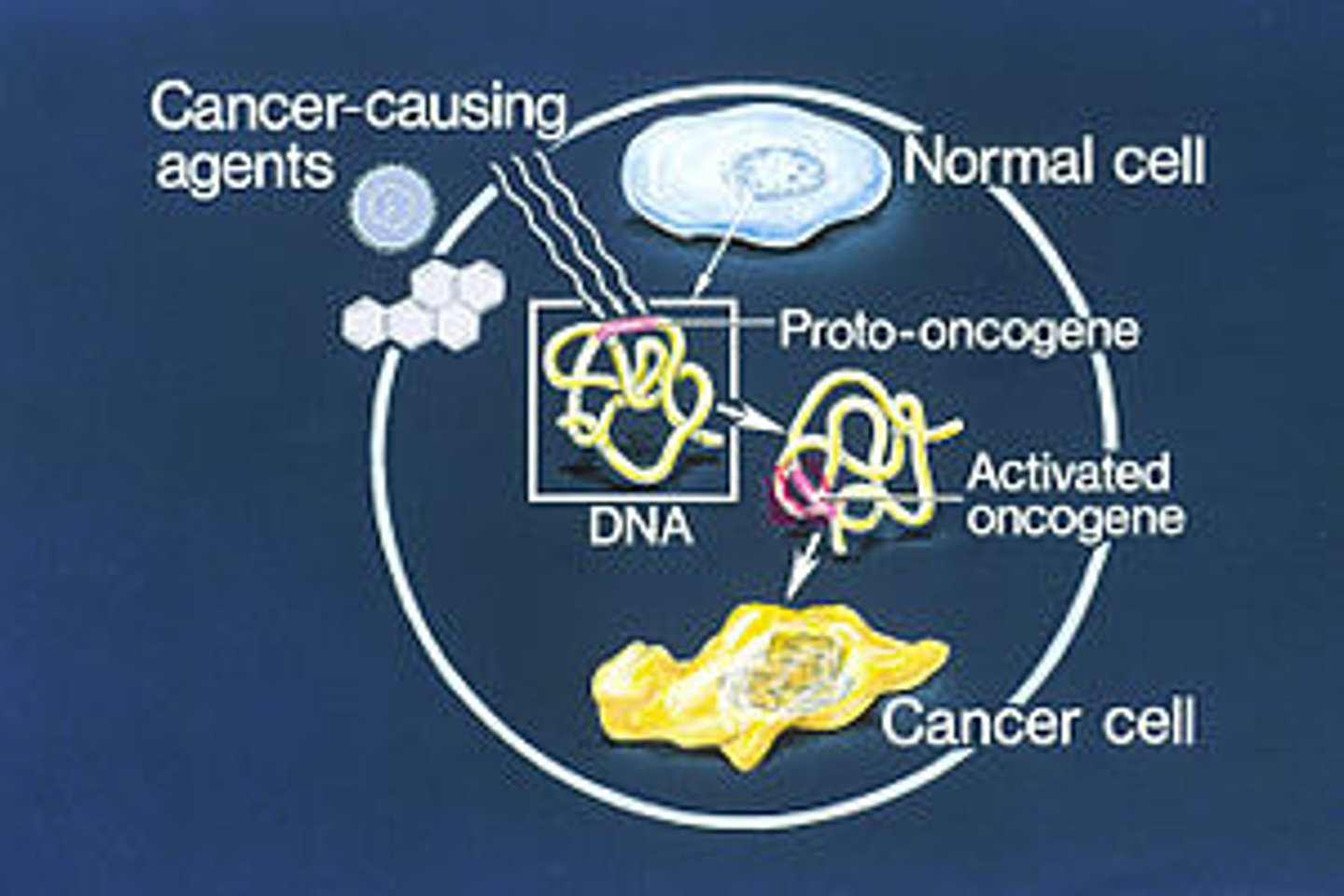
Compare oncogenes and proto-oncogenes.
A proto-oncogene is a gene that codes for a protein involved in cell growth. When a proto-oncogene is mutated it is called an oncogene and causes uncontrollable cell growth.
What are mitogens?
Mitogens are chemical substances that encourage the cell to start cell division.
Which of the following are mechanisms by which a proto-oncogene may become an oncogene?
I. Deletion/point mutation
II. Gene amplification and/or increased mRNA stability
III. Chromosomal rearrangement
(A) I and II Only
(B) I and III Only
(C) II and III Only
(D) I, II, and III
(D) I, II, and III
The following are mechanisms by which a proto-oncogene may become an oncogene:
I. Deletion/point mutation
II. Gene amplification and/or increased mRNA stability
III. Chromosomal rearrangement
How might a deletion or point mutation in a proto-oncogene cause cancer?
A deletion/point mutation may cause the proto-oncogene to begin producing hyperactive protein in normal amounts or produce a normal protein that is over expressed. In either case, because the protein is involved in cell growth, its increased activity/presence will result in too much cell growth (aka cancer).
How might gene amplification and/or increased mRNA stability of a proto-oncogene cause cancer?
If a gene amplification or increased mRNA stability occurs then the proto-oncogene's protein will be produced in higher than normal levels, resulting in too much cell growth (aka cancer).
How might chromosomal rearrangement cause cancer?
Chromosomal rearrangement may result in the proto-oncogene landing in a location within the chromosome where it will be overexpressed, resulting in too much cell growth (aka cancer).
How might the mutation of a Tyrosine Kinase cause cancer?
Tyrosine Kinases are often involved in secondary signaling pathways. The Tyrosine kinase will often activate a protein, which will then activate another protein, and another, and another, and another until proteins that increase the expression of a gene are activated. If Tyrosine Kinases are mutated it could result in the overexpression of these genes, which ,if they happen to be proto-oncogenes, could cause cancer!
Compare tumor suppressor genes and proto-oncogenes.
Tumor suppressor genes produce protein enzymes that either halt the cell cycle and/or promote apoptosis (decreasing or halting cell growth/division).
Proto-oncogenes are the opposite in that they produce proteins that enhance or promote cell growth/division.
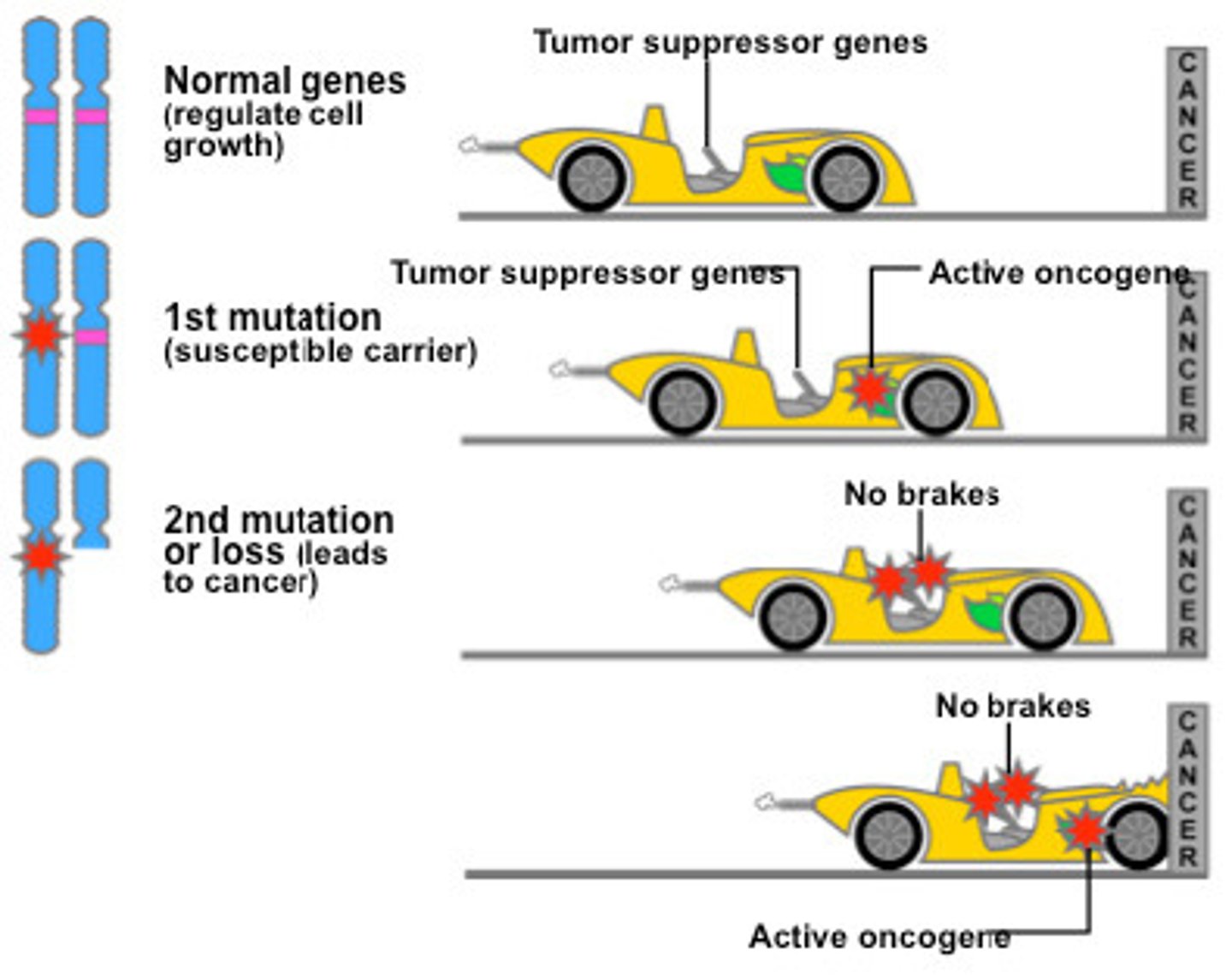
Some tumor suppressor genes code for proteins that can recognize DNA damage and repair it. How might the mutation of this gene cause cancer?
If this type of tumor suppressor gene is mutated, it result in the continued mutation of DNA. Cancer typically entails uncontrolled cell growth and DNA mutations causing the cells to be abnormal and dangerous.
Some tumor suppressor genes code for proteins that can recognize DNA damage and initiate apoptosis. How might the mutation of this gene cause cancer?
A cell will initiate apoptosis if it has an invader or if it thinks it may be cancerous. This prevents the cell from growing out of control. Without apoptosis, cancer will likely result.
Some tumor suppressor genes code for proteins that act as repressors for proto-oncogenes. How might the mutation of this kind of tumor suppressor gene cause cancer?
If the repressor does not function properly, the proto-oncogene will get turned on permanently, resulting in uncontrollable cell growth (aka cancer).
What is the Two-Hit Hypothesis?
The Two-Hit Hypothesis states that both alleles of a tumor suppressor gene must mutate for cancer to progress. The reason for this is that as long as you have one allele to produce the proteins needed to moderate/halt the cell cycle, that is good enough.
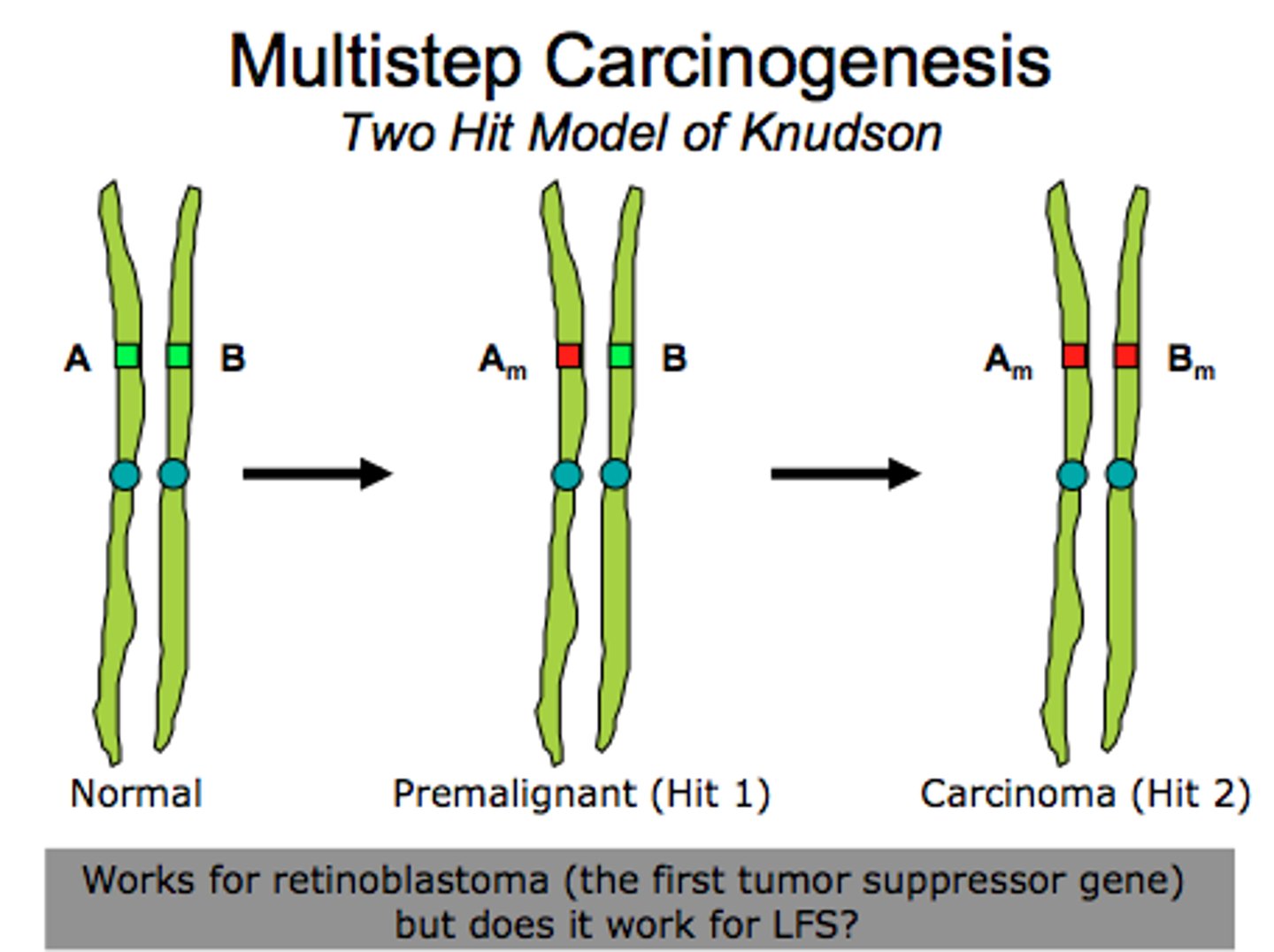
A mutation of one allele yields the cancerous phenotype. Which gene could this be?
(A) An Oncogene
(B) Tumor Suppresor Gene
(C) A and B
(D) None of the Above
(A) An Oncogene
A mutation of one oncogene allele yields the cancerous phenotype because oncogenes are dominant alleles. A significant exception to this is p53, which is a tumor suppressor gene that only requires one allele to be mutated to yield a cancerous phenotype. However, this is the only exception, so the answer is still generally "an oncogene"
What does the p53 tumor suppressor gene do when active?
The p53 tumor suppressor gene may prevent the cell from moving from the G1 phase to the S phase of the cell cycle when there is DNA damage. At this point, it may activate DNA repair proteins or initiate apoptosis.
The p53 tumor suppressor gene indirectly inhibits cyclin-CDK by activating what protein?
(A) p10
(B) p21
(C) p33
(D) p47
(B) p21
The p53 indirectly inhibits the cyclin-dependant kinase complex (cyclin-CDK) by activating protein called p21 which in turn then binds to and inactivates cyclin-CDK.
One exception to the Two Hit rule for tumor suppressor genes is the p53 rule known as the "dominant negative." Describe dominant negative.
Dominant negative is when a mutated protein from the mutated allele inhibits the function of the normal protein from the normal allele. This is common with p53.
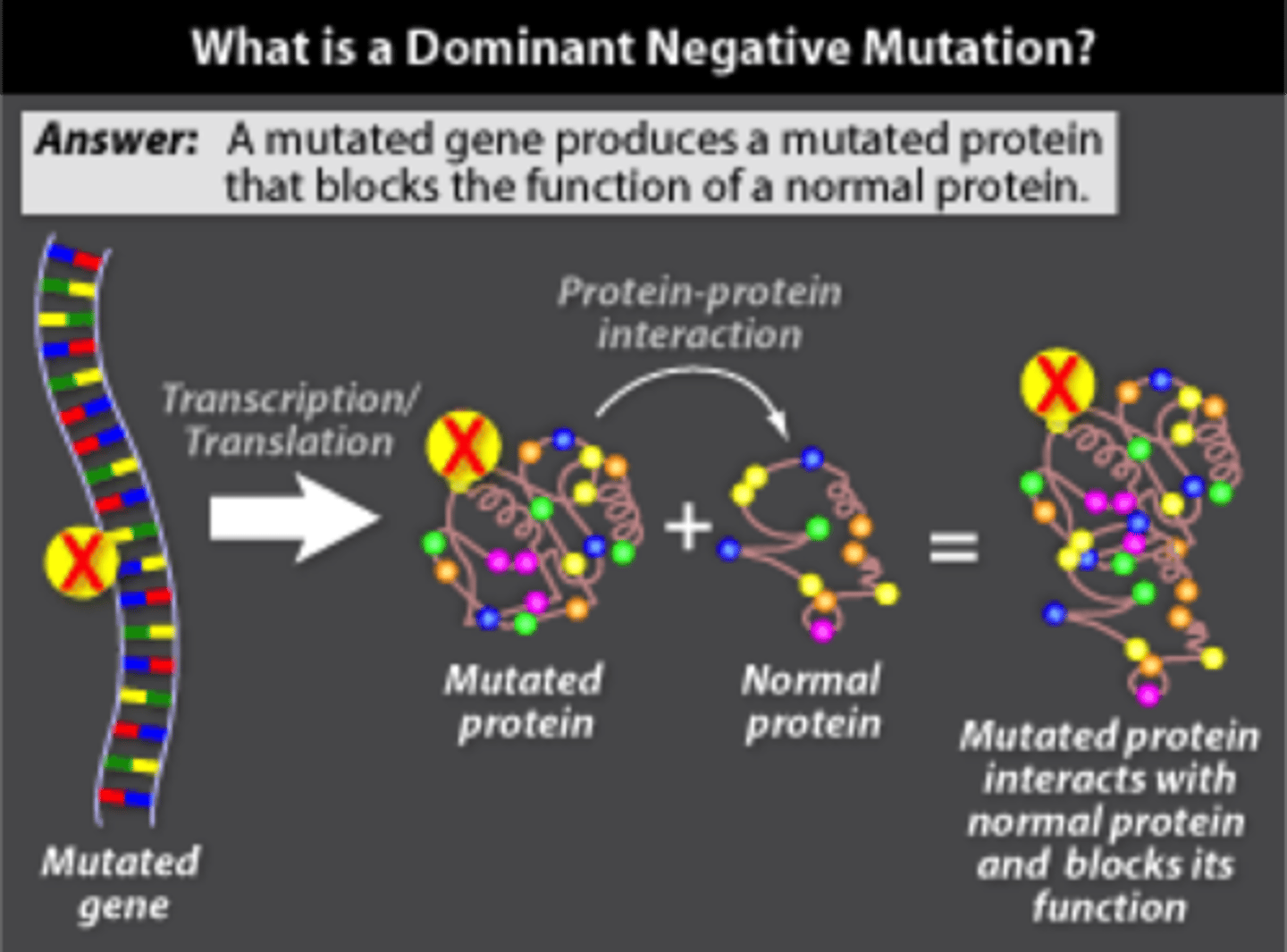
CRB Which of the following would be ways to always increase the expression of a gene?
I. Increase the Gene Dosage (amount of copies of a gene in the the cell)
II. Increase SNPs.
III. Increase Copy Number Variation
(A) I only
(B) I and III only
(C) II and III only
(D) I, II and III
(A) I only
Increasing the Gene Dosage will always increase the Expression of a gene.
CRB Why could increasing the Copy Number Variation sometimes not increase the expression of a gene?
To increase the Copy Number Variation, there are 2 possibilities:
1. Increase the Copy Number of a gene
2. Delete copies of a Gene.
Clearly, the 2nd option would decrease the expression of the gene and still increase Copy Number Variation.
CRB There are Endosymbiotic theories on how organelles originated. Which of the following does NOT support this claim?
(A) Mitochondria and Chloroplasts have an inner membrane that could have been their own plasma membrane.
(B) Mitochondria and Chloroplasts both are seen undergoing mitosis in cells.
(C) Mitochondria and Chloroplasts each contain unique genetic material.
(D) Mitochondria and Chloroplasts are both capable of transcribing and translating their own genome.
(B) Mitochondria and Chloroplasts both are seen undergoing mitosis in cells.
In actuality, their replication, transcription and translation are under nuclear gene control.
CRB Where are the proteins that are used in the Electron Transport Chain coded for?
(A) Mitochondrial Genome only
(B) The Eukaryotic Cell's Genome Only
(C) Parts are in either the Mitochondrial Genome or Eukaryotic Genome.
(D) This has not yet been elucidated.
(C) Parts are in either the Mitochondiral Genome or Eukaryotic Genome.
Some proteins in the ETC are coded for in the Mitochondrial Genome, whereas many others are coded for in the Eukaryotic Genome.
CRB Which of the following statements about mitochondrial DNA (mtDNA) are true?
I. mtDNA mutations cause 85% of all mitochondrial diseases.
II. mtDNA mutations can lead to different phenotypes
III. 99% of mtDNA is inherited from the mother.
(A) II only
(B) I and II only
(C) II and III only
(D) I, II and III
(C) II and III only
Each of the following statements are true:
I. mtDNA mutations cause 15% of all mitochondrial diseases.
II. mtDNA mutations can lead to different phenotypes
III. 99% of mtDNA is inherited from the mother.