207 Exam 2
1/131
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
132 Terms
antibiotics
compounds produced by one species of microbe that can kill or inhibit the growth of other microbes (now typically chemically synthesized)
what complicates screening for new antibiotics
some drugs only become antibiotics once partially metabolized in our bodies (e.g. sulfa drugs)
selective toxicity
selectively kill or inhibit the pathogen but not the host
target:
processes only occurring in target species (e.g. peptidoglycan bs)
structural differences shared protein/complexes (e.g.
large subunit ribosome)
other physiological differences (e.g., sulfanilamide mechanism)
sulfanilamide mechanism
inhibits biosynthesis of a vitamin key to nucleic acid biosynthesis in all life
humans don’t synthesize this vitamin, instead uptake from diet, so are not affected
pathogenic bacteria cannot take up the vitamin so have
to synthesize it
antibiotics at high concentrations
at high concentrations antibiotics with selective toxicity can have side effects
allergies can be a real concern (antibiotics are foreign substances to our bodies, so immune system can overreact to them)
spectrum of activity
broad vs narrow
bactericidal
kills bacteria
bacteriostatic
inhibits growth
minimal inhibitory concentration (MIC)
the lowest concentration of the drug that prevents growth
Varies for different bacterial species
Test by serial dilution of antibiotic
How do you determine if and and what concentration the antibiotic has bactericidal activity?

cell wall inhibitors
cell membrane inhibitors
metabolic inhibitors

DNA replication inhibitors

RNA polymerase inhibitors

protein synthesis inhibitors
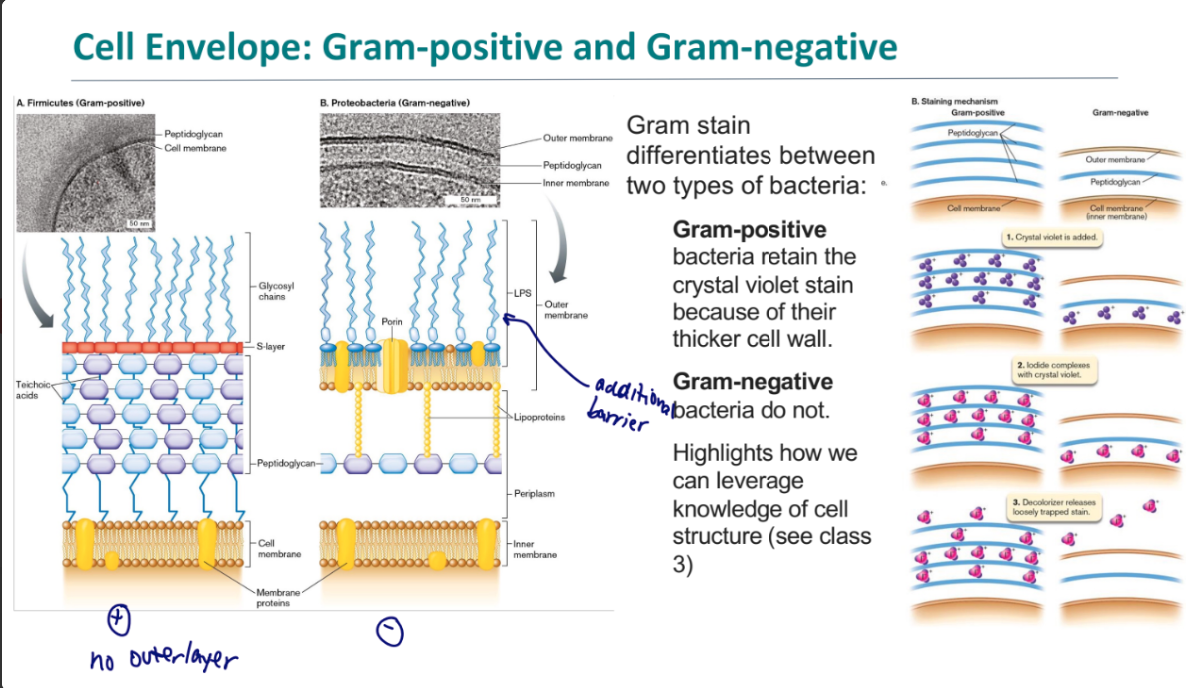
gram postitive vs gram negative
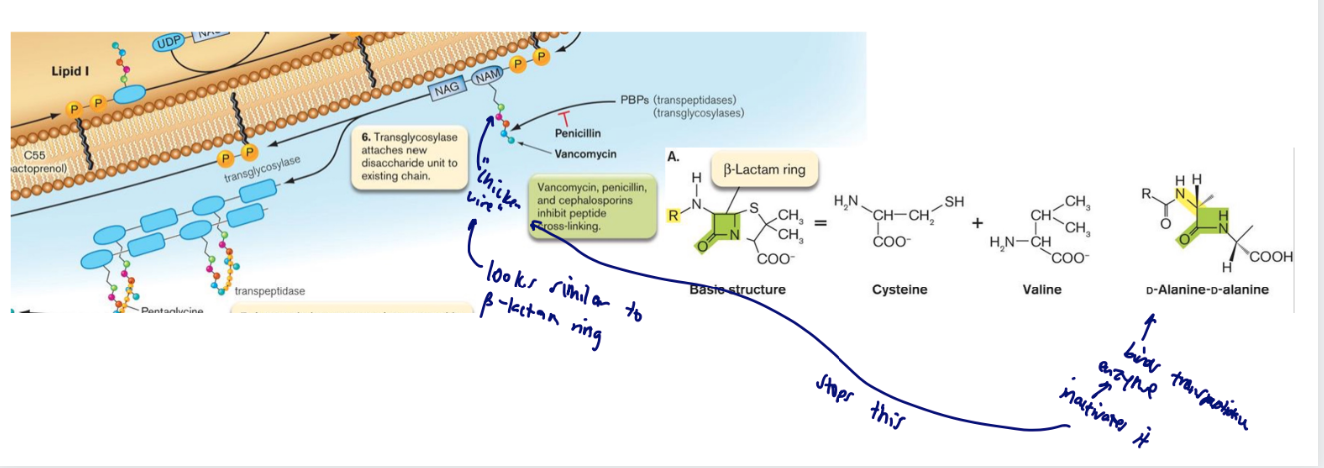
beta lactam antibiotics
Penicillin is an antibiotic derived from cysteine and valine
condensed by fungal enzymes to form a beta-lactam ring structure
The beta- lactam ring chemically resembles the D-Ala-D-Ala peptide crosslink within peptidoglycan
Allows penicillin to bind to and inhibit the transpeptidase enzyme that cross-links peptidoglycan chains.
ampicillin and amoxixillin
versions of penicillin that can cross OM of gram negative
more hydrophilic
Cephalosporins
class of natural beta-lactams modified to combat penicillin-resistant pathogens
ways bacteria develop resistance to penicillin and beta lactams
beta-lactamase enzyme that degrades beta-lactam ab
altered transpeptidase that no longer binds beta-lactam ab
Methicillin-resistant Staphylococcus aureus (MRSA)
Methicillin used initially as not sensitive to resistance due to beta-lactamase enzyme that degrades beta-lactam ab
arose due to mutations leading to transpeptidase no longer binding to beta-lactam ab like methicillin
Vancomycin now often last line of defense against MRSA
now also increasingly VRSA (still very rare, 16th case id’ed in 2021)
interacts w/ peptide bond instead of the enzyme
New antibiotics like teixobactin may offer hope (targets multiple pathways at once, including cell wall biosynthesis, making resistance less likely)
Gramicidin
Cyclic peptide produced by Bacillus brevis
Inserts into membranes as a dimer and forms a leaky cation channel that disrupts ion concentration gradients
Used only topically, as it also targets our cells’ plasma membrane
Why is a combined use of erythromycin, which inhibits protein synthesis, and penicillin counterproductive?
E blocks growth of peptidoglycan, which P needs to stop new cross links
Why would a combination of erythromycin and penicillin sometimes be a good idea?
when you need to suspend growth but not kill cells
if they contain toxins (endo or exotoxins)
E slows the growth, then P comes in
secondary metaboliotes
often have no apparent primary use in the producing organism
antibiotics
only useful in certain conditions
Not essential for survival under standard conditions
May enhance ability to compete favorably with others
antibiotic resistance mechanisms
Only finish synthesis when exported from cell
Make enzymes to disable antibiotics
why do microbes make antibiotics
Hypothesis 1: Biological warfare-gaining competitive advantage over other microbes or killing them to access cellular resources
Hypothesis 2: At subinhibitory concentrations typically found in nature, they may act as signaling molecules regulating community interactions
how do bacteria keep antibiotics out of cell
Destroy the antibiotic before it enters the cell.
The beta-lactamase enzyme specifically destroys penicillins
Decrease membrane permeability across the outer membrane
Gram-negative bacteria can express outer membrane porins with pores too narrow to allow drug penetration.
Pump the antibiotic out of the cell via specific transporterS
Membrane pumps bail drugs out of cell faster than they can enter
Multidrug resistance (MDR) efflux pumps can export
many different kinds of antibiotics (work similarly to ABC export systems)
Can be problematic and contribute to drug resistance because they often work with little regard to structure.
resistance to many antibiotics
how do bacteria prevent antibiotics from binding to target
Modify the target so that it no longer binds the antibiotic.
Mutations in key penicillin-binding proteins and ribosomal proteins confer resistance to methicillin and streptomycin, respectively.
Add modifying groups that inactivate the antibiotic and make it less able to bind its target.
This increases the MIC
how do bacteria dislodge an antibiotic already bound to its target
Ribosome protection (or rescue)
Gram-positive organisms can produce proteins that bind to ribosomes and dislodge macrolide antibiotics bound near the peptidyltransferase site.
how did resistance start
The presence of drug does not cause resistance, but it will kill off or inhibit the growth of competing bacteria that are sensitive, while resistant microbe grows to high numbers.
De novo antibiotic resistance develops through gene duplication and/or mutations
Antibiotic resistance also can be acquired via horizontal gene transfer (conjugation, transduction, and transformation).
antibiotics in animal feed
Collateral damage of antibiotics use: disturbing the microbial balance of power in the gut (C. diff, and links to IBD, vitamin deficiency, obesity, asthma)
antibiotics stweardships
coordinated interventions that improve and measure antibiotic use
Do not use antibiotics to treat viral infections
Do not use an antibiotic if a patient’s microbiome includes a strain that is resistant to the drug. (avoid risk of HGT)
Know which antibiotic resistant strains are prevalent in the community or hospital before prescribing.
Consider how long the patient needs to take the antibiotic: leverage competition with sensitive bacteria that are more fit without ab present (trade-off)?
De-escalate antibiotic usage whenever possible: transition from broad to narrow spectrum when possible
directly countering drug resistance
Dummy target compounds overwhelm resistance enzymes
Alter antibiotic’s structure so that it sterically hinders access of bacterial modifying enzymes
finding new antibiotics
Screening of microbes, plants, and animals– incredible diversity remains untapped
Chemical synthesis of new compounds
Genome sequence analysis to identify potential bacterial molecular targets
Interfering with quorum-sensing mechanisms
CRISPR-based strategies for reversing antibiotic resistance
quorum sensing
phage therapy and problems
very narrow spectrum
could gain resistance to this too
anti fungal agents
Fungal infections are much more difficult to treat than bacterial infections.
Fungi are eukaryotes, and so selective toxicity issues arise.
Fungi have an efficient drug detoxification system that modifies and inactivates many drugs.
Fungal infections can be divided into two main groups.
Superficial mycoses: treated topically
Systemic mycoses: treated internally
rapamycin
Key drug in current medicine, initial as antifungal, but mostly as key drug as immunosuppressant in transplantation, heart stents, and in cancer treatment.
Derived from Streptomyces hygroscopicus, originating from soil sample from Rapa Nui (Easter Island) taken by Canadian exhibition in 1970s
Improbable path to becoming a billion-dollar drug
ways genomes differ
Gene content
differences between different E. coli strains: pathogenic vs not, targeting GI vs urogenital systems
Sequence composition differences
Between botulism toxin proteins affecting different animal
Genome organization differences
between V. cholerae and closely related environmental
Vibrio species
why is DNA ideal storage
stable
mutable
replicable
genome
All genetic information that defines an organism
Genes
Stretches of DNA information that can be “sent” out as RNA
structural gene
produces a functional RNA, which usually encodes a protein
regulatory sequence
regulates the expression of a structural gene.
Does not encode an RNA or protein
Includes promoters & binding sites for regulatory proteins
variation in genome organization
Number, size, shape of chromosome/plasmids
More DNA = trade-off between cost of replication/expression and additional functional capabilities.
non coding DNA
It is typically > 90% of eukaryotic genomes, but < 15% of prokaryotic genomes
Non-coding DNA of eukarya includes introns and pseudogenes (inactivated genes).
Some archaeal genes have introns too, but rare
introns not in bacteria
DNA functions
DNA is more stable than RNA (2
Nucleotides joined together via bonds between the 5ˈ phosphate group of one nucleotide and the 3ˈ OH group of another.
Nucleotide base complementary between two strands of DNA: leveraged for DNA repair
hydrostatic interactions (A:T, G:C) stable under physiological conditions (need for pH and ionic homeostasis inside cell)
Extremophiles (pH, temperature) have additional DNA binding proteins or additional supercoiling to stabilize DNA)
Antiparallel orientation to allow base-pairing → implications for replication process
how is DNA compacted
Organized in domains, each domain supercoiled by topoisomerases
anchored by histone-like proteins
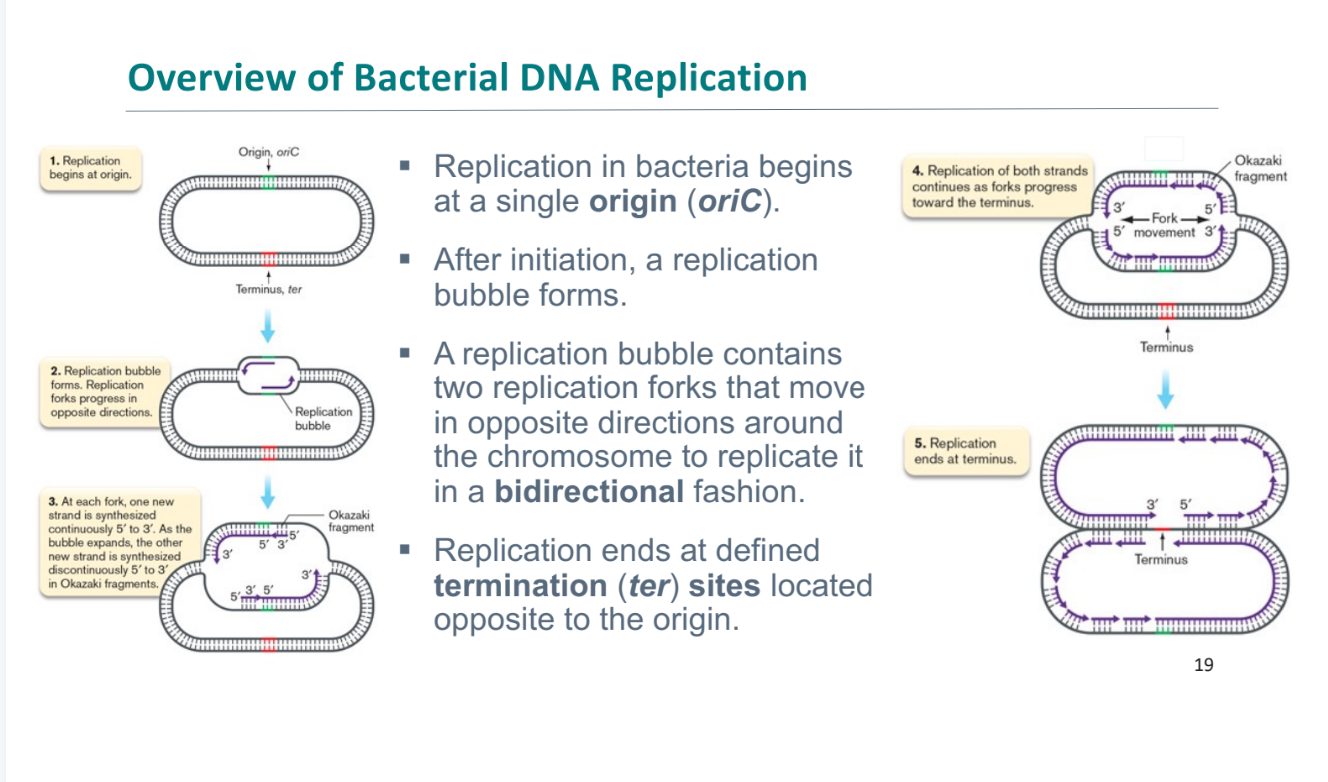
where does bacterial replication begin
oriC
what happens after initiation of bacterial replication
replication bubble
continues until termination sequence (ter)
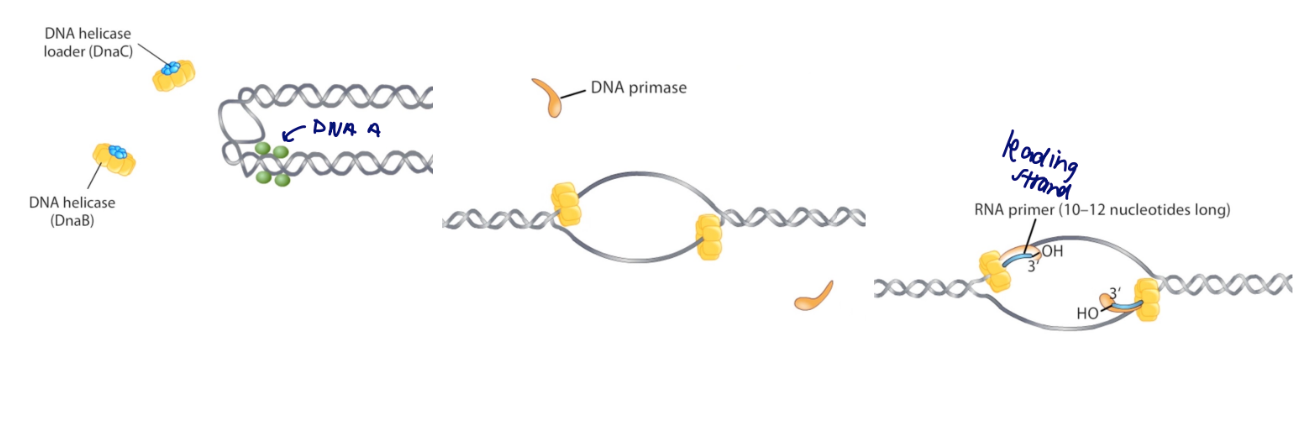
initiating bacterial replication steps
precisely timed in function of cell growth (div and rep are linked)
DnaA bound to ATP accumulates during growth, binds to region near oriC
triggers the initiation of replication by looping and partially unwinding the DNA (replication bubble)
allows replisome to assemble
Within the replication bubble, DnaC (helicase loader) loads
DnaB (helicase) onto each single stranded template.
DnaB recruits DnaG (primase)
primase synthesizes a short RNA primer against each template strand
completes with DNA pol III and sliding clamp
difference in replication in archaea and eukaryotes
archaea can have many Oris (not triggered by DNA A)
eukaryotes have many Oris but they form in specific phases in the cell cycle (meiosis or mitosis, not continuously)
archaea DNA replication
DNA polymerase unique to them
the rest of their replication machinery related to the eukaryotic proteins, rather than to the bacterial versions.
DNA elongation
The replisome ensures that the leading and lagging strands are synthesized simultaneously
5’ to 3’ direction
the problem(lagging) strand loops out after passing through its polymerase.
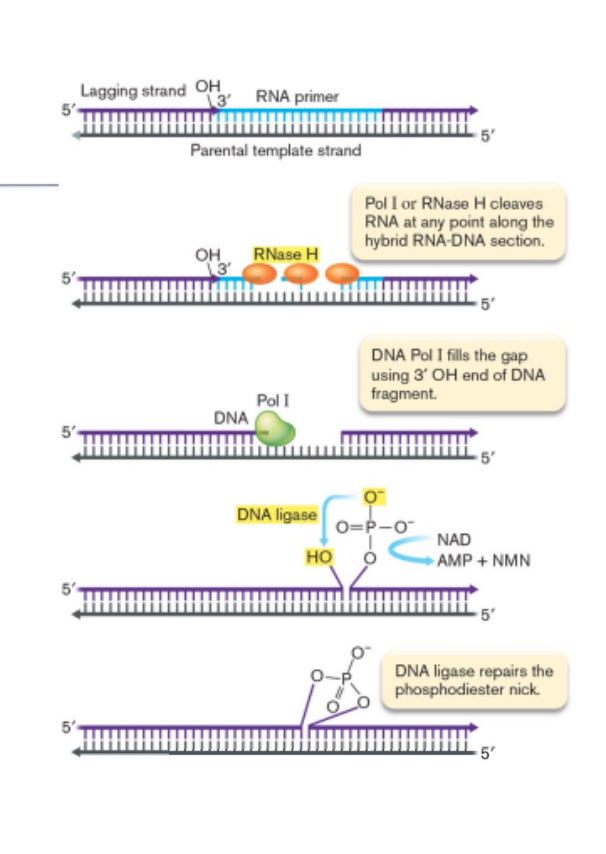
filling in the gaps of leading and lagging strand
RNA primers removed by RNase H or DNA pol I
DNA pol I makes DNA patch
DNA ligase repairs the remaining phosphodiester nicks
what is a benefit of bidirectional replication
faster
less exposed ssDNA at a time
plasmids
found in archaea, bacteria, and eukaryotic microbes
Typically much smaller than chromosomes
Usually circular
Copy number per cell varies widely
Contain nonessential genes that often play critical roles in certain situations (e.g., antibiotic resistance)
Can be transferred between cells
plasmid maintenance strategies
Some plasmids ensure their inheritance by carrying genes whose functions benefit the host microbe under certain conditions (e.g., antibiotic resistance, pathogenesis factors, symbiosis proteins).
High-copy-number plasmids flood the host cell cytoplasm with copies that give each daughter cell a very high likelihood of receiving at least one copy by chance alone.
Low-copy-number plasmids evolved dedicated partitioning systems that ensure both daughter cells receive copies of the plasmid. (this requires use of ATP!)
what happens after translation
each polypeptide is properly folded (this involves chaperone proteins)
placed at the correct cellular or extracellular location (for example using the secretions systems we covered earlier)
making of a protein diagram
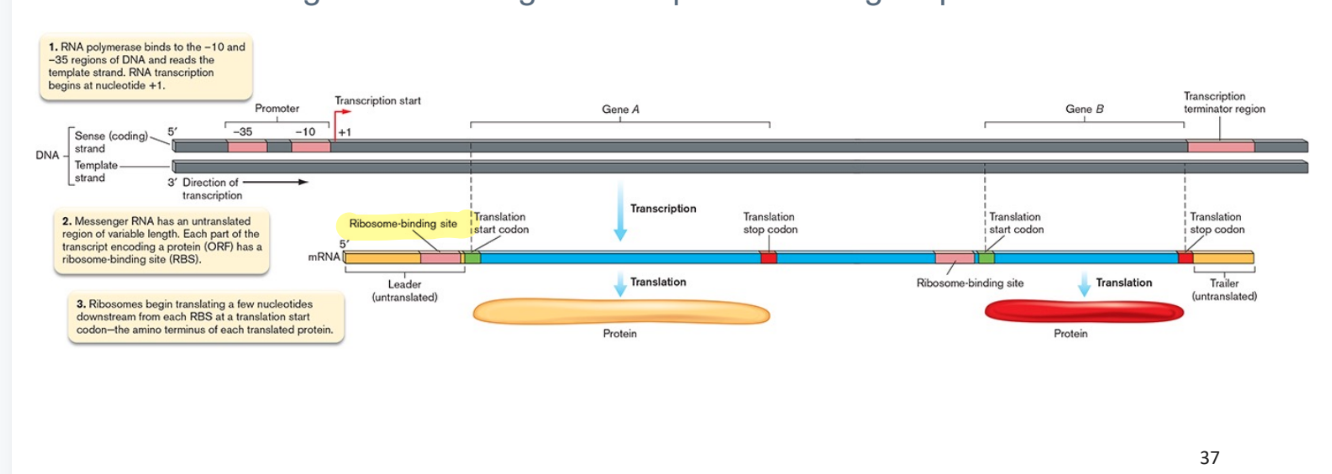
monocystronic
RNA produced from a single gene
all eukaryotic genes
polycistronic
In bacteria and Archaea, genes may be organized in operons (multiple genes transcribed in single transcript), encoding multiple proteins
RNA polymerase holoenzyme
Core polymerase (many subunits)
Required for the elongation phase
Sigma factor
Required for the initiation phase: different
sigma factors allow for coordination of expression of different sets of genes
recognizes promoter
euk & arc have core enzyme and no sigma factor, and
instead use independent proteins called transcription factors
housekeeping sigma factor
Recognizes, based on electrostatic interactions the promotor consensus sequences at the -10 and -35 positions
promoter consensus sequence
Consensus sequence represents most common nucleotide at each position.
Individual promotors diverge from this consensus sequence
Some nucleotides more (yellow) and less (red) conserved across sequences.
A bacterium needs to transcribe a gene both in normal conditions as well as when undergoing heat shock. How is this possible?
different promoters
initiation of transcription
RNA pol holoenzyme binds to the promoter.
This is followed by melting of the helix and synthesis of the first nucleotide of the RNA
termination of transcription
RNA pol detaches from the DNA, after the transcript is made
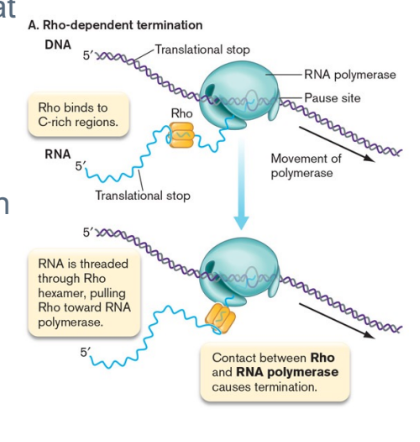
rho dependent termination
Relies on a protein called Rho and a strong pause site at the 3′ end of the gene
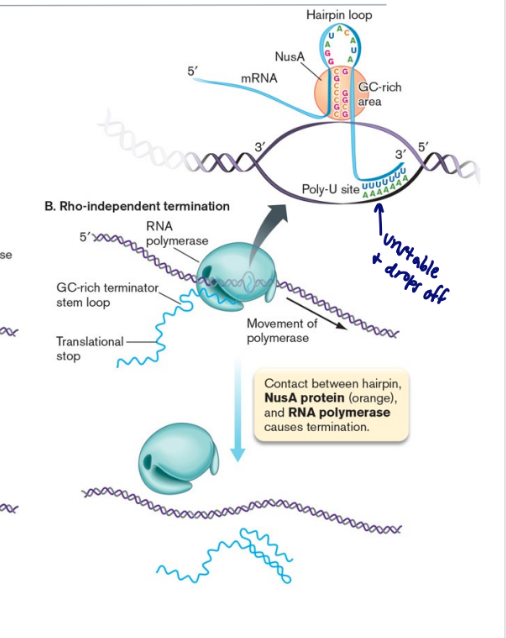
rho independent termination
Requires a GC-rich region of RNA, as well as 4–8 consecutive U residues
antibiotics that affect transcription
Kill or retard the growth of a pathogen
Not harm the host
Rifamycin B - Selectively binds bacterial RNA pol, Blocks RNA exit to inhibit initiation, Polymerase can still bind promoter
Actinomycin D - Non-selectively intercalates between GC base pairs in DNA, Inhibits transcription elongation
classes of RNA and stability
codons
Consists of nucleotide triplets
There are 64 possible codons:
61 specify amino acids
Includes the start codons
3 are stop codons
The code is degenerate or redundant.
Multiple codons can encode the same amino acid.
operates universally across species
very few exceptions
tRNA
convert language of RNA into that of proteins.
two functional regions:
Anticodon: Hydrogen bonds with the mRNA codon specifying an amino acid
3’ (acceptor) end: binds the amino acid
contain a large number of unusual, modified bases.
same tRNA can recognize multiple codons
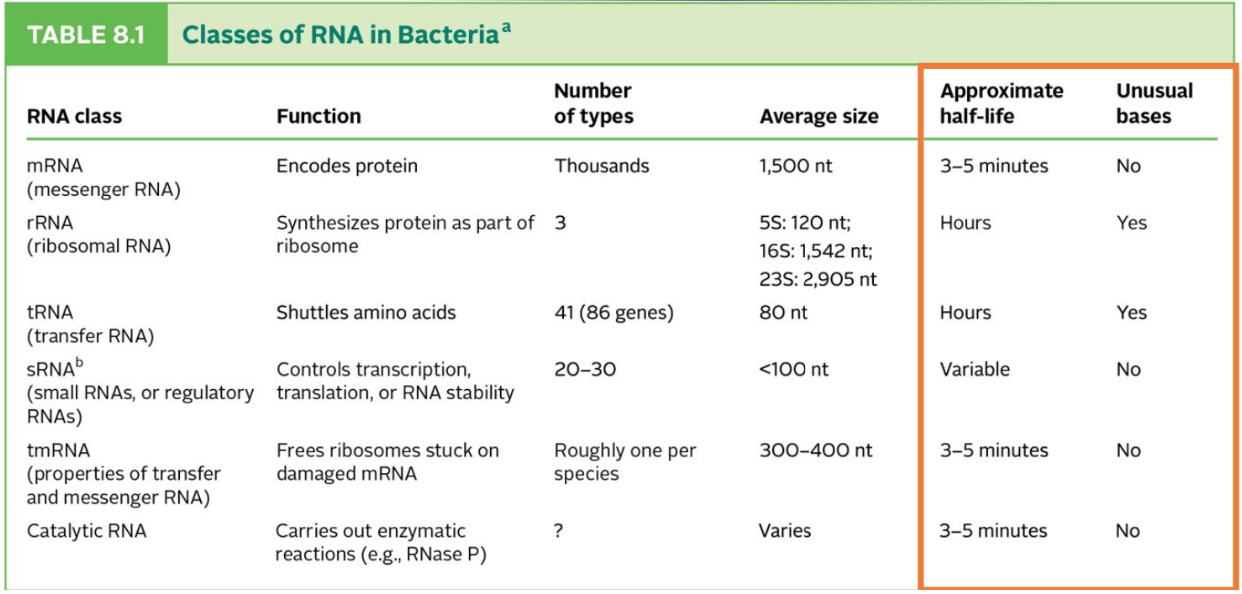
attaching AAs to tRNAs
Each tRNA must be charged with the proper amino acid before it encounters ribosome.
Ribosome cannot check if tRNA has correct amino acid on it.
carried out by a set of enzymes called aminoacyl-tRNA synthetases
Each cell has 20 of these “match & attach” proteins, one for each amino acid.
Each aminoacyl-tRNA synthetase must recognize its own tRNA but not bind to any other tRNA
each tRNA has its own set of interaction sites that match only the proper synthetase
prokaryotic ribosome
the subunits are 30S and 50S and combine to form the 70S ribosome
enzymatic activity - peptidyltransferase
ribozyme
part of 23S rRNA of large subunit
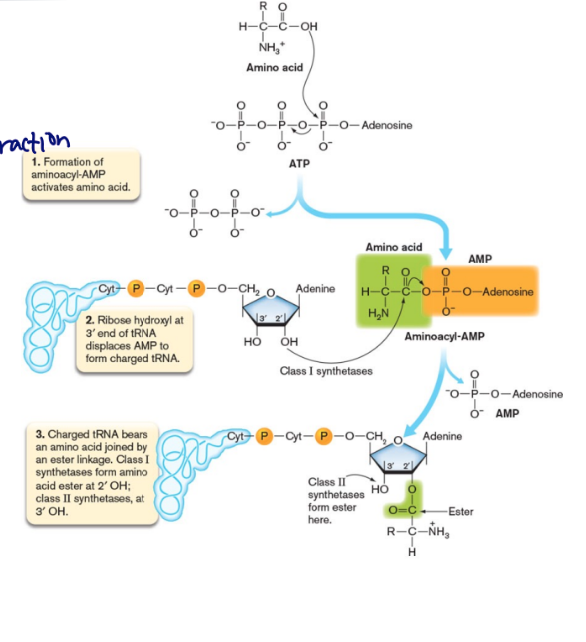
How does ribosome “know” which of 3 reading frames to use
The upstream, untranslated leader RNA contains a purine-rich ribosome binding site with the consensus 5′-AGGAGGU-3′.
This Shine-Dalgarno sequence is complementary to a sequence at the 3′ end of 16S rRNA of the 30S subunit
only in Bacteria and Archaea, Eukarya have 5’ cap instead
initiation of translation
brings the two ribosomal subunits together, placing the first amino acid in position
requires protein factors and GTP
Dissociated ribosome units needed
16S rRNA and mRNA interact at RBS
Met-tRNA binds the ribosome
Full ribosome assembles (GTP hydrolyzes)
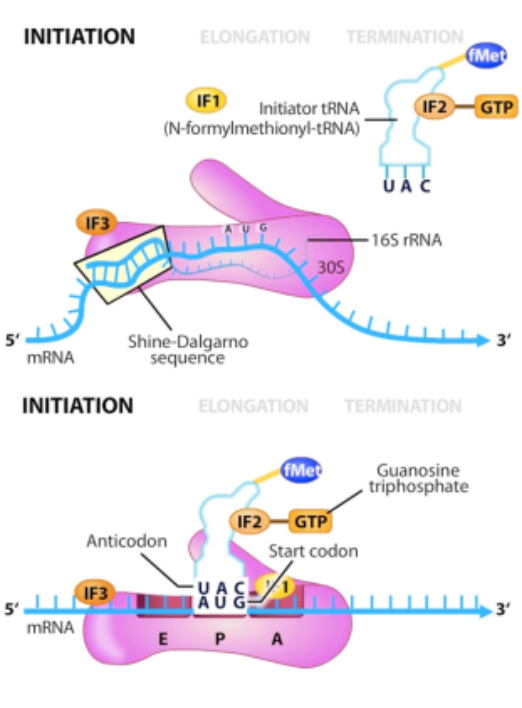
elongation of translation
sequentially adds amino acids as directed by mRNA transcript
requires protein factors and GTP
tRNA-AA loads onto ribosome
Ribozyme activity (23S rRNA) binds new AA to existing peptide (GTP hydrolysis)
Empty tRNA removed upon addition of next tRNA-AA
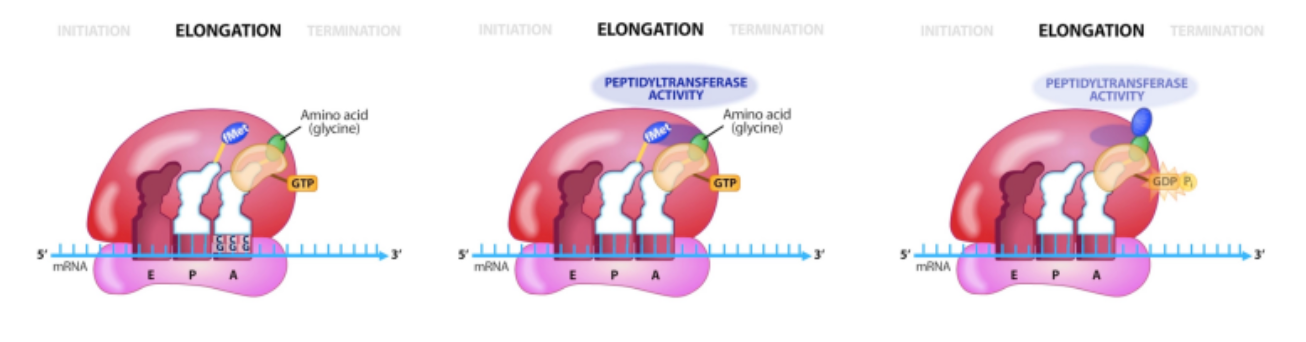
termination of translation
releases the completed protein and recycles ribosomal
subunits
requires protein factors and GTP
No tRNA with anticodon complementary to stop codons
Release factor proteins enters ribosome instead, eventually leading to dissociation of the two ribosome subunits at the expense of GTP hydrolysis
targets of antibiotics
protein synthesis based on differences in structure of proteins and rRNA involved relative to eukarya
additional proteins (initiation factors etc.)
Archaea and Bacteria are more similar but differences in translation factors as well
protein folding and secretion
Often, a protein must be modified after translation either to achieve an appropriate 3D structure or to regulate its activity.
Post-translational modifications can affect activity of proteins-important in regulation (see next class)
A healthy cell “cleans house” by degrading damaged or unneeded proteins
Proteins destined for the bacterial cell membrane or envelope regions require special export systems.
Proteins meant for the cell membrane are tagged with hydrophobic N-terminal signal sequences of 15-30 amino acids.
types of gene regulation
Alteration of DNA sequence – flipping a DNA segment
Control of transcription – Repressors, activators, sigma factors, sRNAs
Control of mRNA stability – RNase activity, sRNAs
Translational control – Hiding RBS sites, or other mRNA sequences
Post-translational control – cleavage, phosphorylation, methylation, etc.
In general, ____ control is the most drastic and least reversible, whereas control at the ____ is the most rapid and most reversible
DNA sequence level, protein level
transcriptional control
proteins bind at regulatory sequences (operators or repressors) to control initiation of transcription at promoters
environmental changes in metabolites (ligands) or outside factors can affect activity
ligands alter the DN binding affinity of regulatory proteins at the promoter
repressors
prevent gene expression
bind to operators
inducer - binds in absence of ligand
corepressor - binds in presence of ligand
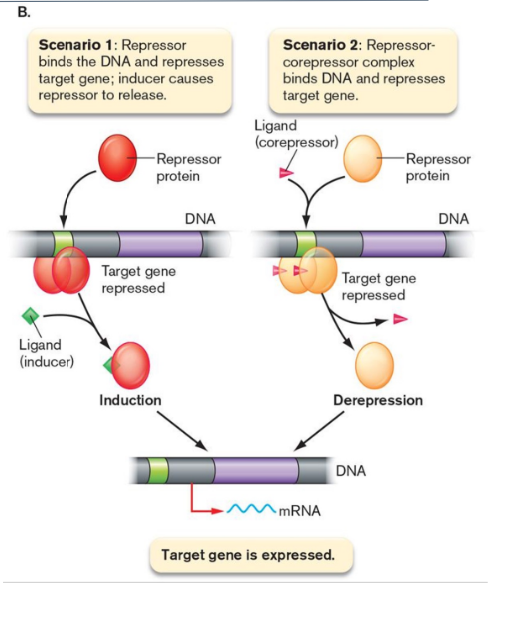
activators
stimulate gene expression
contacts RNA polymerase positioned at a nearby promoter
most are inducers - bind poorly to DNA sequences unless they are bound to their ligand
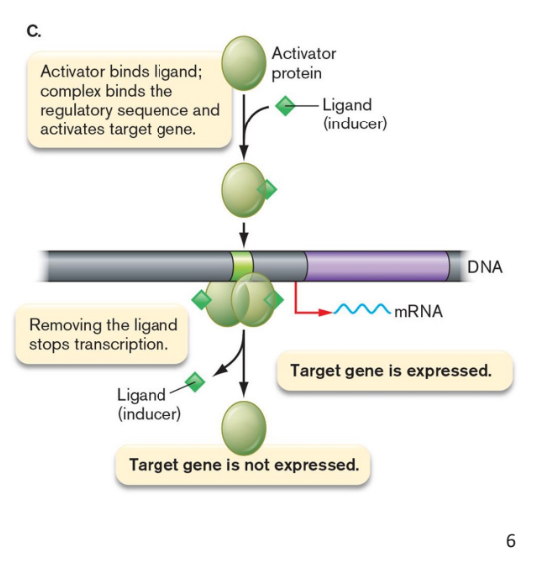
2 component signal transduction system
helps the intracellular proteins interact with the external environment
Sensor kinase in cell membrane
Binds to environmental signal-
Activates itself via phosphorylation
Response regulator in cytoplasm
Takes phosphate from sensor
Binds chromosome
Alters transcription rate for several genes
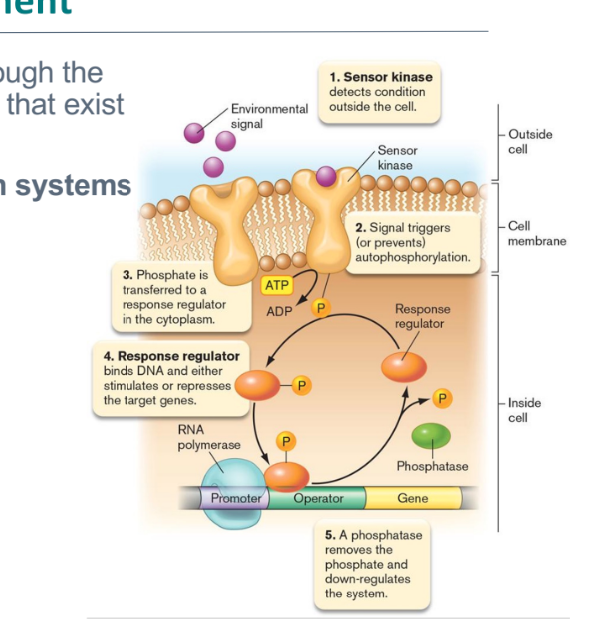
catabolism regulation
often involves induction when substrate is present (activator or no repressor)
catabolite repression
catabolite repression
an operon enabling the catabolism of one nutrient is repressed by the presence of a more favorable nutrient.
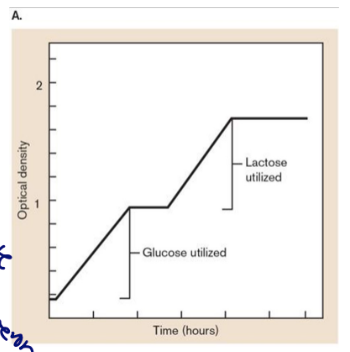
glucose and lactose
diauxic growth (pic)
anabolism regulation
different from cat. repression
typically are regulated by inactive repressors (aporepressors)
bind the end product of the pathway (corepressor).
When an aporepressor binds its corepressor, the complex binds to an operator sequence upstream of a target gene or operon to turn transcription off (repression) by blocking access to the RNA polymerase
Negative feedback mechanism
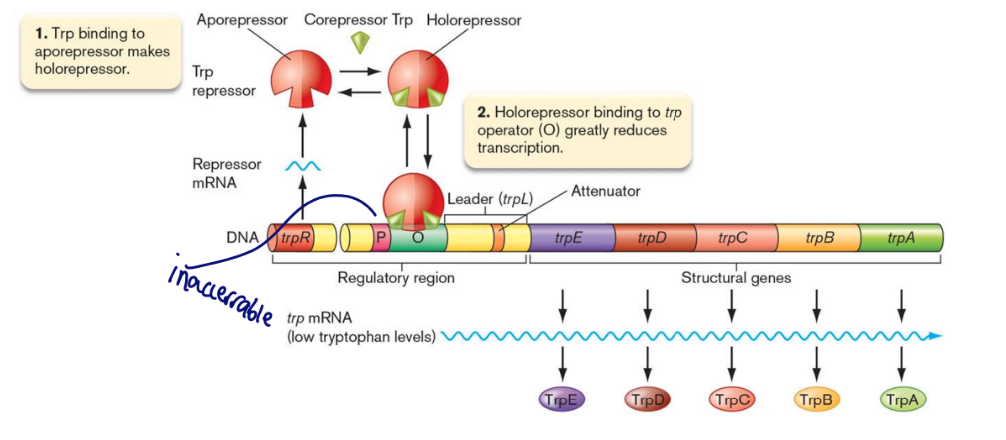
trp operon
trp operon example
repression of anabolic pathways
When trp levels exceed cellular needs, excess Tryptophan (the corepressor) binds to inactive TrpR (the aporepressor).
The complex then binds to an operator sequence upstream of the trp structural genes and represses expression by blocking RNA pol.
what are 2 different ways to control expression of multiple pathways at once (rewatch lecture)
sigma factors → similar promoters
same operon and operator
regulation after transcription
Regulatory sequences in the mRNA can cause premature termination of transcription (e.g., attenuation).
Some mRNA sequences prevent their own translation into protein (e.g., riboswitches).
Other regulatory RNAs influence the fates of transcribed mRNAs (e.g., untranslated RNAs).
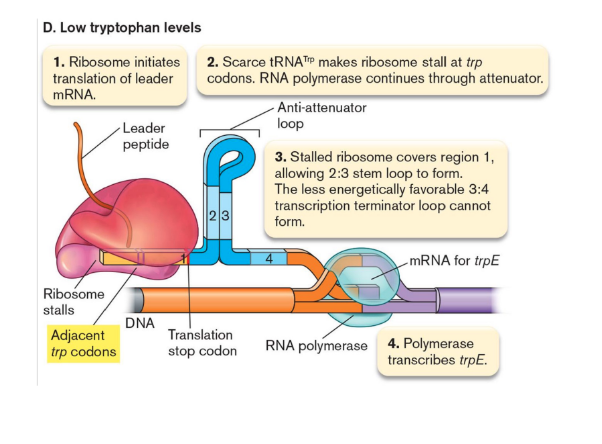
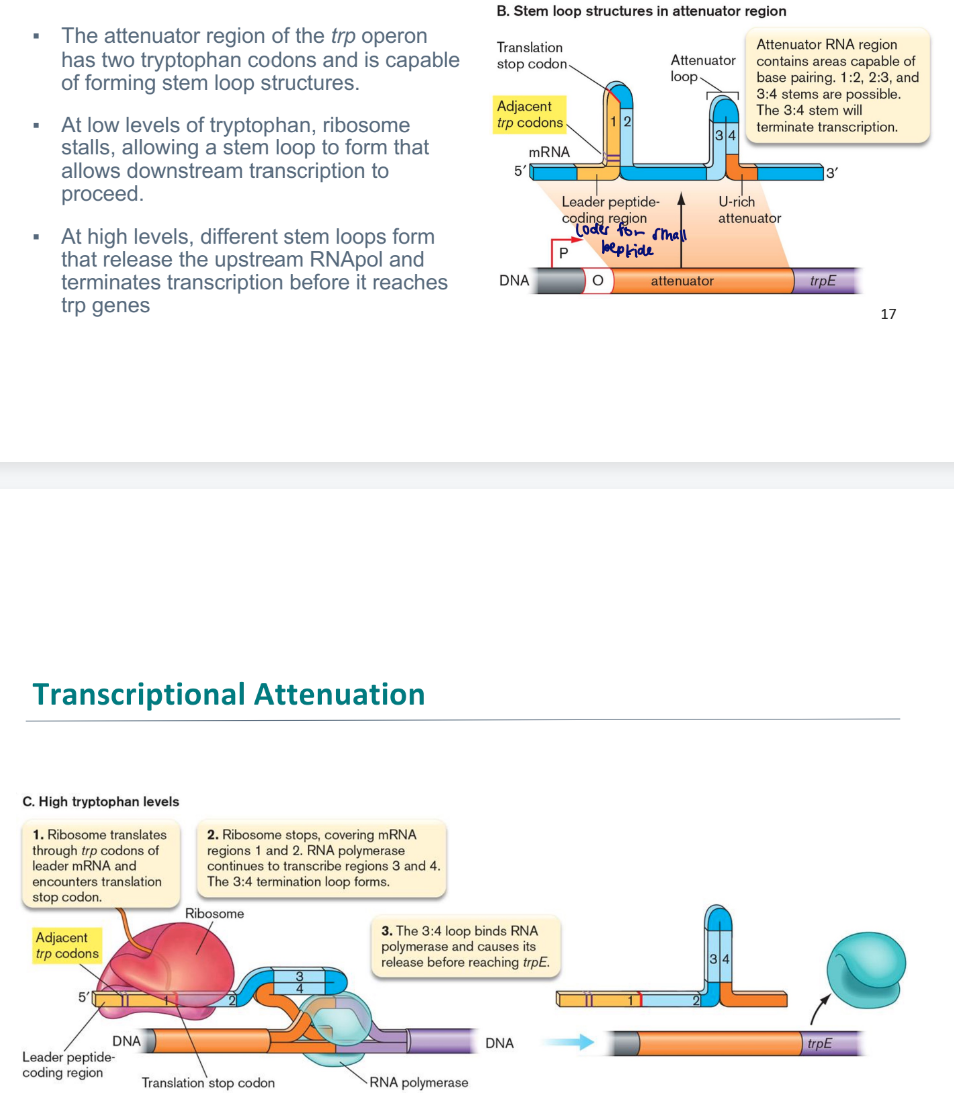
transcriptional attenuation
translation of a leader peptide affects transcription of an operon’s downstream structural genes.
At low levels of tryptophan, ribosome stalls, allowing a stem loop to form that allows downstream transcription to proceed.
At high levels, different stem loops form that release the upstream RNApol and terminates transcription before it reaches trp genes
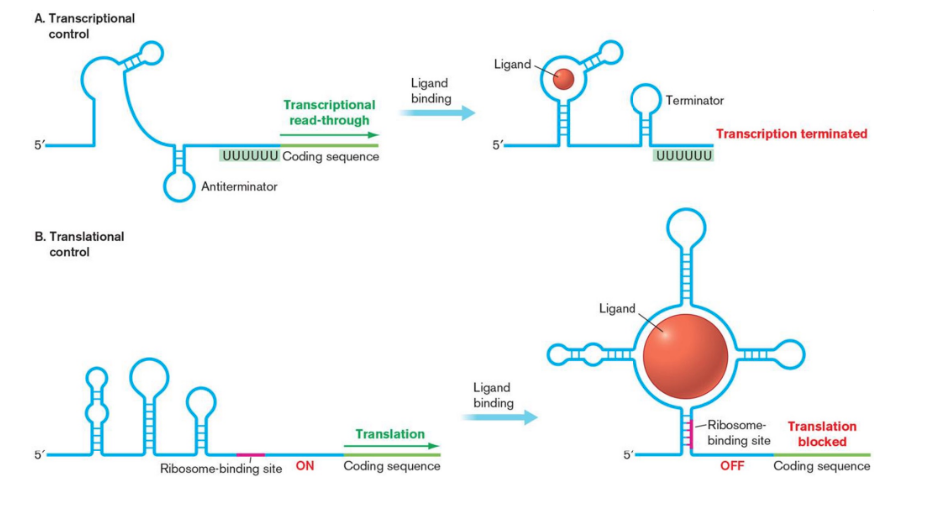
riboswitches
usually found in the 5' untranslated region of mRNA that control gene expression by folding into three-dimensional structures that bind specific metabolites to sense their abundance in the cell.
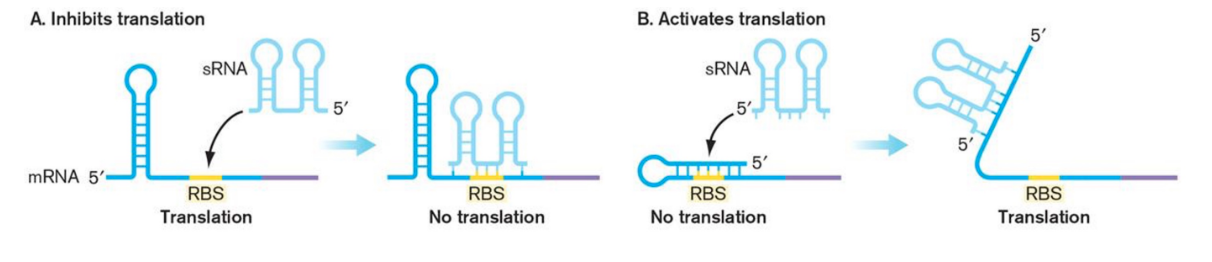
Untranslated Small Regulatory RNAs
Attenuators & riboswitches are part of mRNA transcript they control
Untranslated Regulatory RNA molecules are transcribed independently and typically affect gene expression post-transcriptionally.
Small RNAs (sRNAs) represent one of the most economical ways to regulate gene expression.
They do not require protein synthesis.
They diffuse rapidly.
They typically act on preexisting messages
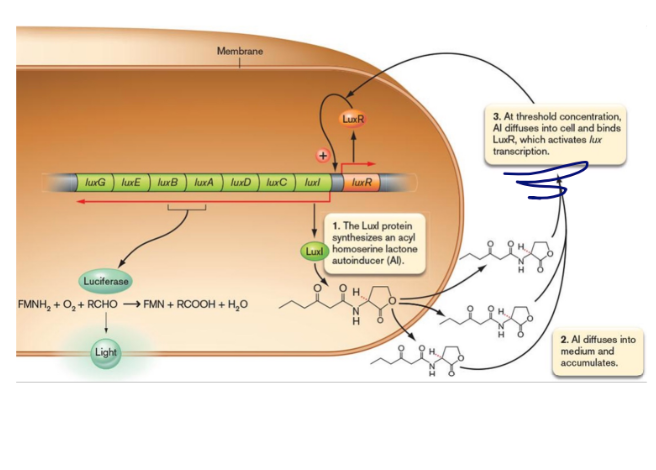
quorum sensing
The process where bacterial cells work together at high
density.
Discovered in Aliivibrio fischeri a bioluminescent bacterium that colonizes the light organ of the Hawaiian squid
many pathogens use it to control expression of virulence genes
dont turn them on until there are many others around
Pseudomonas aeruginosa (CF), Staphylococcus, Yersinia pestis, Vibrio cholerae,
autoindicator
At a certain extracellular concentration, the secreted autoinducer reenters cells.
Binds to a regulatory molecule. In the case of vibrio fischeri is LuxR
LuxR-autoinducer complex then activates transcription of the luciferase target genes
circadian clocks
can anticipate based on pattern
photocynthetic bacteria
Controlled by KaiABC proteins, that together cycle KaiC between P and NP state on a 24 hr cycle.