AP Biology Ultimate Flashcard Set
1/231
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
232 Terms
Water moves
High to low water potential
Low to high osmolarity
Low to high solute concentration
Hypertonic solution
draws water out of cell
Hypotonic solution
cell swells and could lyse
Isotonic solution
In equilibrium
1st law of thermodynamics
Energy cannot be created or destroyed, only transferred or transformed
e.g. photosynthesis (radiant to chemical), consuming to climb tree (chemical to kinetic)
2nd law of thermodynamics
Energy transformation increases the entropy of the universe
e.g. trophic levels 90% heat loss only 10% efficiency for endothermic individuals
Exergonic versus endergonic
exergonic releases energy e.g. cellular respiration; change in free energy < 0
endergonic absorbs energy e.g. photosynthesis; change in free energy > 0
3 types of work cells perform
Mechanical (beaten cilia, chromosome movement, contraction of muscle cells via microfilaments)
Transport (pumping substances across membranes)
Chemical (molecule e.g. protein synthesis)
ATP
Adenosine Triphosphate: Adenine + Ribose + 3 phosphate groups
Obtain energy by breaking bond between 2nd and 3rd phosphate group
Enzymes
proteins that end in -ase that catalyze reactions by lowering activation energy
Enzyme-Substrate specificity
The structure of enzymes includes the active site which specifically interacts with substrate molecules (shape, size, charge)
Enzyme-substrate compatibility
the substrate’s charge and shape must be compatible with active site of enzyme
Enzyme-substrate complex
when both enzyme and substrate are together
Induced fit
enzymes modify shape of their active site to allow the substrate to bind better
Enzyme catabolism vs anabolism
substrate is broken down vs. substrates are built into complex molecules
3 factors that affect enzyme shape and thus function
temperature, pH, and chemical
Denaturation
when enzymes protein structure is disrupted —> can’t catalyze anymore or is less efficient
sometimes REVERSIBLE
The effect of heat on enzyme function
Increases rate of enzyme activity due to higher speed of movement and increased collision frequency between enzymes and substrates; then denatures at a point
The effect of pH on enzyme function
Causes hydrogen bonds in enzyme structure to break
Cofactors and Coenzymes
Cofactors: nonprotein molecules that assist enzyme function
Coenzymes: organic cofactors i.e. vitamins
Enzyme inhibition (2 types; not active/allosteric)
Permanent: inhibitor binds with covalent e.g. toxins
Reversible: inhibitor binds with weak interactions
The effect of relative substrate and product concentration on enzyme activity
Increased substrate concentration causes increased reaction rate (substrate saturation)
Increased product concentration can act as inhibitor to earlier enzymes in same pathway [FEEDBACK INHIBITION]
Competitive enzyme inhibitors
Binds to active site
Can be reversed with increased substrate concentrations to compete back!
Noncompetitive/Allosteric enzyme inhibitors
Binds to allosteric site, changing shape of active site and preventing substrate binding
Noncompetitive/Allosteric activators
Binds to allosteric site, stabilizes open active site
Allosteric cooperativity
Binds to active site, stabilizes other open active sites
Feedback inhibition
When the end product of metabolic pathway acts as inhibitor to early enzyme in the same pathway
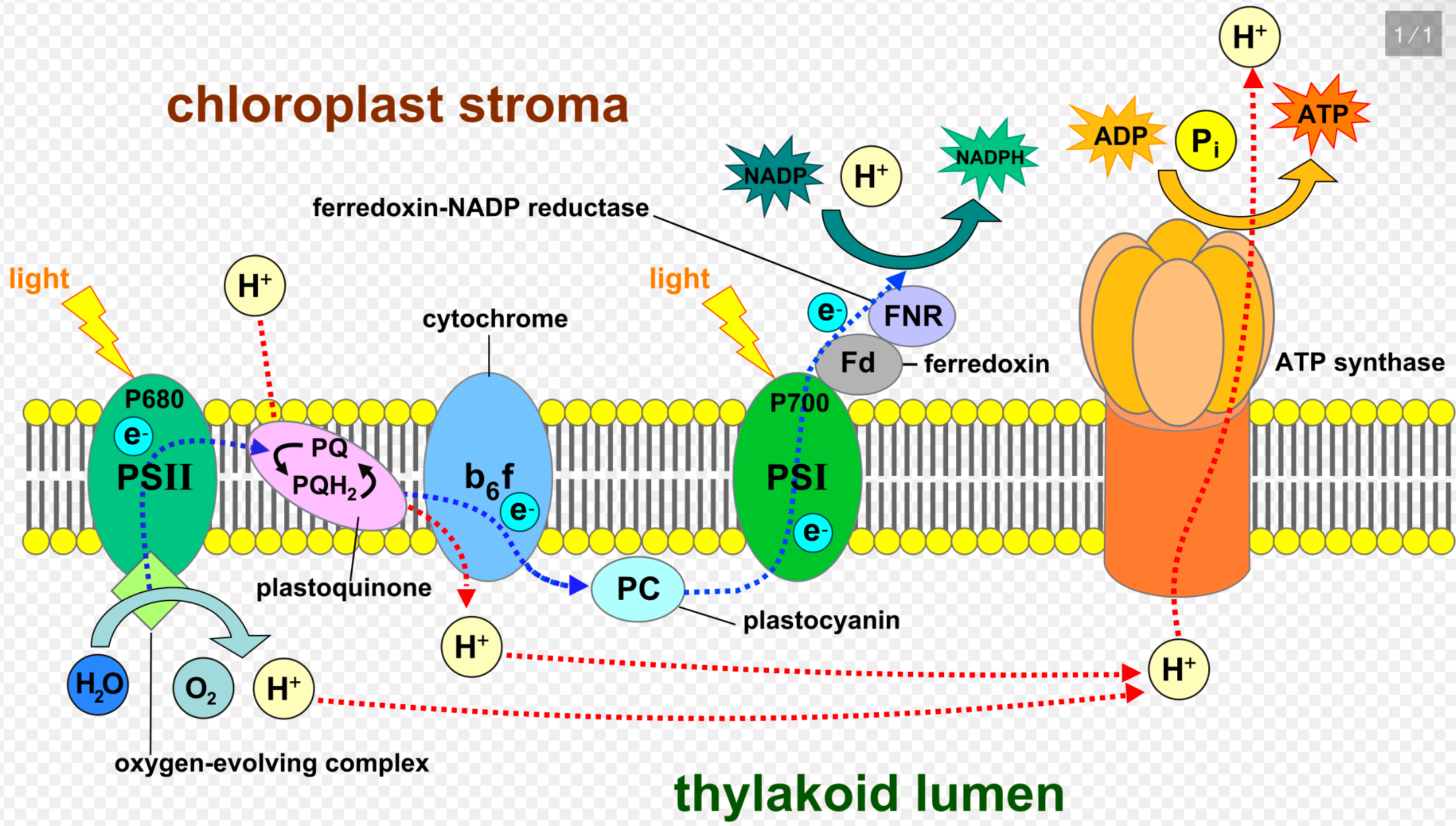
Light reactions
Light hits thylakoid
Electrons get excited, PSII —> PS1
ETC: water splits in PSII and donates e-
Fall of electrons drives H+ into lumen against gradient (active transport)
O2 dissipates from water
H+ flows through ATP synthase to stroma to make ATP
NADP+ electron carrier gets them from splitting of water ETC and the H+ from outside (stroma), becoming NADPH
So reactants: light, H2O, NADP+
Products: NADPH, O2, and ATP
Light-independent reactions/Calvin Cycle
Carbon fixation (CO2 incorporated into calvin 1 at a time)
Reduction (ATP and NADPH donates e- to reduce to G3P)
Regeneration of RuBP
So reactants: ATP, NADPH, CO2
Products: ADP, NADP+, G3P (3-carbon sugar used to synthesize glucose)
Photorespiration (C3 Plants)
when plants close their stomata to stop water loss; O2 is fixated instead of CO2
C4 Plants, CAM Plants
Alternate method to carbon fixation: spatial separation (partial closing of stomata), temporal separation
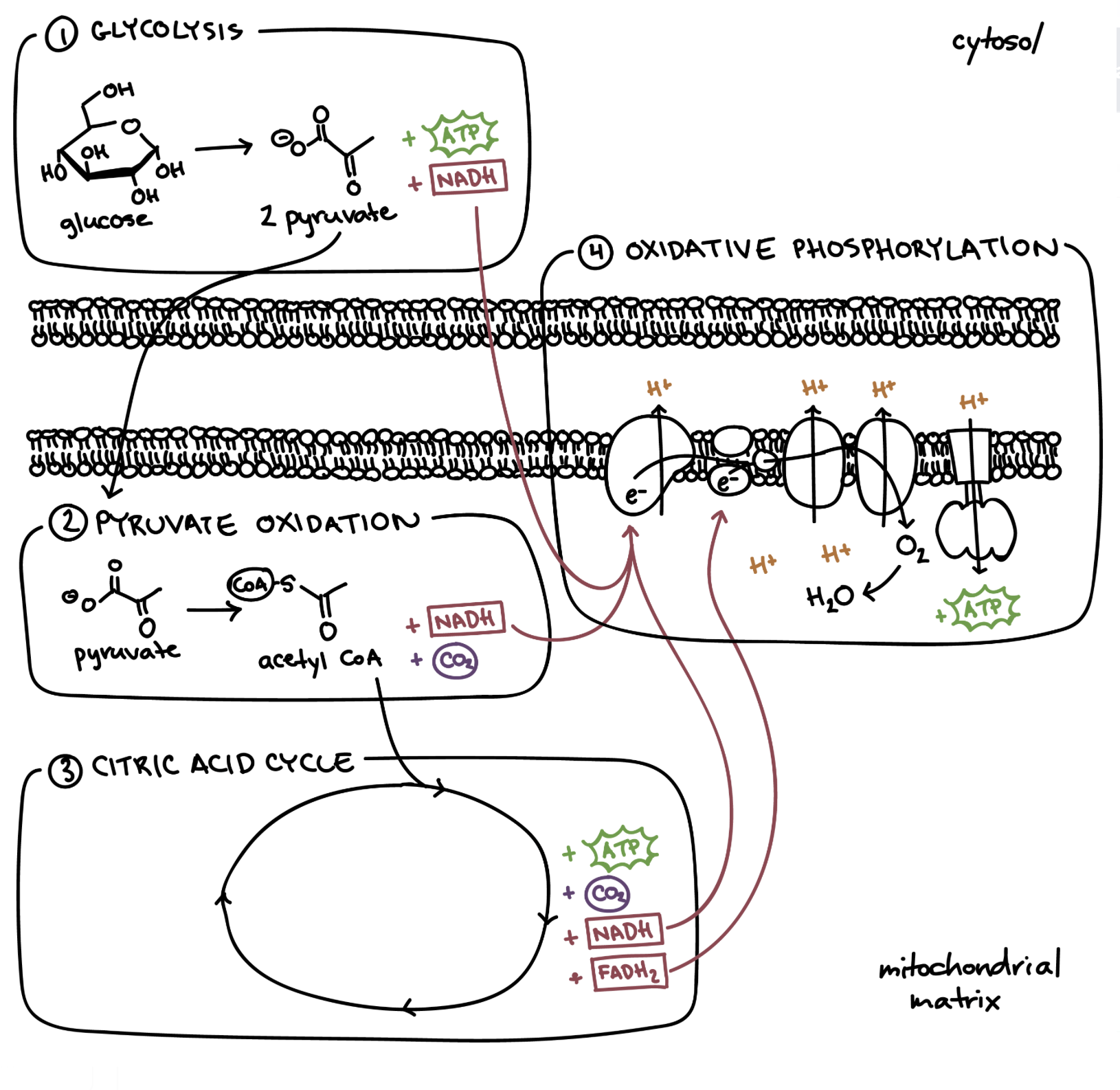
Steps of Cellular Respiration
Glycolysis (cytosol = the goo)
Pyruvate oxidation/Krebs/Citric Acid cycle (mitochondrial matrix = the room)
Oxidative Phosphorylation (cristae = the folds)
ETC (e- fall drives H+ gradient creation)
Chemiosmosis (H+ flow to create ATP)
Glycolysis
In cytosol; glucose splits into 2 pyruvate
Reactants: glucose, ATP and ADP, NAD+
Products: pyruvate, net ATP, NADH and extra H+, H2O
Pyruvate oxidation/Krebs/Citric Acid cycle
In mitochondrial matrix; if oxygen present:
pyruvate oxidized into acetyl coA; creates acetyl coA, NADH, and CO2
acetyl coA converted into citrate; creates citrate, NADH, FADH2, and CO2
Reactants: acetyl coA —> citrate, NAD+
Products: NADH, FADH2, ATP, CO2
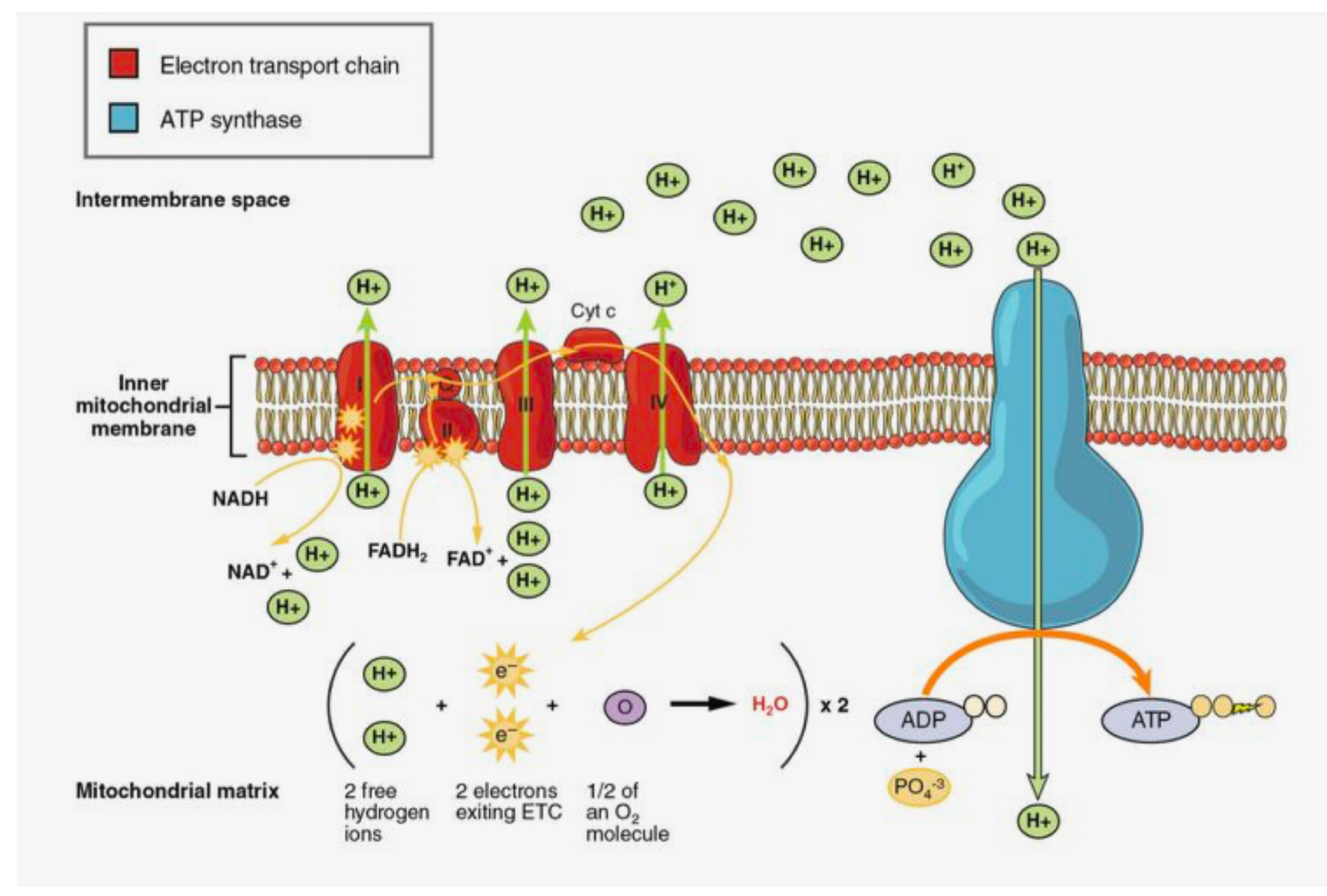
Oxidative Phosphorylation (ETC part)
In inner membrane/cristae;
NADH and FADH2 donate e-s to ETC from inner side and H+ to freely roam inside mitochondrial matrix
H+ + electrons all used up from the cycle + O2 produces water inside matrix
H+ pumped out into the inter membrane space
Flow back in so ATP synthase can create ATP inside matrix
Reactants: NADH, FADH2, O2
Products: NAD+, FADH, H2O, ATP
Cytoskeleton (Microtubules vs Microfilaments vs Intermediate filaments)
Microtubules: cell rigidity/structure; form mitotic spindle in cell division
Microfilaments/actin filaments: cell movement e.g. cytokinesis, muscle contraction
Intermediate filaments: anchor organelles in place
Respiration w/o oxygen
Anaerobic (w/ ETC)
Fermentation (w/o ETC) to produce ATP
Alcohol (NAD+ reduced from glycolysis, directly donates e-s to acetaldehyde, forms ethanol waste)
Lactic acid (NAD+ reduced from glycolysis, directly donates e-s to pyruvate, forms lactate waste)
3 Factors of Genetic diversity via Meiosis
Segregation (1 allele)
Independent assortment in metaphase 1
Random fertilization
Chromosome numbers throughout meiosis
Beginning: 2n chromosomes / 2n chromatids
After S phase: 2n chromosomes / 4n chromatids
After Meiosis 1: n chromosomes / 2n chromatids per daughter (2)
After Meiosis 2: n chromosomes / n chromatids per daughter (4)
True breeding
organisms that produce offspring of same variety
Chi square degrees of freedom
n-1 (for genotypes, n = alleles; for phenotypes, n = different phenotypes)
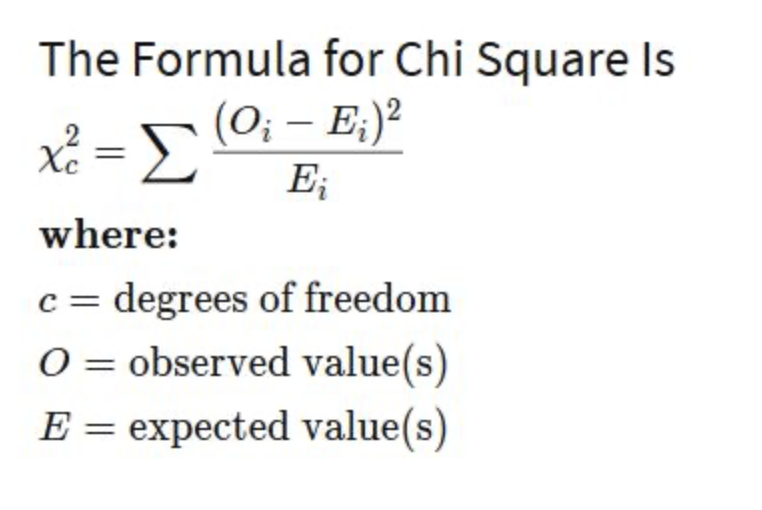
Chi Square
form of statistical test used to compare observed results with expected results
whether data from experiment is a “good fit”
if deviations from expected data are due to random chance or other circumstances
testing the null
x²=sum of ((observed-expected results)²/expected)
Dihybrid cross phenotypic ratio
9:3:3:1
Law of segregation
Each gamete has only 1 allele for given trait (since meiosis divides homologous chromosomes)
Law of independent assortment
Genes for one trait aren’t necessarily inherited with genes for another (eye color and hair color are inherited differently)
A reason why laws of independent assortment/segregation aren’t working
Genes are probably are on the same chromosome and have lower recombination frequency (linked genes)
Pedigree tips
If trait is dominant, one parent must have trait
If trait is X-linked, males are more commonly affected than females
If genders are equally affected, probably autosomal
Monohybrid cross
BB x bb
Dihybrid cross
YYRR x yyrr
Nonmendelian genetics
Traits have varying degrees of dominance (codominance or incomplete)
multiple genes act on a trait (polygenic inheritance)
sex-linked
mitochondrial/chloroplast DNA
Incomplete dominance
red + white = pink flowers (neither allele fully dominant)
Codominance
2 alleles are both expressed
type AB blood = A + B
Multiple alleles
some genes exist in forms with more than two alleles
Epistasis
allele at 1 locus affects allele at another locus. it can mask the effect of another locus e.g. gene for pigment & gene for depositing pigment in hair; if second 1 is albino, then first gene for pigment does not matter.
Linked genes
genes located near each other on same chromosome that tend to inherit together
50% or higher recombination frequency
genes are far apart on same chromosome or on two different
If x² > critical value [from table]
There is a statistically significant difference between observed and expected (reject null!)
If x² < critical value [from table]
there is NOT a statistically significant difference between observed and expected (fail to reject—not accept—null!)
How to calculate map distance between genes
probability that they will segregate as a unit aka recombination frequency
each % point = 1 map distance unit
50% or higher = located on different chromosomes or far apart on same; UNLINKED
Chloroplast and mitochondrial genes inheritance
randomly assorted to gametes and daughter cells (do not follow Mendelian rules)
In animals, mitochondria = from egg so maternally
In plants, mitochondria and chloroplasts = from ovule so maternally
Phenotypic plasticity
when a single genotype produces multiple phenotypes under different environmental conditions
e.g.
height and weight in humans
flower color based on soil pH
seasonal fur color in arctic animals
sex determination in reptiles
Increased UV on melanin production in animals
Presence of opposite mating type on pheromone production in yeast and other fungi
Example of genetic disorders
Aneuploidy (results from X-ray radiation):
Nondisjunction (trisomy 21/down syndrome) when chromosome 21 doesn’t separate properly; extra chromosome
Inversion: when segment of chromosome breaks and flips
Deletion: when it breaks off
Duplications: when it breaks off of one homologous chromosome and attaches to other so other has 2x gene
Translocation: when it breaks off and attaches to a NON-homologous chromosome
Sickle cell anemia: autosomal recessive disease from mutated gene leads to abnormal hemoglobin
X-linked color blindness
Tay-Sachs disease: autosomal recessive disease from mutated gene
Niche partitioning
Decreased competition among species for a niche because they are accessing a resource in different ways
Trophic cascade
Negative effect of removing key species —> population growth change exponentially, interrupted energy flow and limit resource availability
Species richness vs. relative abundance
many different species vs. lots of individuals per species
Removal of keystone species
Ecosystem collapse because of disproportionate influence to their relative abundance in an ecosystem; prey increases in number and prey’s food populations decrease
More biodiversity for an ecosystem
More resilient
Polar water meaning
unequal electron sharing (O has partial negative, H have partial positive)
Hydrogen bonds in water are intermolecular or intramolecular
intermolecular
The strength of hydrogen bonds
weak compared to ionic/covalent
Water properties
Cohesion
High heat of vaporization
High specific heat
Capillary action
Surface tension
Density of ice (less dense than liquid)
Adhesion
Capillary action (plant xylem is polar so H2O can bind); when adhesion > cohesion
Excellent solvency to polar molecules to which it can hydrogen-bond
Transpiration
Plant sweating, releasing water vapor from stomata which makes room for more water to go up
Important for water cycle
Monomer of Protein
Amino acid
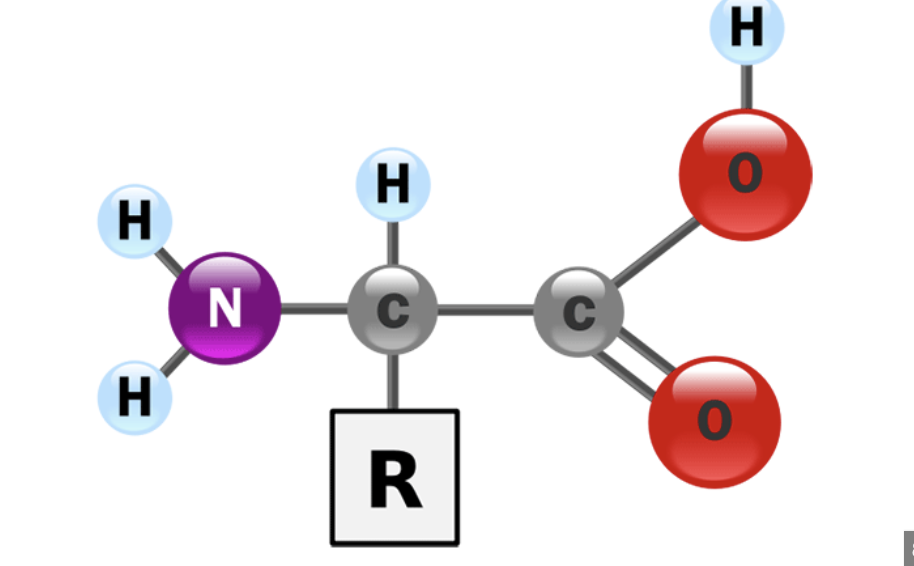
Amino Acid structure
central carbon, variable R side chain (20 diff kinds can be polar/nonpolar/ionic), amine group and carboxyl group on either side of carbon
Peptide bond
Primary structure bond which link amino acids together
Secondary structure
Beta sheets (pleated/folded paper) and alpha helices (corkscrew) because of hydrogen bonds between carbonyl and amine groups of amino acids
Tertiary structure
3D shapes; minimizes free energy
formed by R group interactions
hydrogen bonds between polar R groups
ionic bonds between charged side chains
disulfide bridges (covalent) between sulfur from sulfhydryl (SH) group on cysteines
Quaternary protein structure
Multiple polypeptides interacting with each other; not for all proteins
Carboxyl group
COOH
Carbonyl group
CO
Hydroxyl group
OH
Carbohydrate monomer, dimer, polymer
Monosaccharide: glucose
Disaccharide: sucrose
Polysaccharide: storage (starch/glycogen), structure (cellulose/chitin)
Type of bonds in carbohydrates
Glycosidic bonds
Passive transport (2 types)
Simple diffusion (small non polar molecules or gases)
Facilitated diffusion (hydrophilic uncharged molecules and charged ions via transport proteins)
Osmosis
both are down concentration gradient / does not require ATP
Active Transport
Requires ATP, unfavorable movement can be coupled with favorable movement i.e. cotransport
Sodium/Potassium pump
ATP synthase
Endocytosis/exocytosis
5 carbon sugar types
Ribose (2 OHs)
Deoxyribose (1 OH, 1 H)
Chromosome numbers through mitosis
Beginning: 2n chromosomes / 2n chromatids
After S-phase: 2n chromosomes / 4n chromatids
After mitosis: 2n chromosomes / 2n chromatids per daughter (2)
Biotechnology (4 types)
Used to analyze and manipulate DNA/RNA
PCR (Polymerase Chain Reaction)
DNA fragments are copied then amplified and even run through electrophoresis
Used to identify organisms and perform phylogenetic analyses
Bacterial transformation
Plasmids removed, gene of interest inserted, recombinant DNA reintroduced so gene expressed
DNA Sequencing
Determines order of nucleotides in DNA molecule
Gel Electrophoresis
Separates molecules according to size and charge
DNA is negatively charged, so will move to + side and smaller will move faster
How endothermic organisms generate heat from cellular respiration
Decoupling oxidative phosphorylation from ETC generates heat, which can be used to regulate body temperature
done by proteins allowing leakage of H+ across plasma membrane as heat
Prokaryotes’ chromosome
single, circular, double stranded chromosome in nucleoid
Eukaryotes vs prokaryotes vs plasmids chromosomal structure
Euk: multiple linear chromosomes
Prok: single circular chromosome
Plasmids: circular double-stranded (like mini-DNA)
Plasmids
Small double-stranded, circular DNA molecules in both prokaryotes and eukaryotes (double stranded because DNA, DNA because double stranded)
Purines
Double ring structure; Adenine and Guanine
Pyrimidines
Single ring structure; Cytosine, Uracil, and Thyamine
Direction DNA (replication) and RNA (transcription) are synthesized in
5’ to 3’
Semiconservative process
DNA replicated; one strand of DNA = template for new strand of complementary
Helicase
Unzipper of DNA in replication
Topoisomerase
Relaxes supercoiling in front of replication fork
Ligase
Joins Okazaki fragments on lagging strand
DNA polymerase
synthesizes DNA strands; requires RNA primers to initiate!!