Chapter 10 - Molecular Structure of Chromosomes
1/72
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
73 Terms
Chromosomes
Structures that contain the genetic material and are complexes of DNA and proteins.
Genome
Comprises all the genetic material that an organism possesses. In bacteria, it is typically a single circular chromosome. In eukaryotes, however, it refers to one complete set of nuclear chromosomes.
Mitochondrial / chloroplast
Fill in the blank…
Eukaryotes possess a ( ) genome while plants have a ( ) genome.
To store the information required to produce the traits of an organism through protein-encoding genes.
What is the main function of the genetic material?
1) Synthesis of RNA and cellular proteins
2) Replication of chromosomes
3) Proper segregation of chromosomes
4) Compaction of chromosomes so they can fit in cells
DNA sequences are necessary for what four functions?
Intergenic Regions
Non-transcribed DNA between adjacent genes in bacterial chromosomes.
Circular / million / thousand / protein-encoding / single / copies / one / repetitive
Fill in the blank…
Bacterial chromosomal DNA is usually a ( ) molecule that is a few ( ) nucleotides in length. A typical bacterial chromosome contains a few ( ) different genes. ( )-encoding genes account for the majority of bacterial DNA. Most bacterial species contain ( ) type of chromosome, but it may be present in ( ). ( ) origin of replication is required to initiate DNA replication and ( ) sequences may be interspersed throughout the chromosome.
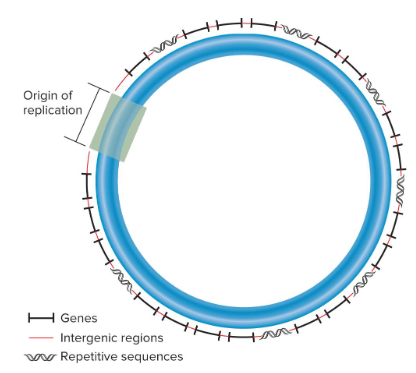
Nucleoid
A region of the bacterial cell in which the bacterial chromosome is found. It is not bounded by a membrane, so DNA is in direct contact with the cytoplasm.
Microdomains
Also called loop domains, these are loops formed in bacterial chromosomal DNA that are about 10,000 bp long and enable DNA to be compacted about 1,000-fold. The number of loops formed varies according to the size of the bacterial chromosome and the species.
Macrodomains
Adjacent microdomains can be further organized into these loop structures of 800-1,000 kbp.
Nucleoid-Associated Proteins (NAPs)
DNA-binding proteins that help bacteria to form microdomains and macrodomains. They facilitate chromosome compaction, organization, and segregation. They can bend the DNA or act as bridges for DNA to bind to other DNA regions. They also play a role in gene regulation.
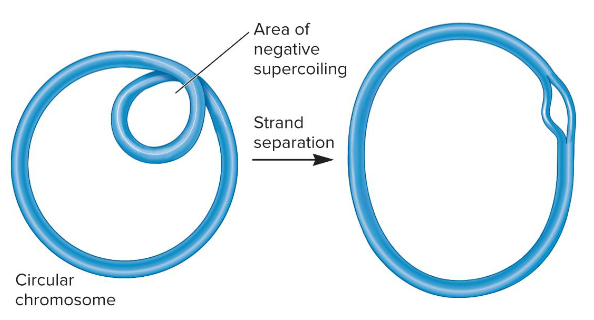
1) Helps in compaction of chromosome
2) In localized regions, creates tension that may be released by DNA strand separation
What are the two major effects of negative supercoiling?
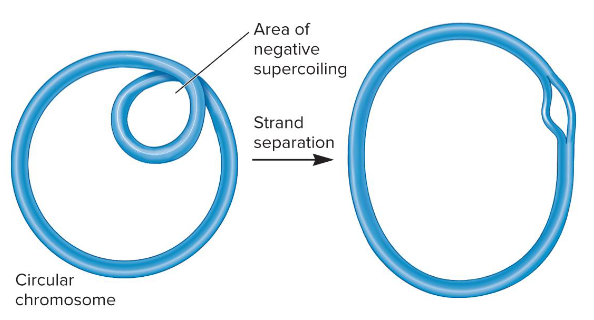
Negatively
Fill in the blank…
The chromosomal DNA in bacteria is ( ) supercoiled.
1) DNA gyrase (DNA topoisomerase 2)
2) DNA topoisomerase 1
The control of supercoiling in bacteria is accomplished by what two enzymes?
The competing action of these two enzymes governs the overall supercoiling of bacterial DNA.
DNA gyrase
An enzyme that introduces negative supercoils using energy from ATP. It can also relax positive supercoils when they occur and can untangle intertwined DNA molecules.
DNA topoisomerase 1
An enzyme that relaxes negative supercoils.
Sets / linear / interspersed
Fill in the blank…
Eukaryotic species contain one or more chromosome ( ). Each set is composed of several different ( ) chromosomes. The chromosomes are usually tens to hundreds of millions of base pairs long and have a few hundred to several thousand genes (genes are ( )).
Short / long / introns
Fill in the blank…
In simpler eukaryotes like yeast, genes are relatively ( ). In more complex eukaryotes, genes are ( ) and tend to have many ( ).
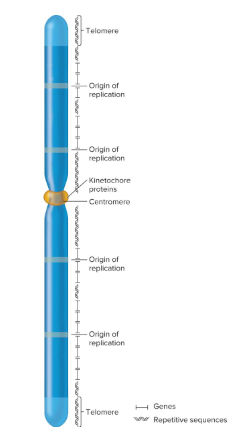
Origins of Replication
Chromosomal sites necessary to initiate DNA replication. Eukaryotes contain many of these sites.
Centromeres
Regions that play a role in the segregation of chromosomes.
Telomeres
Specialized regions at the end of chromosomes that are important in replication and for stability.
1) Origin of replication
2) Centromeres
3) Telomeres
What three types of DNA sequences are required for chromosomal replication and segregation?
Origins of replication / kinetochore / telomeres / repetitive
Fill in the blank…
Each chromosome contains many ( ) that are interspersed. Each chromosome contains a centromere that forms a recognition site for the ( ) proteins during sorting of mitosis and meiosis. ( ) contains specialized sequences located at both ends of the linear chromosome. ( ) sequences are commonly found near centromeric and telomeric regions, but they can be interspersed throughout the chromosome.
Greater / complexity
Fill in the blank…
The total amount of DNA in eukaryotic species is typically much ( ) than that in bacterial cells. Eukaryotic species, however, vary substantially in size. Usually, variation is not related to the ( ) of the species.
Ex) There is a two-fold difference in the size of the genome in two closely related salamander species. The difference is size is not because of extra genes, but rather the accumulation of repetitive DNA sequences that do not encode proteins.
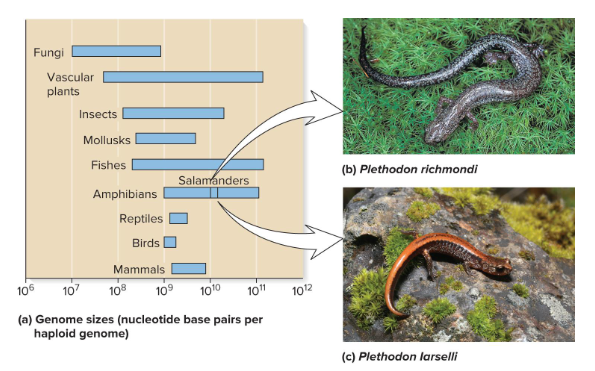
Sequence Complexity
Refers to the number of times a particular base sequence appears in the genome or the instances of repetitive sequences.
There are three main types of repetitive sequences…
1) Unique or non-repetitive
2) Moderately repetitive
3) Highly repetitive
Unique Sequences
Also known as non-repetitive sequences. These are found once or a few times in the genome. They include protein-encoding genes as well as intergenic regions. In humans, these make up 41% of the genome.
Moderately Repetitive Sequences
Repetitive sequences that are found a few hundred to several thousand times in the genome. These include genes for rRNA and histones, sequences that regulate gene expression and translation, as well as transposable elements.
Highly Repetitive Sequences
Repetitive sequences that are found tens of thousands to millions of times. Each copy is relatively short (a few to several hundred nucleotides in length). Some are interspersed throughout the genome while others are clustered together in tandem arrays.
Transposition
The integration of small segments of DNA into a new location in the genome. It can occur at many different locations within the genome. It occurs in bacteria, fungi, plant and animal cells.
Transposable Elements (TEs)
Small, mobile segments of DNA also called jumping genes. They were first identified by Barbara McClintock during her studies of corn and many instances of these have been found since. They can move by two general types of pathways.
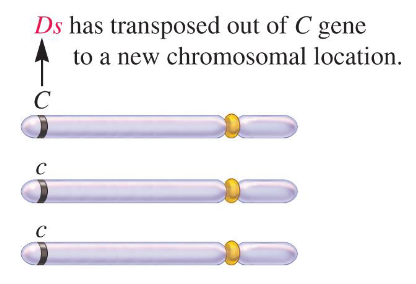
Mutable Site
Also called a mutable locus, this is a location where transposable elements have been inserted into the chromosome. In McClintock’s experiment, this location was named Ds for dissociation because chromosomal breakage occurred here.
Middle / C / function
Fill in the blank…
McClintock found that Ds was in the ( ) of a gene affecting kernel color, where C provided a dark red color while c produced a colorless kernel. The kernels are triploid and she found that that kernel with Ds was colorless with red sectors. She proposed two things…
1) The colorless background of the kernel was due to the transposition of Ds into the ( ) allele, inactivating it.
2) In a few cells, Ds occasionally transposed out of the allele during growth, rejoining the two parts of the allele and restoring its ( ). This produced red sectors of the kernel.
Simple Translocation
The transposable elements moves to a new target site.
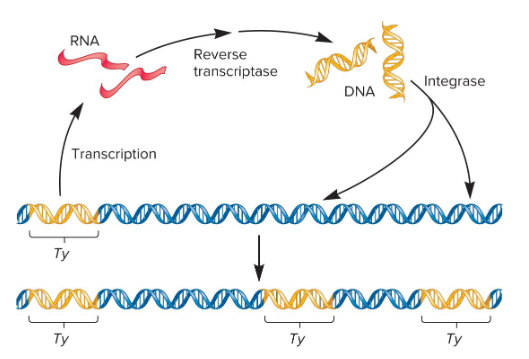
Retrotransposition
The transposable elements move via an RNA intermediate.
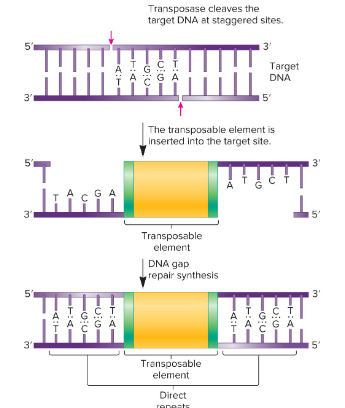
Direct Repeats (DRs)
Identical base sequences flanking all transposable elements that are oriented in the same direction and repeated. They are found next to the inverted repeats of the insertion element.
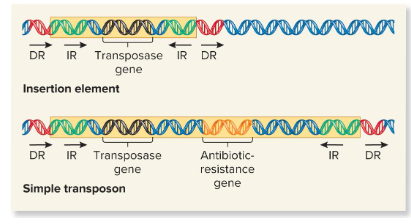
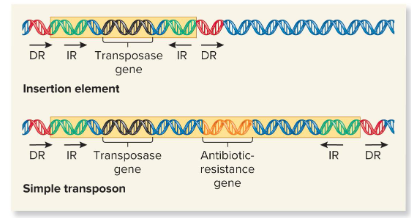
Insertion element
These are the simplest transposable elements, which are flanked by inverted repeats, or DNA sequences that are identical but run in opposite directions of the TEs and are 9-40 bp. It may also contain a gene for the enzyme transposase.
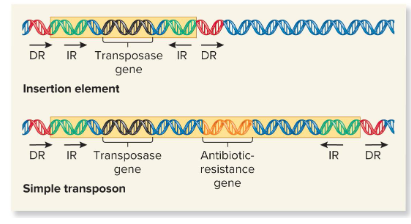
Transposase
An enzyme found in transposable elements that catalyzes the transposition event. It catalyzes the removal of a transposable element and reinsertion at another location. It recognizes the inverted repeats at the ends of a TE and brings them close together.
Simple Transposon
Transposable elements that carry one or more genes not required for transposition.
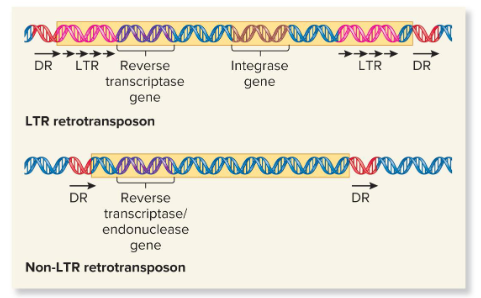
LTR Retrotransposons
Transposans that are evolutionarily related to known retroviruses and retain the ability to move around the genome, but can’t produce mature viral particles. They contain long terminal repeats (LTRs) at both ends and are typically a few hundred base pairs in length.
They encode viral related proteins, like reverse transcriptase and integrase, that are needed for the retrotransposition process.
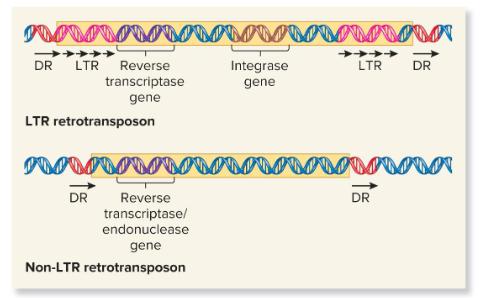
Non-LTR Retrotransposons
Transposons that do not resemble retroviruses in having LTR sequences. They may contain a gene that encodes a protein that functions as both a reverse transcriptase and an endonuclease. Some of these are evolutionarily derived from normal eukaryotic genes.
Autonomous Elements
Transposable elements that contain all of the information necessary for transposition or retrotransposition.
Nonautonomous Element
Transposable elements that lacks a gene, such as one that encodes transposase or reverse transcriptase, which is necessary for transposition.
Ex) Ds locus in corn lacks a transposase gene.
Ac Element
Activator element. It provides a transposase gene that enables Ds (a nonautonomous element) to transpose. Nonautonomous elements like Ds can transpose only when Ac is present at another region in the genome.
1) Transposase subunits bind to inverted repeats
2) Dimerization of transposase subunits causes the TE to loop out.
3) Transposase cleaves outside of the inverted repeats, which excises the transposon from the chromosomal DNA
4) Transposase cleaves the target DNA at staggered sites.
5) The TE is inserted into the target site and DNA gap repair synthesis begins. This produces direct repeats on either side of the TE.
What are the steps of transposition via transposase (simple transposition)?
1) Reverse transcriptase
2) Integrase
What two key enzymes are required for LTR retrotransposan movement (require an RNA intermediate)?
Long Interpersed Elements (LINEs)
Repetitive sequences due to the proliferation of TEs that are usually 1,000-10,000 bp long and occur in 20,000 to 1,000,000 copies per genome.
Short Interspersed Elements (SINEs)
Repetitive sequences due to the proliferation of TEs that are less than 500 bp in length.
Selfish DNA theory
This theory for the significance of transposons says that TEs exist simply because they can. They can proliferate within the host as long as they do not harm the host to the extent that they do not disrupt survival. They don’t have much significance.
Antibiotic resistance / recombination / exon shuffling
Fill in the blank…
Another theory on the significance of TEs is that they confer some advantage. They can…
1) Carry ( ) genes
2) Cause genetic variability through ( )
3) Insert exons into the coding sequences of protein-encoding genes, a process called ( ), which leads to the evolution of genes with more diverse functions.
Overall, TEs can enter the genome of an organism and proliferate quickly, having a variety of effects on chromosome structure and gene expression.
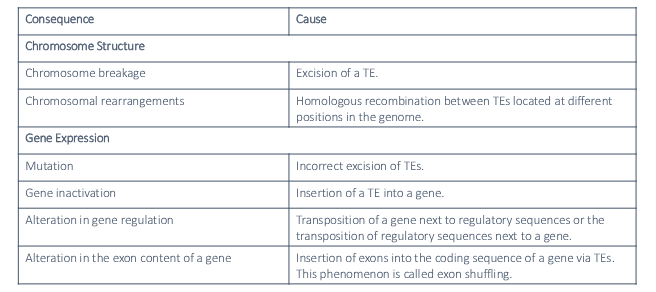
1) Excision of TE
2) Homologous recombination between TEs located at different positions in genome
3) Incorrect excision of TEs
4) Insertion of a TE into a gene
5) Transposition of a gene next to regulatory sequences or the transposition of regulatory sequences next to a gene
6) Exon shuffling
Each consequence is caused by what action involving TEs?
1) Chromosome breakage
2) Chromosome rearrangement
3) Mutation
4) Gene inactivation
5) Alteration in gene regulation
6) Alteration in exon content of a gene
1) Radiation
2) Chemical mutagens
3) Hormones
Since many outcomes of TEs are likely harmful, transposition is highly regulated and occurs in only a few individuals under certain conditions. What agents can stimulate the movement of TEs?
Hybrid Dysgenesis
This occurred when D.melanogaster M strain females were crossed with P strain males. The offspring had a variety of abnormalities with high rates of mutation and chromosome breakage. This outcome occurred because the P elements could transpose freely when they first entered the egg. The egg did not contain any regulatory factors to prevent transposition and the sperm did not contribute any cytoplasm.
Chromatin
The compaction of linear DNA in eukaryotic chromosomes involves interactions between DNA and several different proteins. This DNA-protein complex is known as this. The proteins bound to the DNA in this structure is subject to change during the life of the cell, affecting the complex’s compaction.
Nucleosome
The repeating structural subunit within eukaryotic chromatin. It is composed of a double-stranded segment of DNA wrapped around an octamer of histone proteins. 146 bp of DNA make 1.65 negative superhelical turns around the octamer.
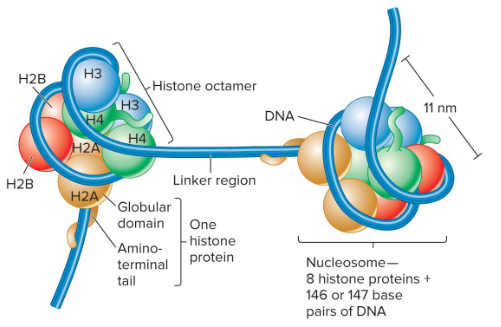
Histone Octamer
A component of nucleosome that is composed of two copies each of four different histone proteins.
Histone Proteins
Components of nucleosomes that are basic. They contain many positively-charged amino acids like lysine and arginine that bind to the negatively-charged phosphates along the DNA backbone. They have a globular domain and a flexible, charged amino terminus or tail.
Core Histones
H2A, H2B, H3, and H4. Two of each of these make up the histone octamer.
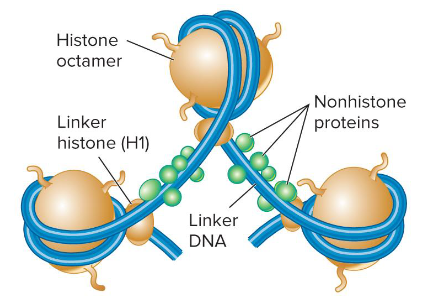
Linker Histone
H1
It binds to DNA in the linker region less tightly than the core histones. It helps to organize adjacent nucleosomes.
1) Digest DNA with the enzyme DNase 1
2) Accurately measure the molecular mass of the resulting DNA fragments using gel electrophoresis
Reason → Linker DNA is more accessible to DNase 1 than the DNA bound to core histones. Thus, the cuts made by DNase 1 should occur in the linker DNA and make DNA pieces that are about 200 bp long
How did Noll perform an experiment to test Kornberg’s nucleosome structure model? What was his rationale for doing it this way?
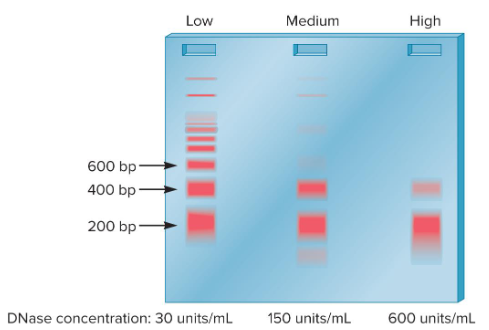
At low or medium concentrations, DNase 1 did not cut at every linker region and longer pieces were observed in multiples of 200 bp
At high concentration, all chromosomal DNA was digested into fragments of ~200 bp
What were the results of Noll’s experiment?
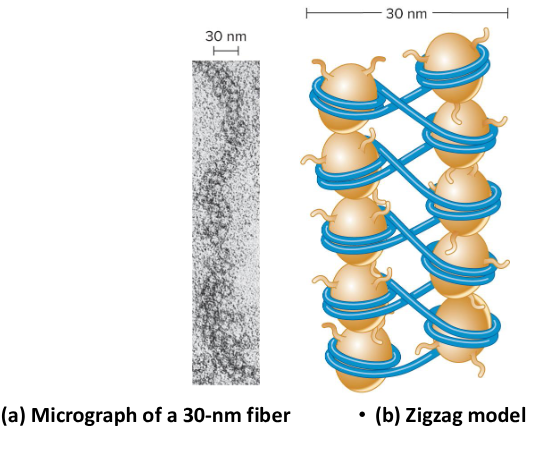
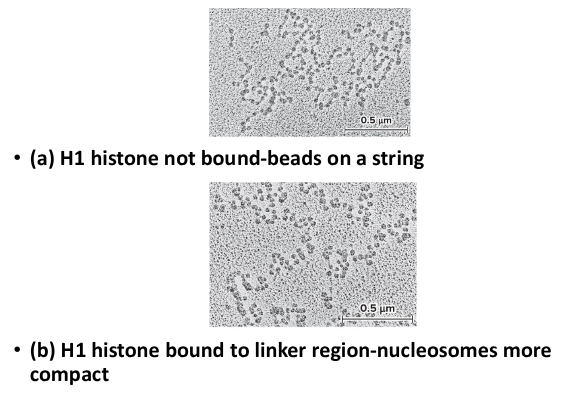
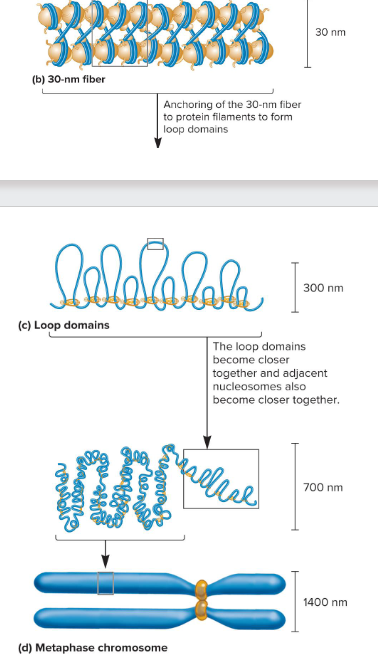
30 nm fiber
This formed when nucleosomes associate with each other to form a more compact structure. The H1 histone play a role in this concentration, remaining bound at low salt concentrations and forcing the beads to associate together into a more compact morphology. At higher salt concentrations, H1 is removed and the histones remain in the beads-on-string morphology.
1) Nucleosome formation
2) 30 nm fiber
3) Loop domains
What are the three levels of chromosome compaction?
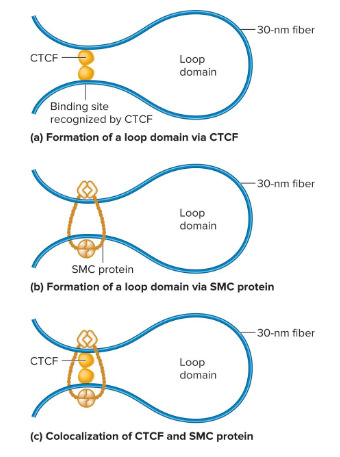
CCCTC Binding Factor (CTCF)
A protein that binds to 3 regularly spaced repeats of the sequence CCCTC. Two of these that are different bind to the DNA and then bind to each other to form a loop.
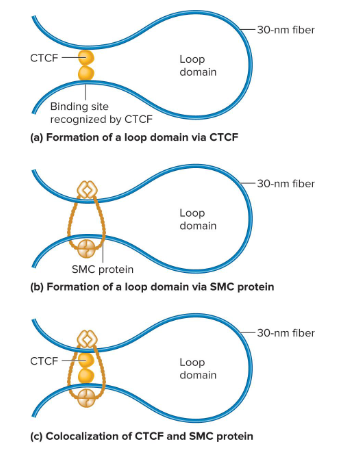
SMC Proteins
A protein that aids in loop formation by forming a dimer that can wrap itself around two DNA segments and form a loop. They may also wrap around sites that are bound by CTCF dimers.
Heterochromatin
Tightly compacted regions of chromosomes that are transcriptionally inactive. Loop domains are compacted even farther.
Euchromatin
Less condensed regions of chromosomes that are transcriptionally active. Here, the 30 nm fiber forms loop domains.
Constitutive Heterochromatin
Regions that are always heterochromatic. They are permanently inactive with regard to transcription and contain highly repetitive sequences.
Facultative Heterochromatin
Regions that can interconverts between euchromatin and heterochromatin.
M / prophase / transcription
Fill in the blank…
As cells enter ( ) phase, the level of compaction changes drastically. By the end of ( ), sister chromatids are entirely condensed and undergo little gene ( ).
Condensin
A multiprotein complex that plays a critical role in chromosome condensation.
Cohesin
A multiprotein complex that plays a big role in sister chromatid alignment.
SMC proteins
Structural Maintenance of Chromosomes proteins. They are found in both condensin and cohesin. They use energy from ATP to catalyze changes in chromosome structure.
1) In interphase, condensin 2 enters the nucleus and plays a role in condensation during early prophase while condensin 1 stays in the cytoplasm
2) At the end of prophase, the nuclear envelope dissociates and both condensins bind to chromosome and compact the loops
3) From S phase until the middle of prophase, cohesin is found along the length of each sister chromatid and holds the chromatids together.
What are the steps of condensin and cohesin working together for chromosome compaction?