Biochemistry I Lesson 2: Enzymes
1/91
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
92 Terms
DNA replication, the TCA cycle and the expression of genes are examples of reactions that can be sped up by which type of proteins?
(A) Transport
(B) Globular
(C) Denatured
(D) Catalytic
(D) Catalytic
DNA replication, the TCA cycle and the expression of genes are examples of reactions that can be sped up by the use of Enzymes, which are also known as Catalytic Proteins.
CRB Which of the following can Catalysts and Enzymes change for a reaction?
I. Gibbs Free Energy
II. Equilibrium
III. Rate of Reaction
(A) III only
(B) I and II only
(C) II and III only
(D) I, II and III
(A) III only
Catalysts and Enzymes only change the Rate of the Reactions! They do NOT affect the equilibrium position or the Gibbs Free Energy.
CRB Compare Enzymes and Catalysts.
Catalysts are any material that are not used up in a reaction, but can speed up the process of a reaction.
Enzymes are Biological Catalysts made of proteins. Also note that there are Ribozymes (RNA that can have catalytic functions similar to protein enzymes).
What is the primary role of enzymes?
(A) Increase the stability of the products.
(B) Increase the stability of the reactants.
(C) Increase the rate of a reaction.
(D) Decrease the change in Gibb's Free Energy from reactants to products.
(C) Increase the rate of a reaction.
An enzymes primary role is to increase the rate of a reaction. This is often done via increasing the stability of intermediates in the reaction.
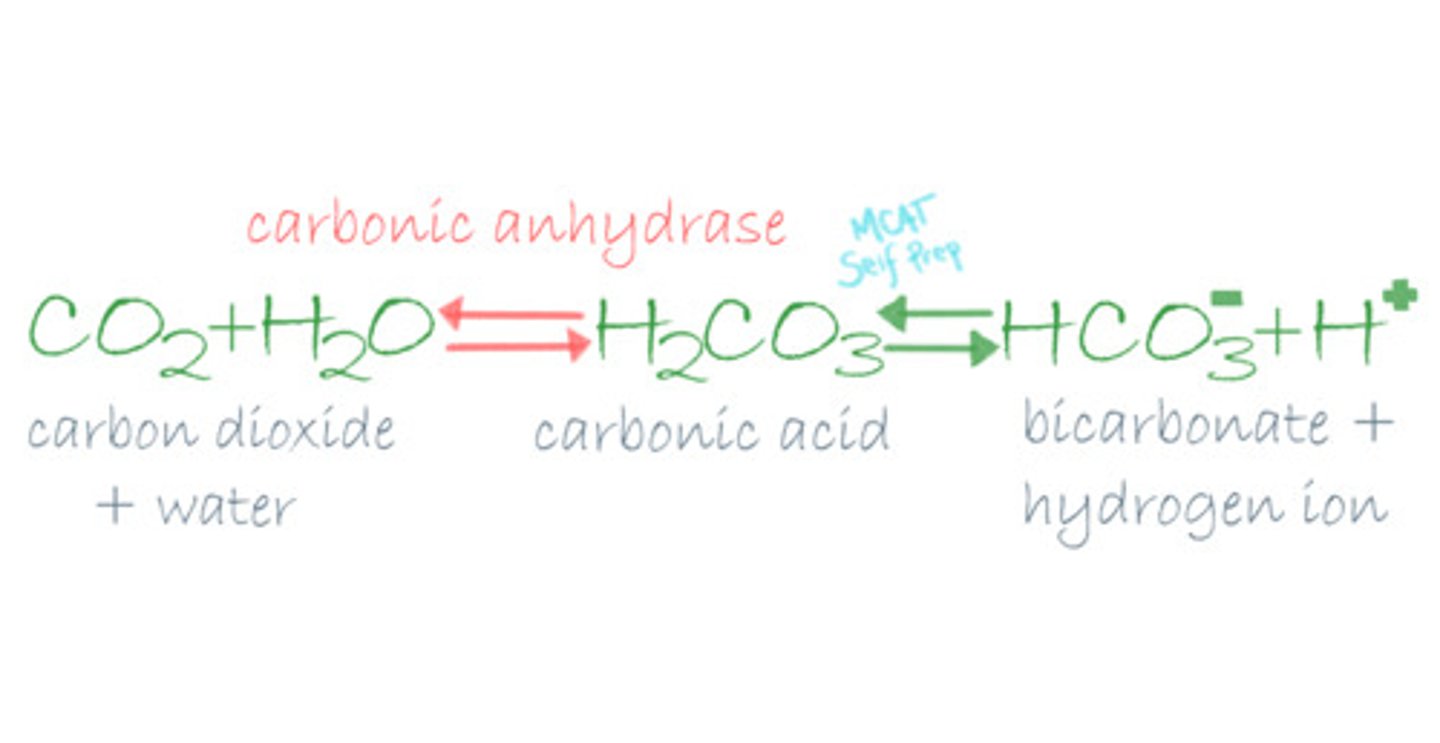
True or False? The following reaction requires the enzyme Carbonic Anhydrase in order to proceed: H2O + CO2 <--> H2CO3
False. This reaction can still occur without Carbonic Anhydrase. Carbonic Anhydrase will simply speed up this reaction. This is why soda will fizz MORE when it hits your tongue.
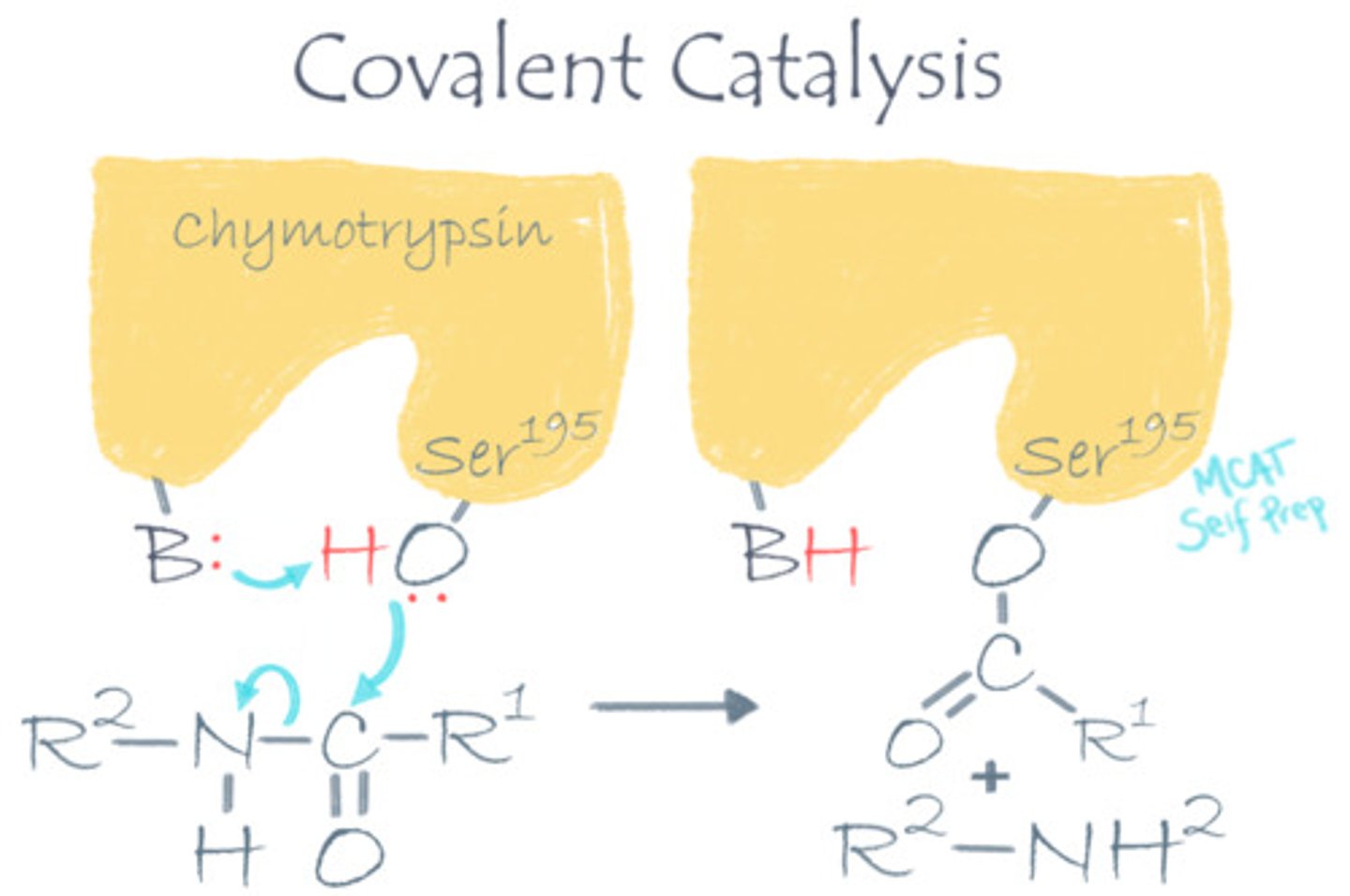
In a decarboxylation reaction where a carboxy (CO2) group is being removed from a molecule, a lot of electrons move around. What kind of catalytic strategy would help this reaction go faster?
(A) Acid/base Catalysis
(B) Electrostatic Catalysis
(C) Covalent Catalysis
(D) Proximity/Orientation Catalysis
(C) Covalent Catalysis
A covalent catalysis would help this reaction go faster because the enzyme can act as an electron carrier (electron sink) by covalently binding to its target molecule, thus helping with the transfer of electrons.
CRB A certain decarboxylation enzyme is very particular with both the substrates that it will react with and the type of reaction that it will promote. Does this meet the criteria to be an example of Enzyme Specificity?
Because a given enzyme is being selective in both the substrates it will work with and the type of reaction it is promoting, this is a prime example of Enzyme Specificity.
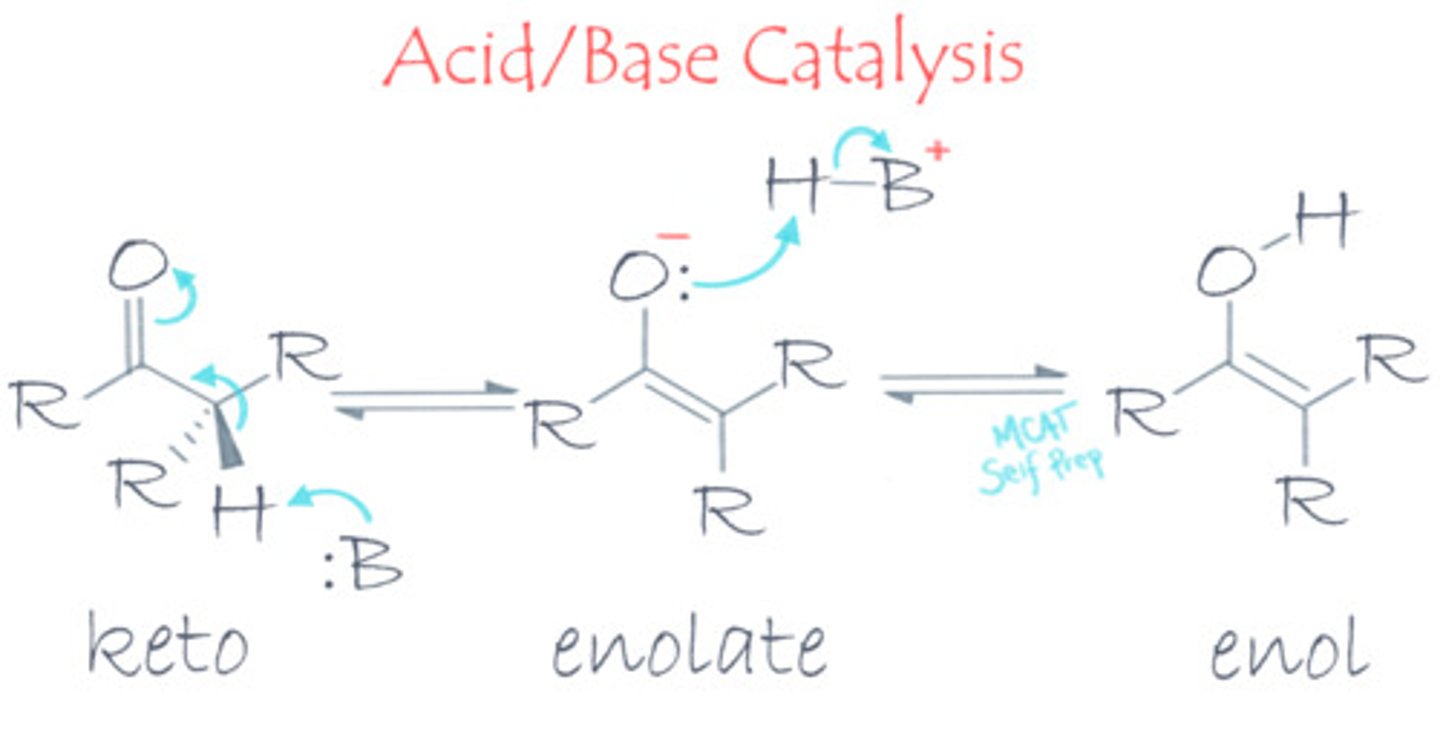
If you look at the keto-enol tautomerization reaction, a proton is moved from a carbon atom to an oxygen atom. What kind of catalysis would help this reaction go faster? Why?
(A) Acid/base Catalysis
(B) Electrostatic Catalysis
(C) Covalent Catalysis
(D) Proximity/Orientation Catalysis
(A) Acid/Base Catalysis
Acid/base catalysis would help this reaction go faster because the enzyme can function as a proton carrier. The enzyme can direct the proton's movement much more efficiently than if the proton were free floating.
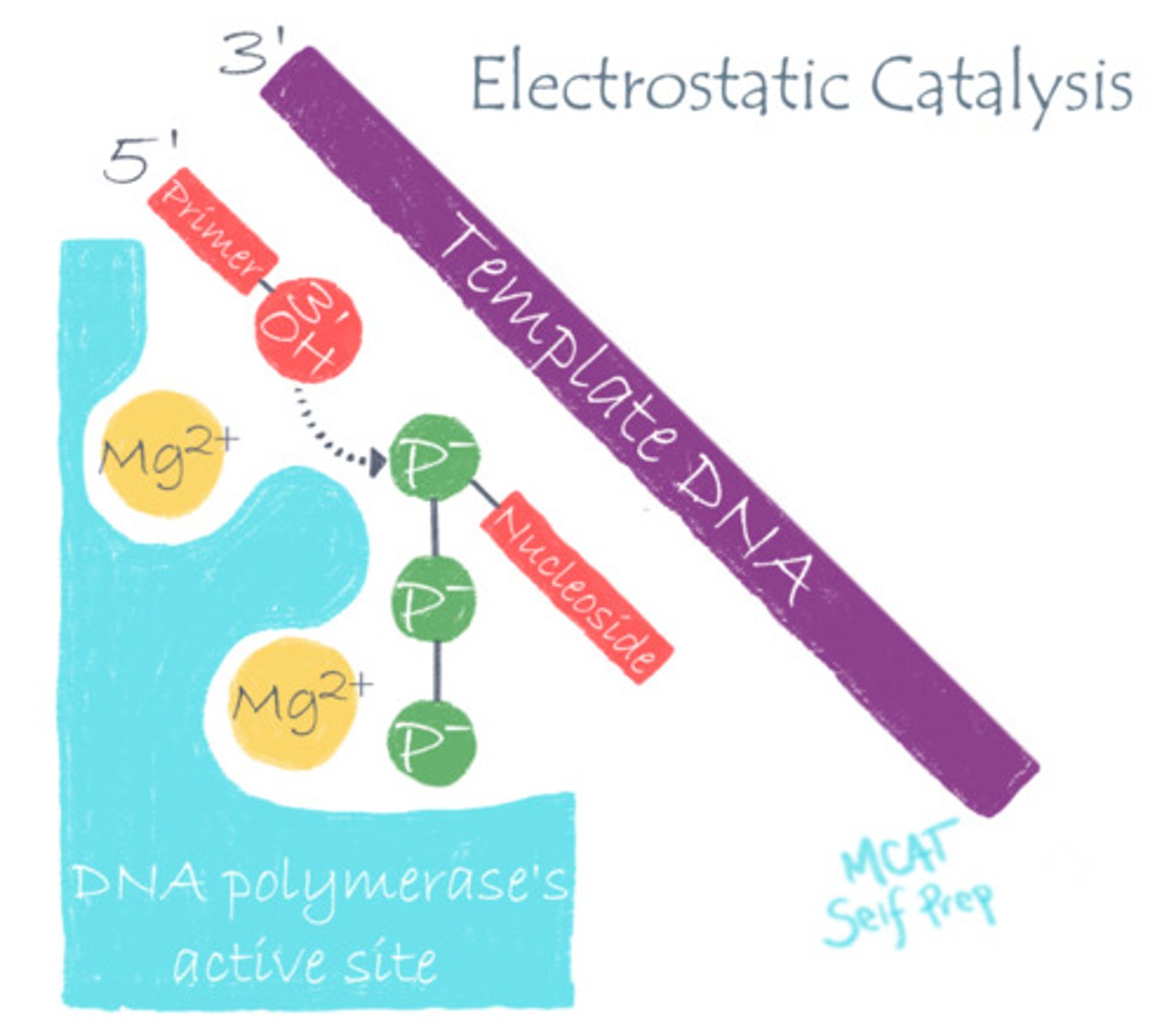
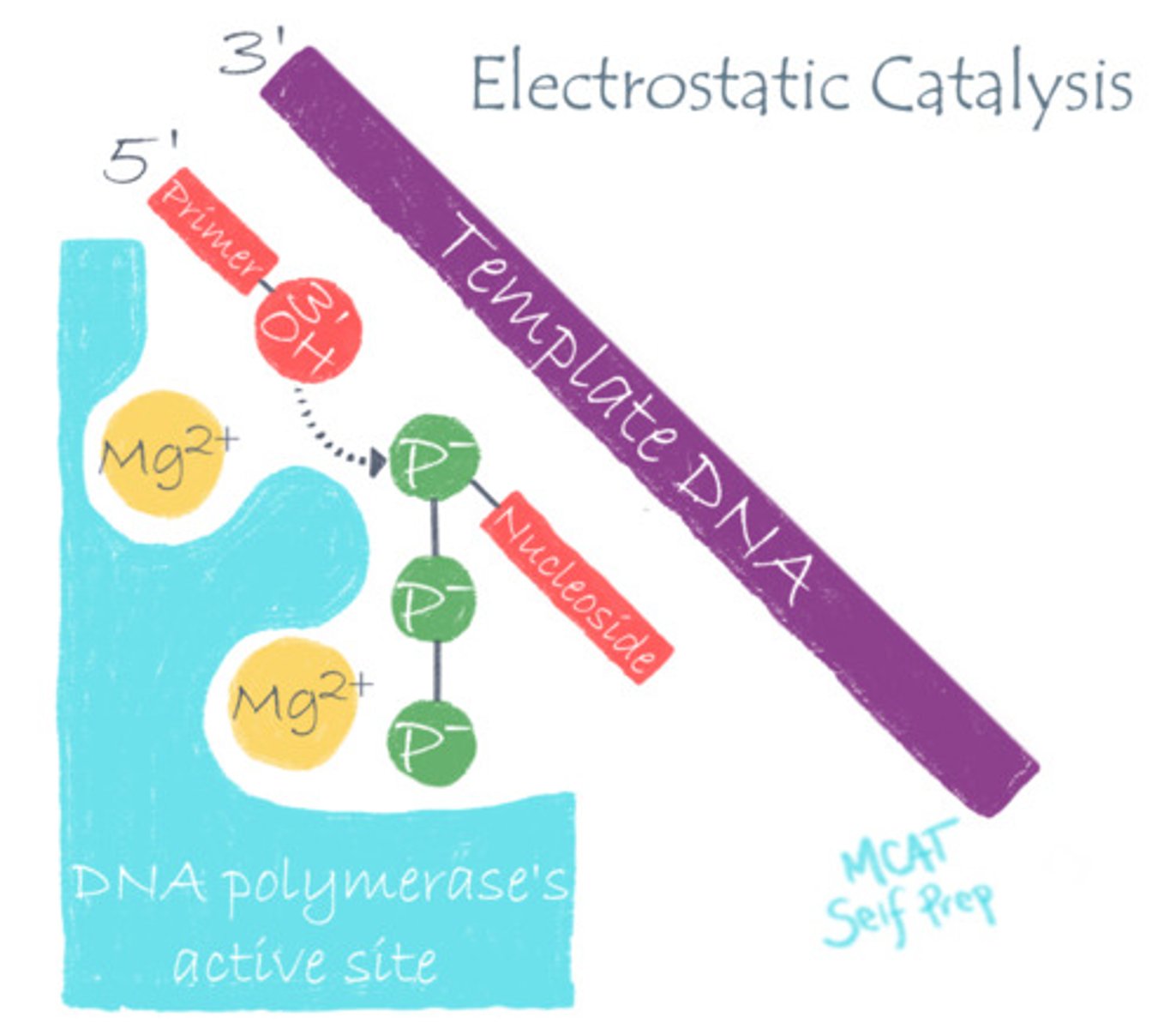
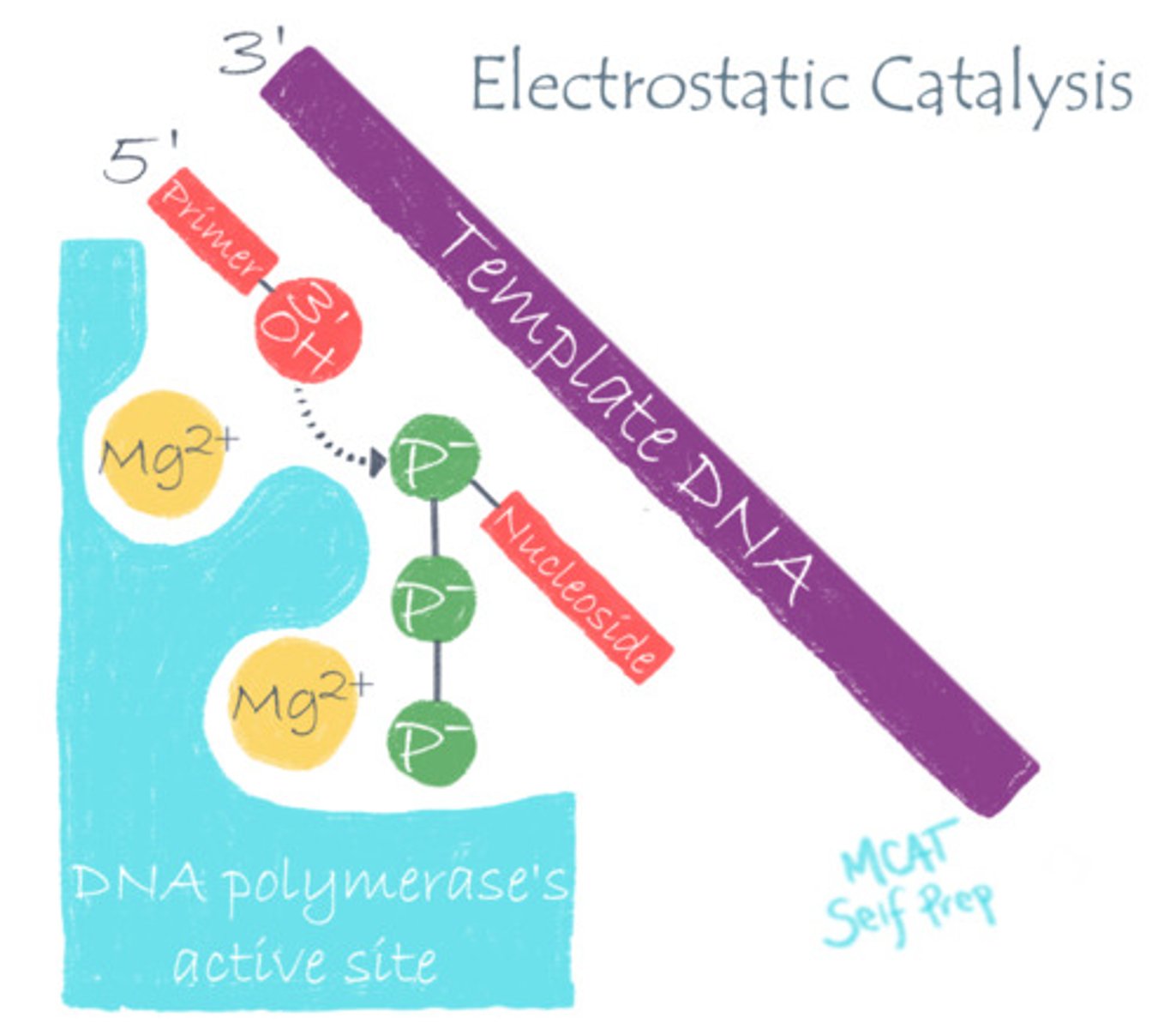
DNA polymerase stabilizes DNA's negative charge via what catalytic strategy?
(A) Acid/base Catalysis
(B) Electrostatic Catalysis
(C) Covalent Catalysis
(D) Proximity/Orientation Catalysis
(B) Electrostatic Catalysis
DNA polymerase utilizes electrostatic catalysis by using positively charged Mg to stabilize the negative phosphate charge on DNA, making it easier to work with.
Changing the pH will most likely influence enzymes involved in which of the following types of reactions?
(A) Acid/base Catalysis
(B) Electrostatic Catalysis
(C) Covalent Catalysis
(D) Proximity/Orientation Catalysis
(A) Acid/base Catalysis
Changing the pH will often affect enzymes involved in acid/base catalysis reactions, involving stabilizing charges, or transfer of electrons.
Describe how enzymes utilize both proximity and orientation effects to speed up reactions
Enzymes are able to bring 2 molecules close together (proximity effects) in the correct orientation (orientation effects) thus increasing successful molecule collisions, generating faster reactions.
What is the difference between enzymes and non-enzymatic proteins?
Non-enzymatic proteins do not catalyze reactions, whereas enzymes do catalyze reactions. Both are examples of proteins.
True or False? Proteins that function as receptors/ion channels, transport proteins, and antibodies are classified as non-enzymatic proteins.
True. Proteins that function as receptors/ion channels, transport proteins, and antibodies are classified as non-enzymatic proteins.
Amylase is most likely a(n):
(A) Enzyme
(B) Transport protein
(C) Ion channel
(D) Motor protein
(A) Enzyme
Amylase is most likely an enzyme. Proteins with names ending in "-ase" are most likely enzymatic proteins. Amylase is an enzyme responsible for breaking down polysaccharides into monosaccharides.
True or False? Some proteins may function as both enzymatic and non-enzymatic proteins.
True. Not all proteins are strictly enzymatic or not. Some proteins have characteristics of both enzymatic and non-enzymatic proteins, and can serve non-enzymatic functions.
You just ate a piece of pizza, causing an increase in blood glucose levels. The pancreas responds by releasing insulin, which will bind to a cell, causing the cell to absorb this excess glucose. What type of protein is detecting and binding the insulin in this scenario?
(A) Transport Protein
(B) Motor Protein
(C) Receptor Protein
(D) Antibody Protein
(C) Receptor Protein
A receptor protein will bind a ligand/signaling molecule (i.e. insulin) and will induce a chemical cellular response.
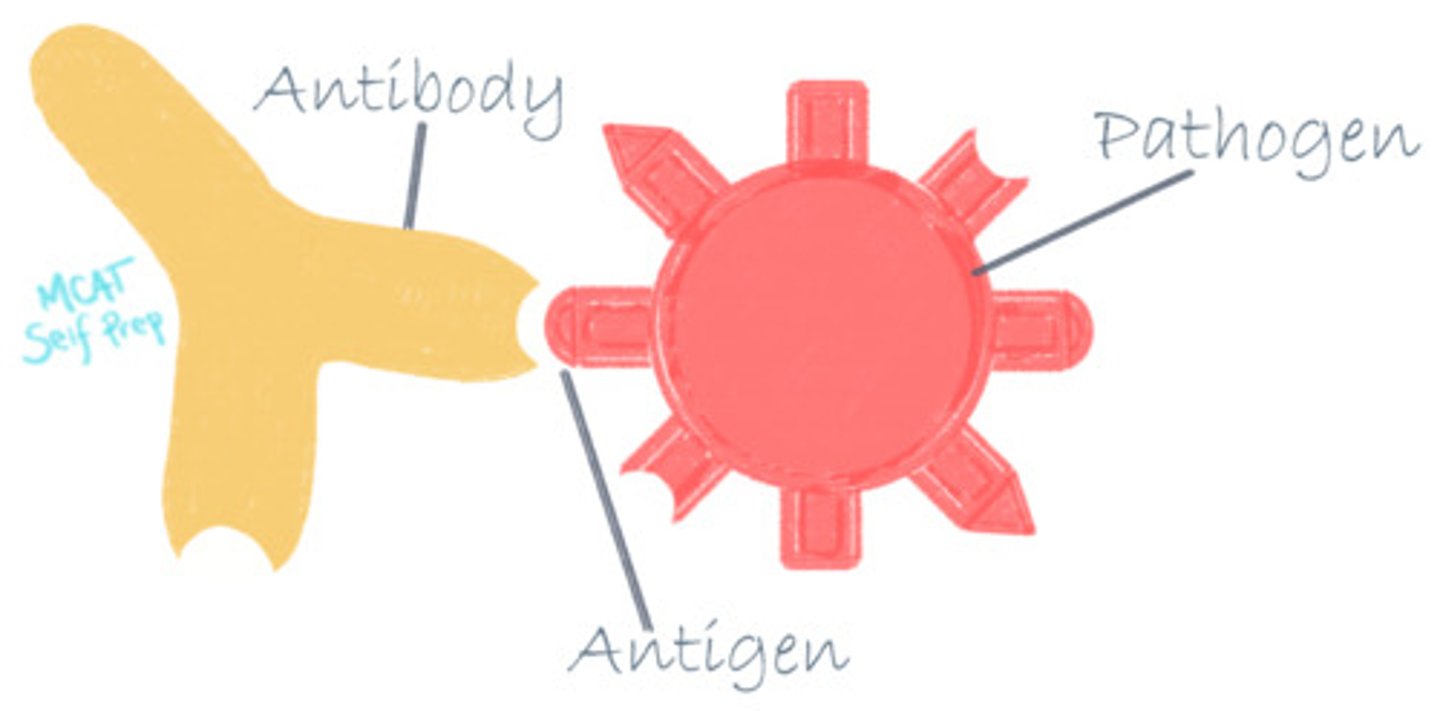
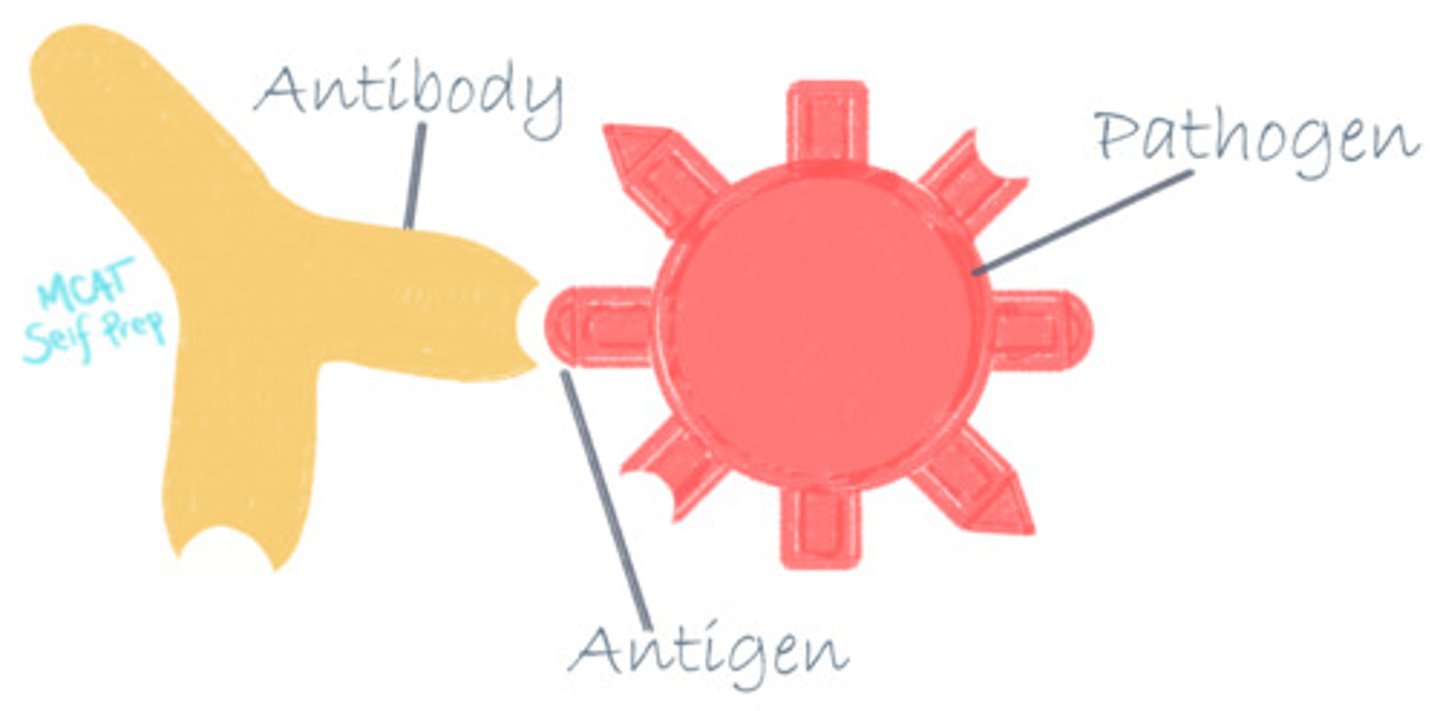
Describe the relationship between antibodies and antigens
Antigens are the ligands of antibodies. If an antigen is not recognized as belonging to the host, the antibody can recognize and flag this protein for elimination or degradation.
CRB True or false? Antibodies are also called Immunoglobulins, and are the most prominent proteins in the immune system.
True. Antibodies are also called Immunoglobulins, and are the most prominent proteins in the immune system.
A protein has a high affinity for its ligand when ligand concentrations are high and a low affinity when ligand concentrations are low. Which protein is this most likely to be?
(A) Transport Protein
(B) Motor Protein
(C) Receptor Protein
(D) Antibody Protein
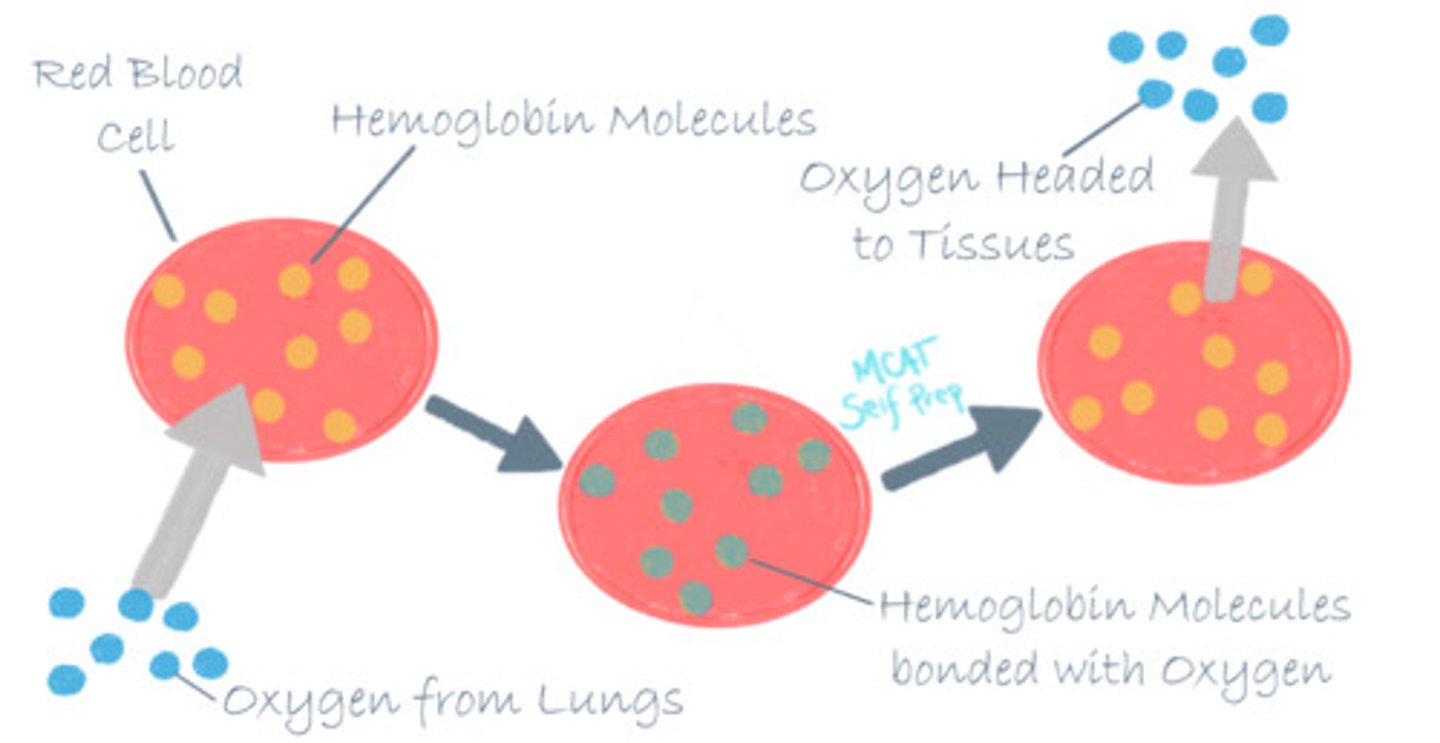
(A) Transport Protein
A great example of a transport protein is hemoglobin, which will bind oxygen where it is present in high concentrations (the lungs) and will have a decreased affinity and release the oxygen where its concentration is lower (other tissues). Other transport proteins can be used to span cellular membranes and control concentration gradients.
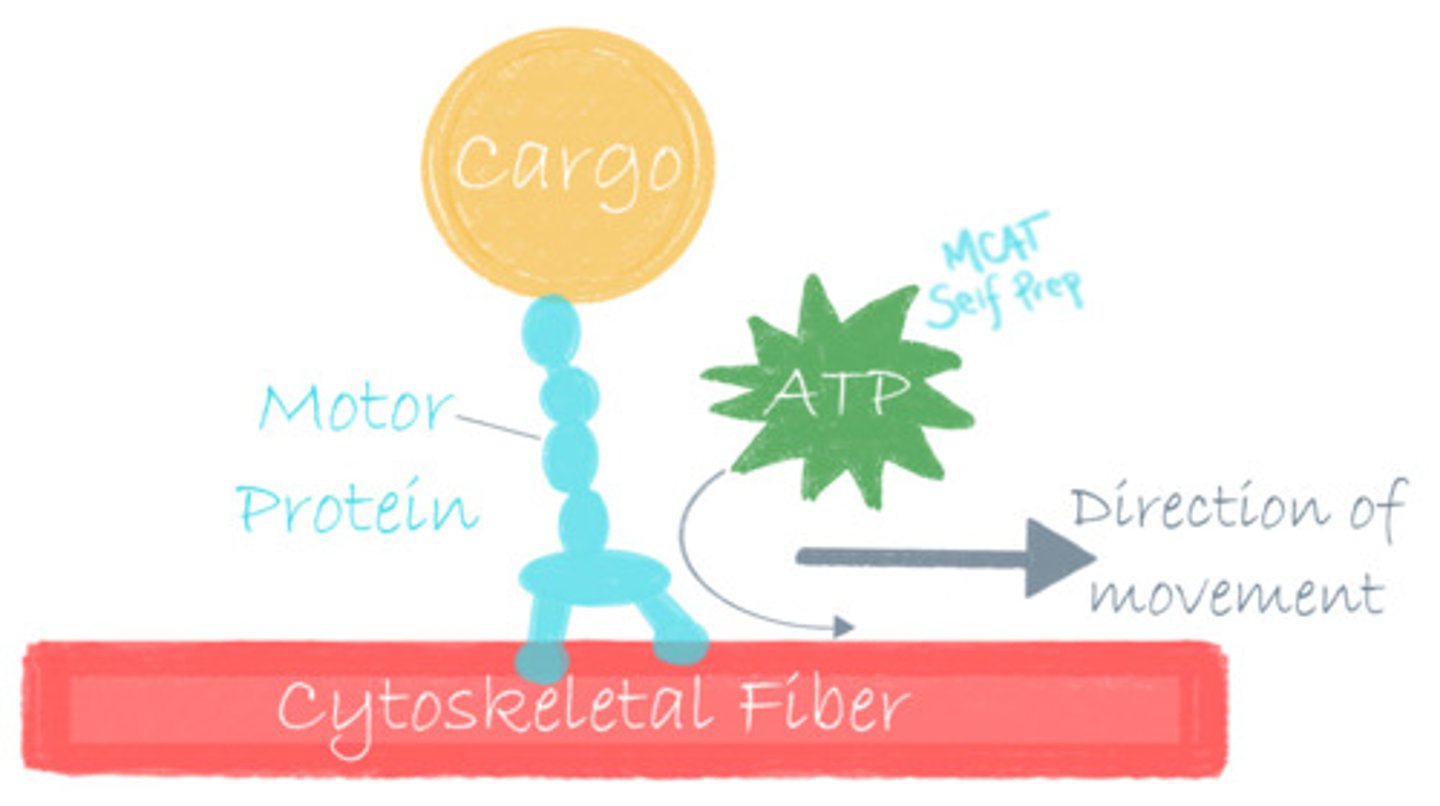
Myosin, kinesin, and dynein are all examples of what kind of protein?
(A) Transport Protein
(B) Motor Protein
(C) Receptor Protein
(D) Antibody Protein
(B) Motor Protein
Myosin, kinesin, and dynein are all examples of motor proteins because they are capable of generating forces and are important for cell motility.
Which motor protein(s) is/are specifically responsible for generating the forces exerted by contracting muscles?
I. Myosin
II. Kinesin
III. Dynein
(A) I Only
(B) III Only
(C) I and II Only
(D) II and III Only
(A) I Only
Myosin is the motor protein responsible for generating the forces exerted by contracting muscles.
Which motor protein(s) is/are specifically responsible for intracellular transport?
I. Myosin
II. Kinesin
III. Dynein
(A) I Only
(B) III Only
(C) I and II Only
(D) II and III Only
(D) II and III Only
Kinesin and dynein are motor proteins that are responsible for intracellular transport.
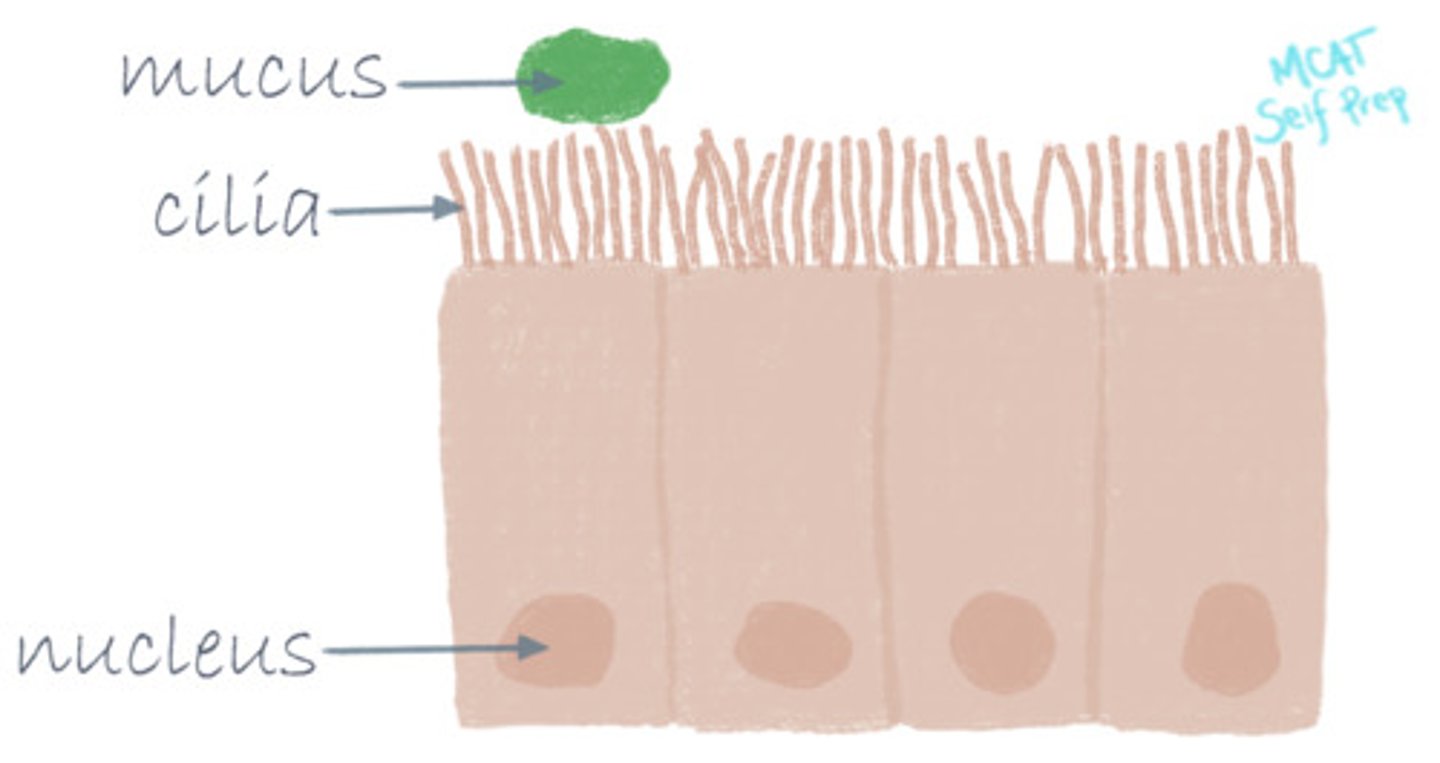
A rare disease called primary ciliary dyskinesia causes the cilia of the cells lining the respiratory tract to malfunction. This leads to an accumulation of mucus and an increased susceptibility to chronic infections like pneumonia and bronchitis. Which motor protein(s) is/are responsible for the motility of cilia?
I. Myosin
II. Kinesin
III. Dynein
(A) I Only
(B) III Only
(C) I and II Only
(D) II and III Only
(B) III Only
Dynein is specifically responsible for the motility of cilia.
Which of the following protein types has the highest affinity for its ligand?
(A) Transport Protein
(B) Motor Protein
(C) Receptor Protein
(D) Antibody Protein
(D) Antibody Protein
Antibodies are the proteins with the highest affinity for its ligand (antigens).
CRB There are also Structural Proteins, which can help to make up the extracellular matrix and cytoskeleton. Which of the following is NOT a Structural Protein?
(A) Collagen
(B) Myelin
(C) Keratin
(D) Actin
(B) Myelin
Common examples of Structural Proteins are Collagen, Elastin, Keratins, Actin and Tubulin.
Myelin is the fatty sheath that surrounds nerve cell axons.
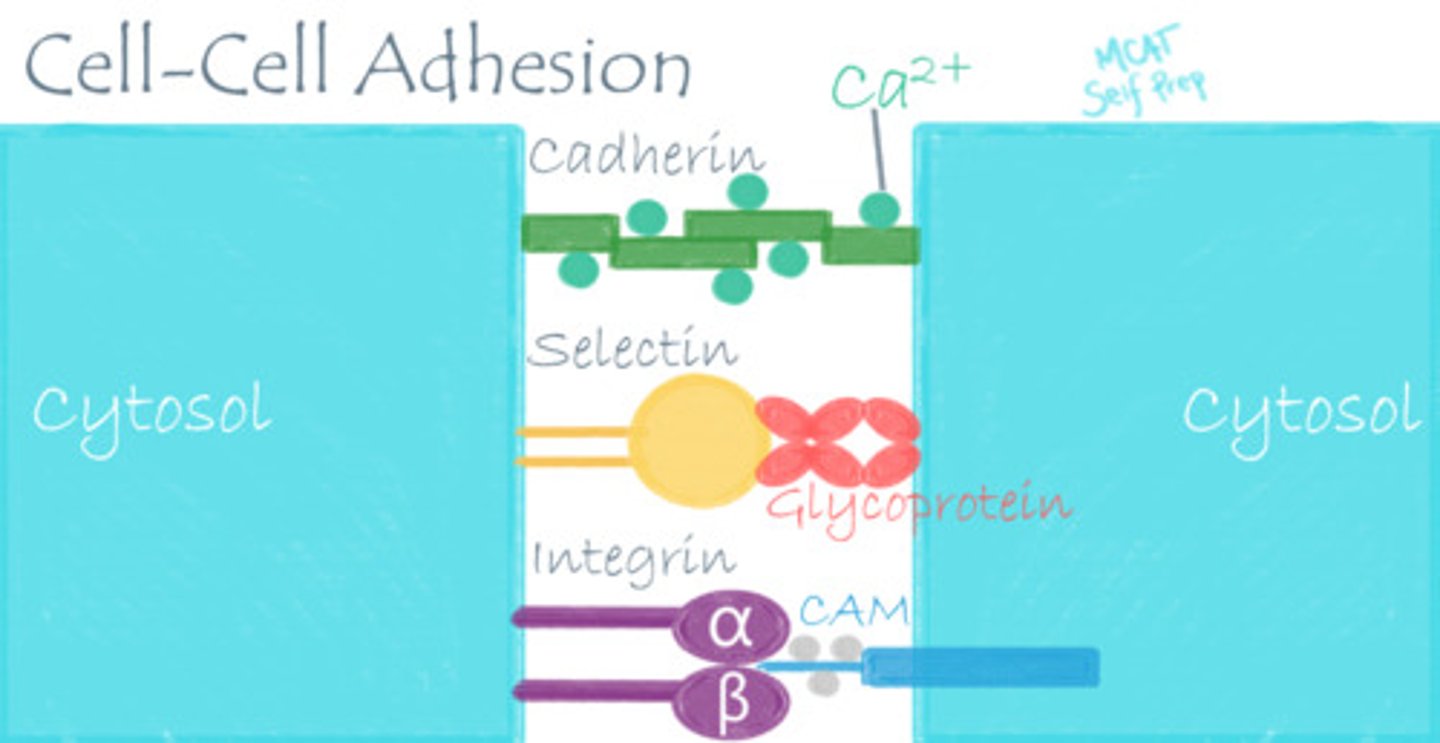
CRB One more major class of Nonenzymatic Proteins are Cell Adhesion Molecules. Which of the following is not one of the three main types of Cell Adhesion Molecules?
(A) Cadherins, which are glycoproteins, are calcium-dependent and hold similar cell types together.
(B) Binding proteins, which include calcium- and DNA-binding proteins.
(C) Integrins, which have alpha- and beta-chains spanning the membranes of cells and can promote cell division.
(D) Selectins, which can bind carbohydrate molecules from other cell surfaces.
(B) Binding proteins, which include calcium- and DNA-binding proteins.
Binding Proteins actually include Transport Proteins, and also include Hemoglobin and can sequester molecules.
Each of the following are a type of Cell Adhesion Molecule:
I. Cadherins, which are glycoproteins, are calcium-dependent and hold similar cell types together.
II. Integrins, which have alpha- and beta-chains spanning the membranes of cells and can promote cell division.
III. Selectins, which can bind carbohydrate molecules from other cell surfaces.
What is the key difference between co-translational modifications and post-translational modifications?
Co-translational modifications are any changes made to the polypeptide while it is being translated.
Post-translational modifications are any changes made to the protein after translation.
During translation, the first amino acid methionine is removed and replaced by an acetyl group. What is this an example of? A co-translational modification or a post-translational modification?
This is an example of a co-translational modification because the removal of methionine and the addition of an acetyl group is happening during the translation process.
Where do most of the post-translational modifications happen in the cell?
(A) ER/Golgi
(B) Cytosol
(C) Nucleus
(D) Lysosome
(A) ER/Golgi
Most of the post-translational modifications happen in the endoplasmic reticulum and the Golgi apparatus of the cell.
What would happen to cells if glycosylation (adding a carbohydrate to a protein) did not work?
I. Cell identification would be rendered more difficult.
II. Cell-cell communication would be rendered more difficult.
III. Receptors on the cell membrane would be rendered less effective.
(A) I and II Only
(B) II and III Only
(C) I and III Only
(D) I, II, and III
(D) I, II, and III
If glycosylation did not work then it would be harder to identify cells and it would be harder for cells to communicate and sense their surrounding environment thus effectively diminishing the functionality of a cell because glycosylation adds carbohydrates to embedded proteins (making them glycoproteins) on the cell that could serve as receptors and ID tags to identify the cell.
What is the role of lipidation and what kind of protein modification is it? A co-translational modification or a post-translational modification?
In lipidation, a lipid is added to a protein in order to help attach proteins to the cell membrane of a cell. Lipidation is a post-translational protein modification.
Which of the following properly characterizes Phosphorylation?
I. It is involved in activating/deactivating enzymes.
II. It promotes the catalysis reaction.
III. It requires relatively little energy.
(A) I Only
(B) II Only
(C) III Only
(D) I and II Only
(D) I and II Only
Phosphorylation is a protein modification that is involved with enzyme activity and helps promote the catalysis reaction. It requires ATP.
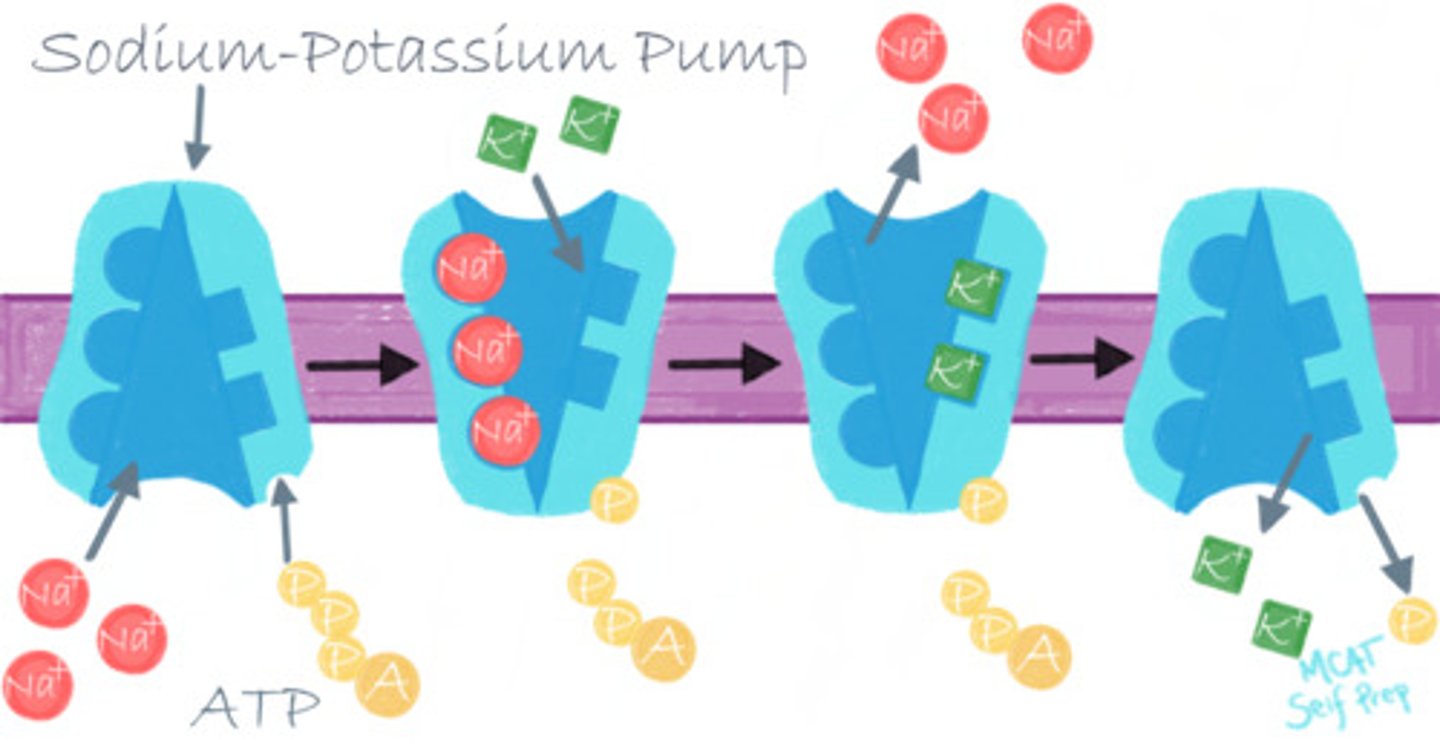
What would happen to the function of the enzyme Na+/K+ ATPase if there was no ATP available?
The Na+/K+ ATPase would not function without ATP as that provides the energy it needs.
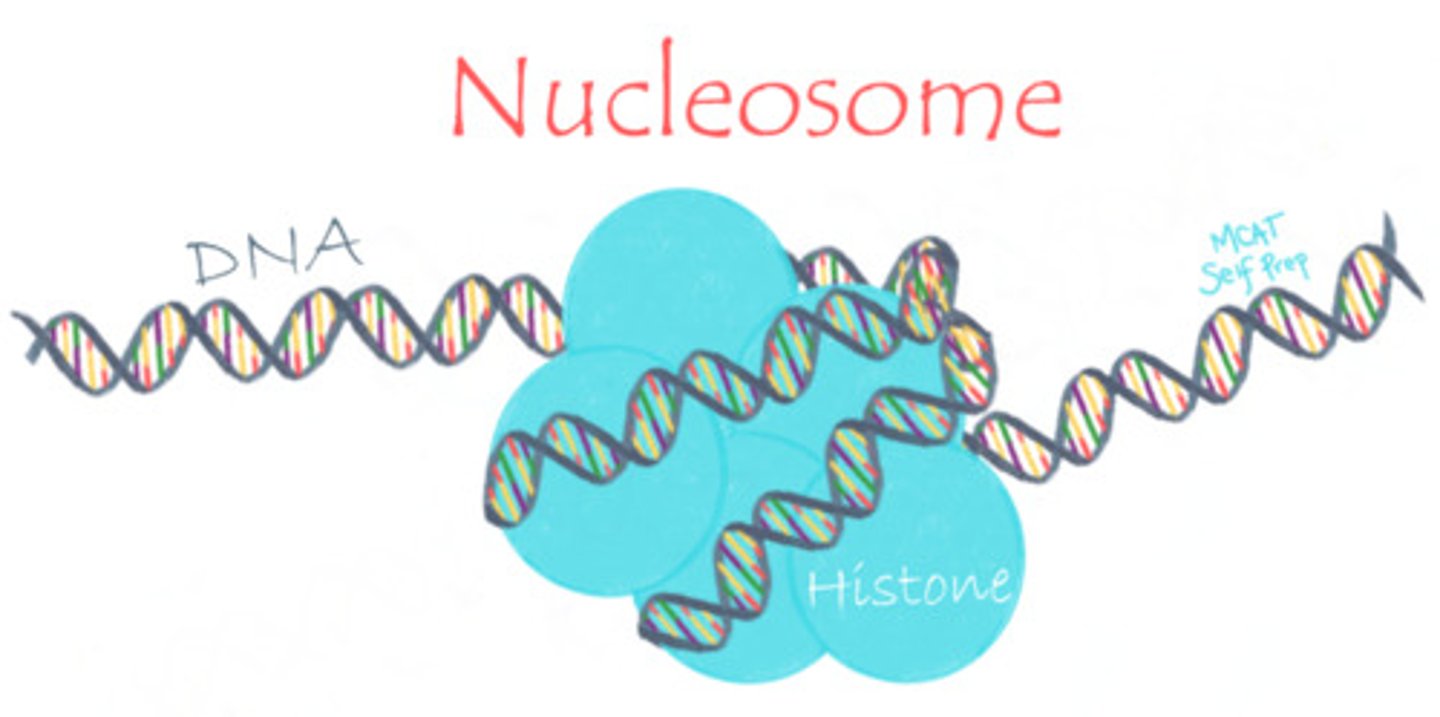
What is the relationship between DNA and histone proteins?
DNA is wrapped around histones. This allows DNA to pack into the chromosome more tightly.
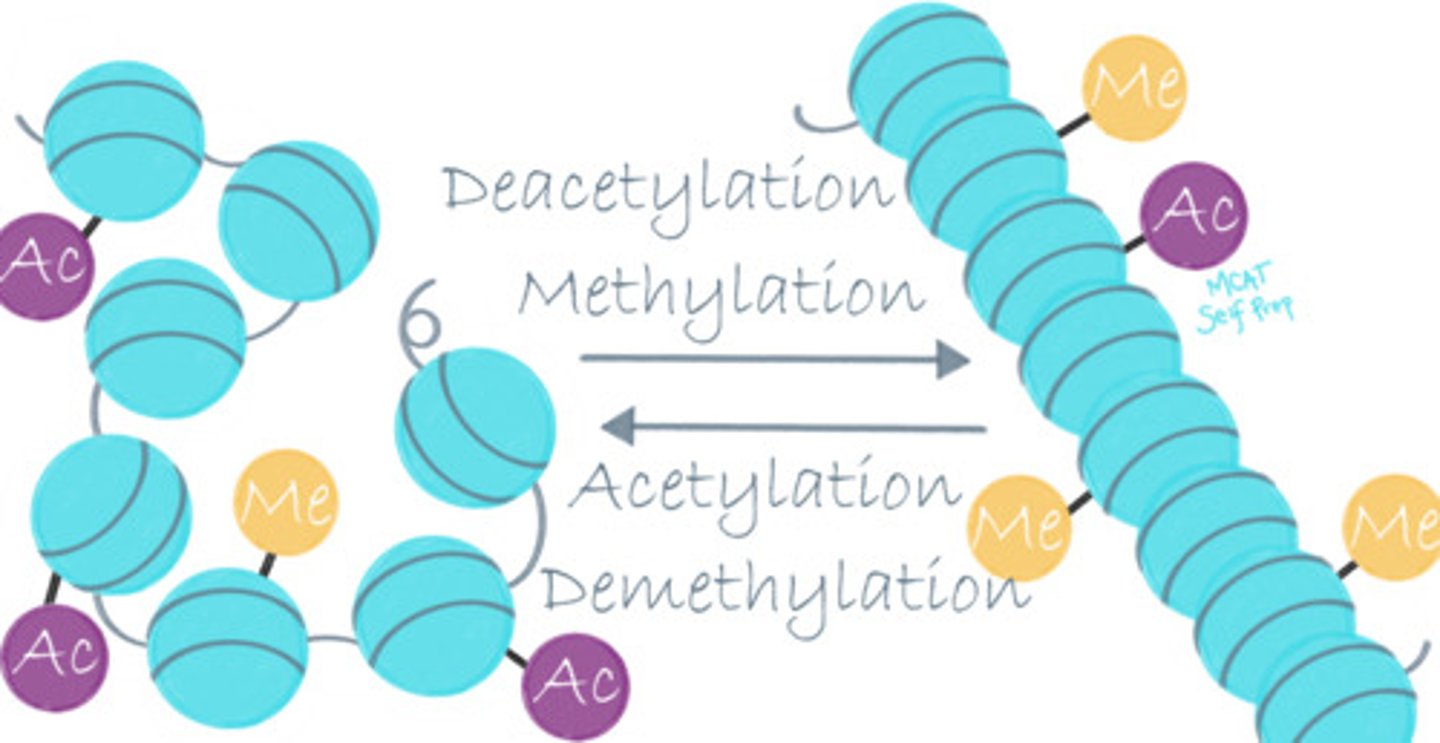
What is the purpose of methylating and demethylating histone proteins?
The purpose of methylating and demethylating histone proteins is to regulate gene expression. Typically, if a histone is methylated then that gene is not expressed. If it is unmethylated, the gene is expressed.
What is the relationship between proteolysis and zymogens?
The proteolysis (cleavage/cutting) of a zymogen (inactive catalytic protein) will convert it into its active, catalytic form.
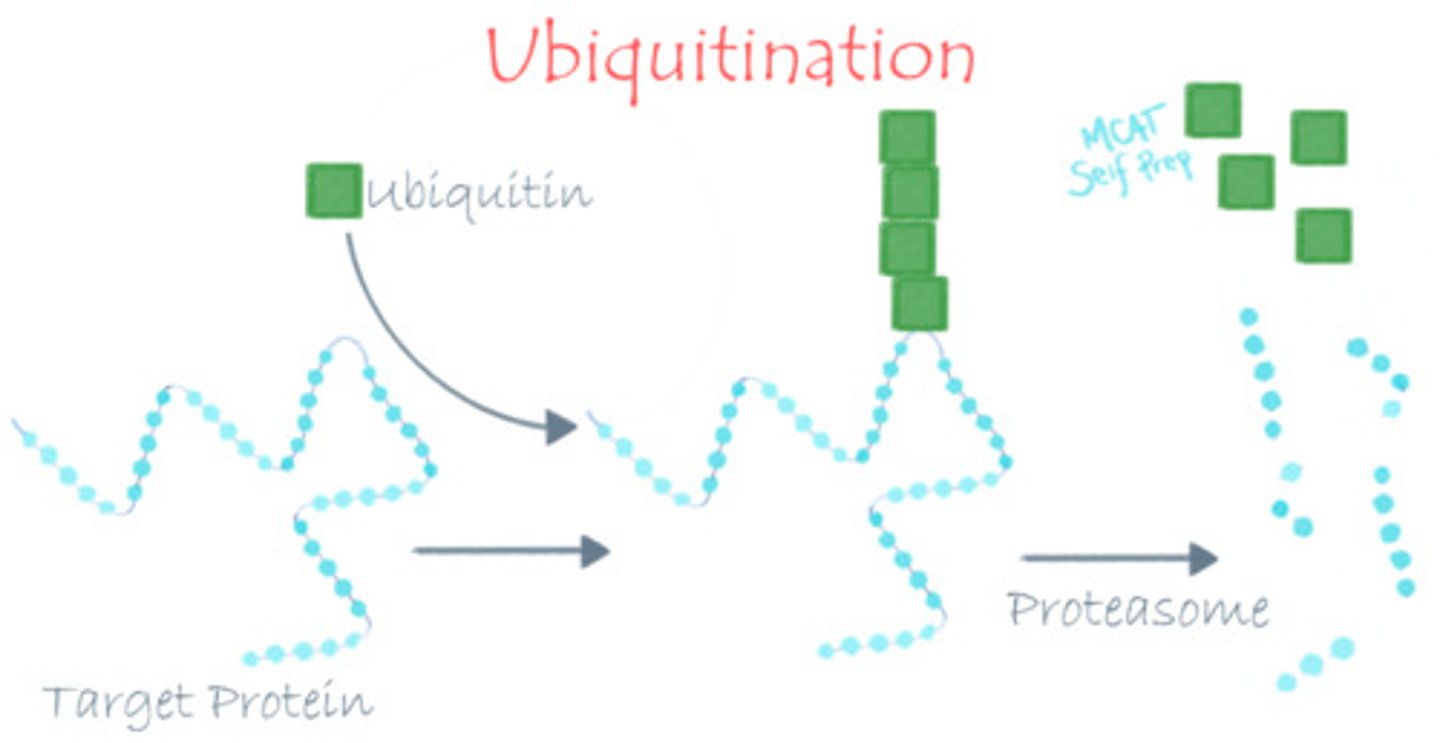
What is the role of ubiquitination?
Ubiquitination is the process of attaching a ubiquitin group to a protein, marking it for degradation.
Which protein modification(s) would you expect to find in proteins that are embedded in the cell membrane?
I. Glycosylation
II. Lipidation
III. Methylation
(A) I Only
(B) I and II Only
(C) II and III Only
(D) I, II, and III
(B) I and II Only
You would expect to find either Glycosylation or Lipidation because these two protein modifications generally happen to proteins that are embedded in the cell membrane.
Glycosylated proteins act as markers for the cell. Lipidation is key for integral membrane proteins to make them more hydrophobic, as the phospholipid bilayer of the cell membrane is also hydrophobic.
How are phosphorylation, methylation, and proteolysis all similar in terms of their role?
Phosphorylation, methylation, and proteolysis are similar in that they all regulate activity by either "activating" or "deactivating" the gene/enzyme.
Acetylation, methylation, and glycosylation examples of:
(A) Pre-transcriptional modification
(B) Post-transcriptional modification
(C) Pre-translational modification
(D) Post-translational modification
(D) Post-translational modification
Acetylation, methylation, and glycosylation are examples of post-translational modifications (modifications to proteins that occur immediately after the proteins are translated).
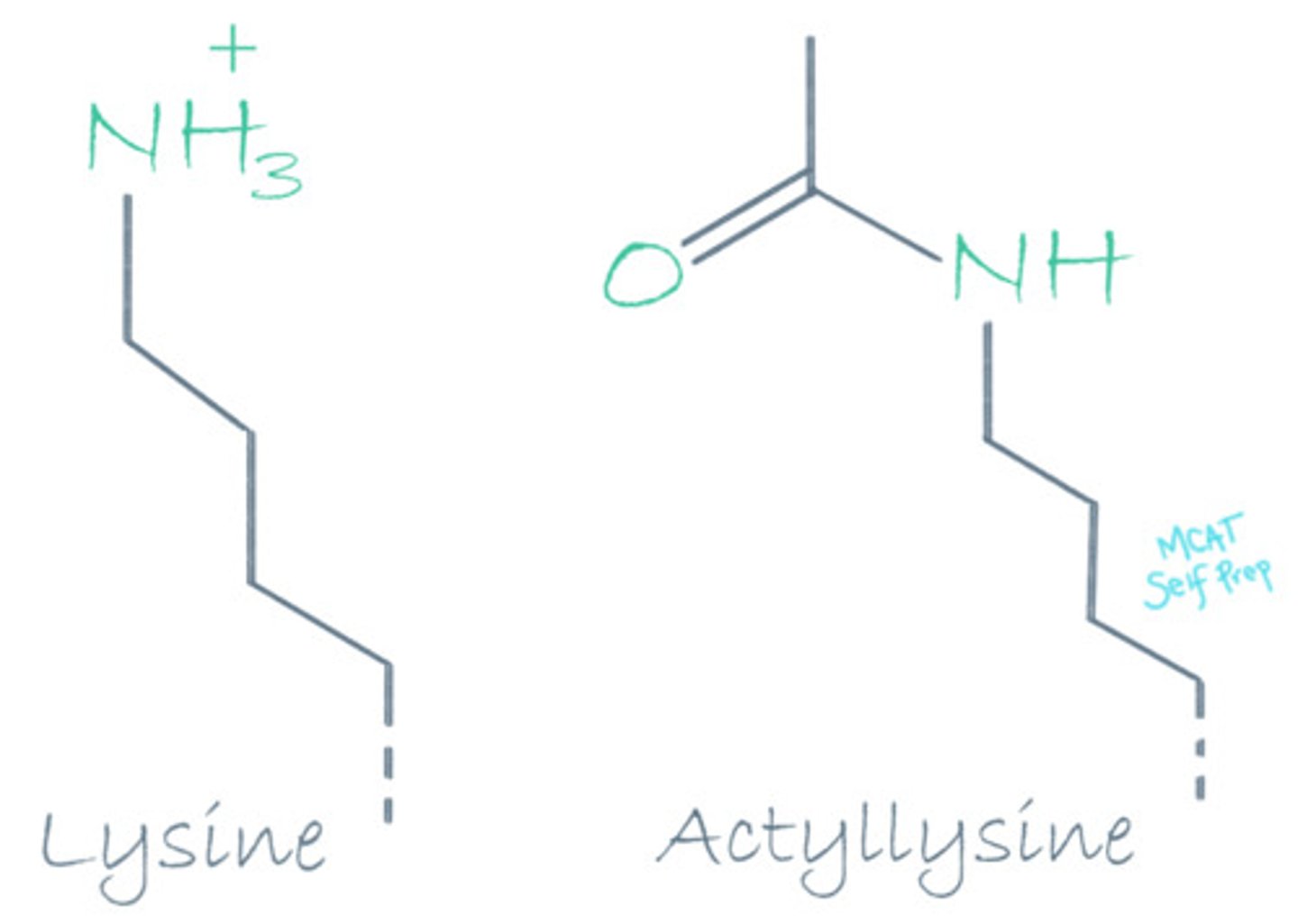
Adding an acetyl group as a post-translational modification is most likely to result in what kind of changes to a Lysine residue's function?
Adding an acetyl group to a Lysine residue will have an electron withdrawing effect that can eliminate the positive charge, affecting the lysine residue's acidity/basicity and its electrostatic interactions.
The pancreas releases trypsinogen into the intestines. Once in the intestine, part of trypsinogen is cleaved to form trypsin, activating the enzyme to break down proteins in the intestine. Trypsinogen is an example of what type of protein?
(A) Non-functional Enzyme
(B) Zymogen
(C) Unzyme
(D) Enzyme
(B) Zymogen
A zymogen is an inactive form of an enzyme that requires a covalent modification to be activated. The enzyme trypsin is secreted as an inactive zymogen that is activated once it reaches the intestines. This is vital, because activated trypsin would digest the pancreas!
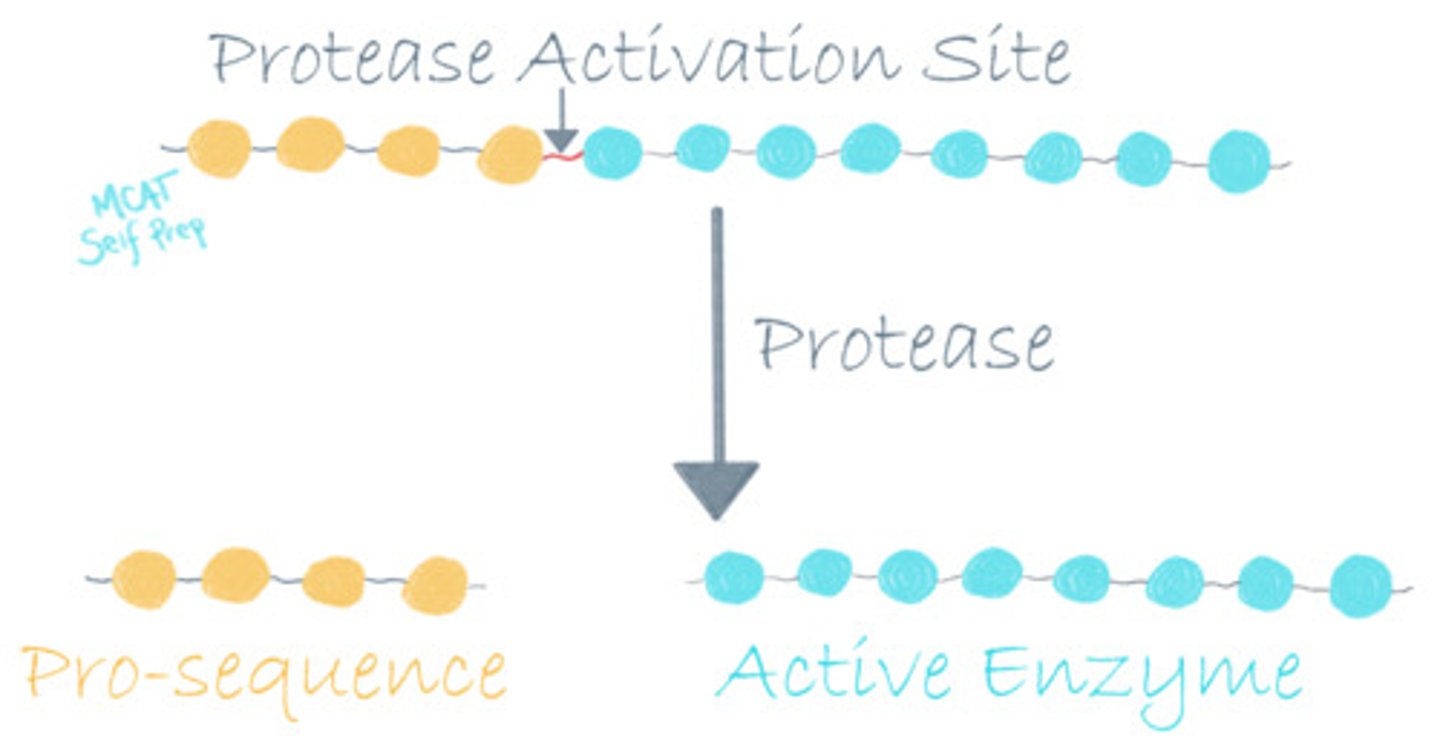
Describe the mechanism of action used by "suicide inhibitors."
Suicide Inhibitors covalently bind to an enzyme, preventing the enzyme from catalyzing any further reactions. Covalent linkages rarely unbind, hence the name "suicide inhibitors".
What is the relationship between a molecule's energy level and its stability?
(A) No correlation
(B) Positive correlation
(C) Negative correlation
(D) Mixed correlation
(C) Negative correlation
A molecule's energy level and its stability are inversely related. If the molecule's energy levels rise, its stability decreases, and vice versa.
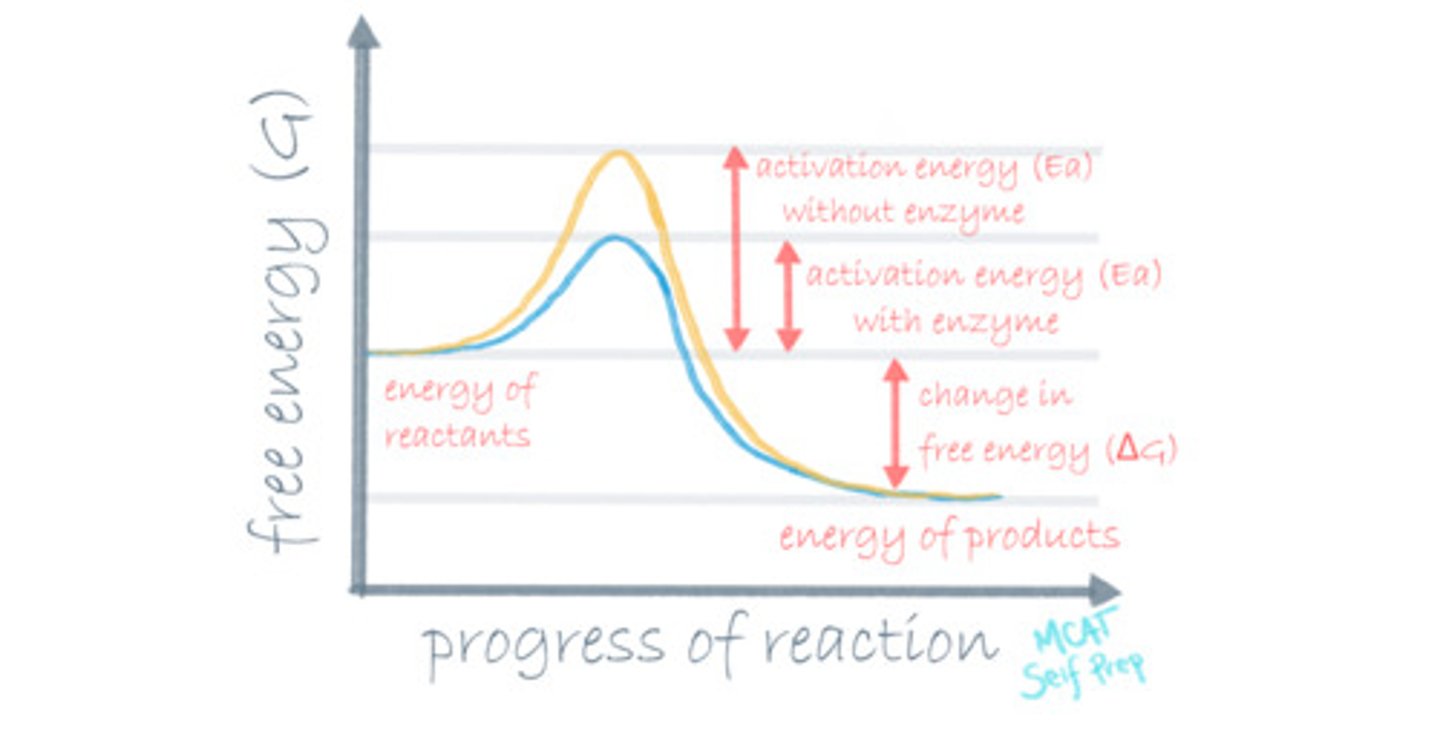
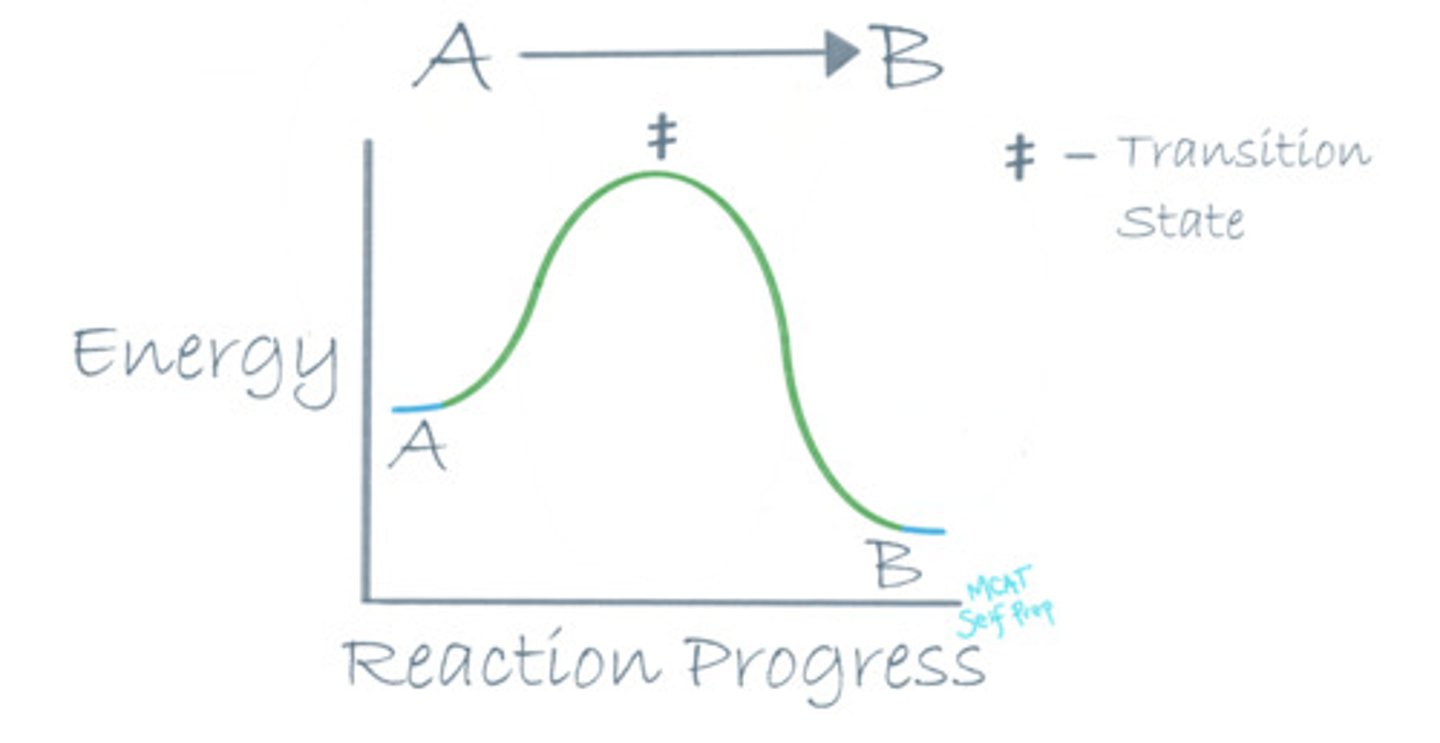
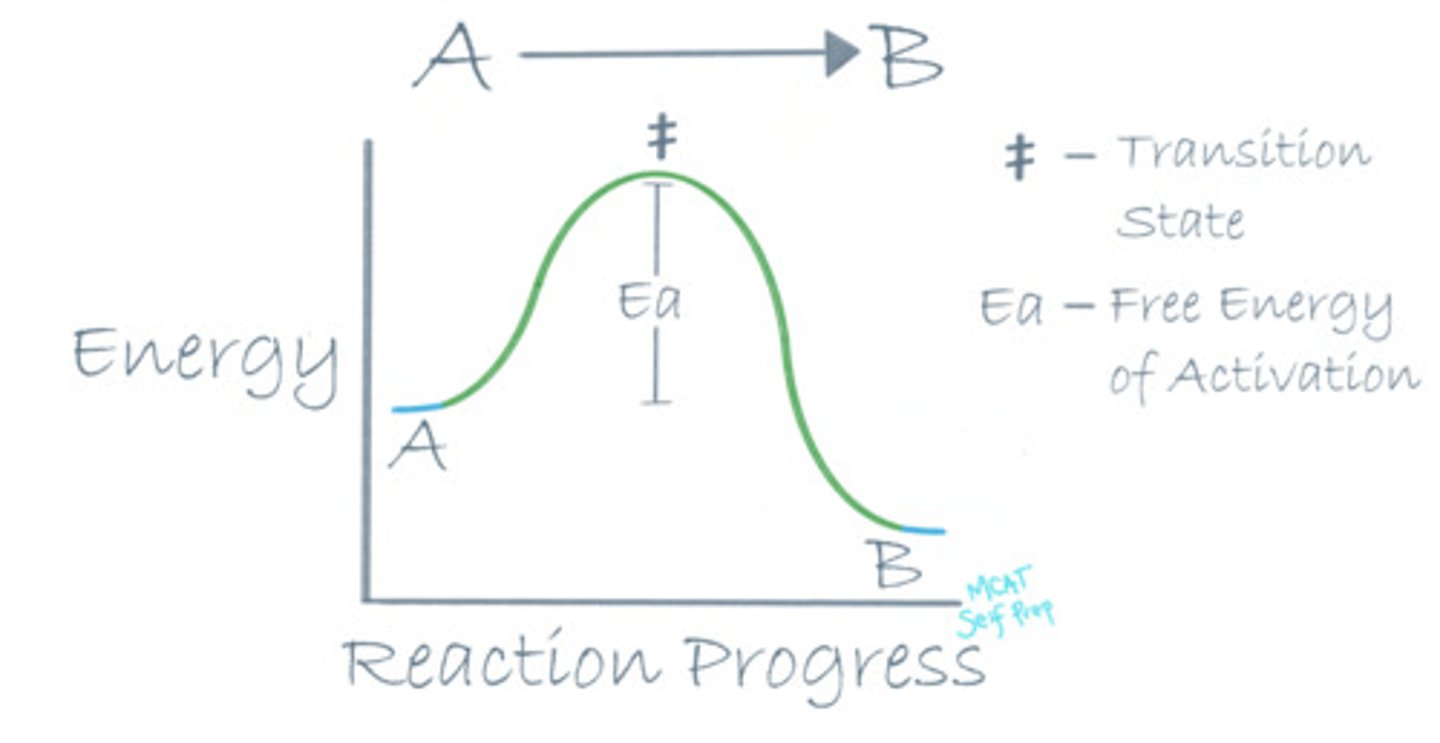
Draw a reaction coordinate diagram for an exothermic reaction. Where is the transition state of the reaction?
For an exothermic reaction, the energy level of the reactants (A) should be higher than the energy level of the products (B), because energy is an additional product of an exothermic reaction.
The transition state on a reaction coordinate diagram is the highest energy point on the path from point A to point B. It is the most unstable point throughout the entire reaction.
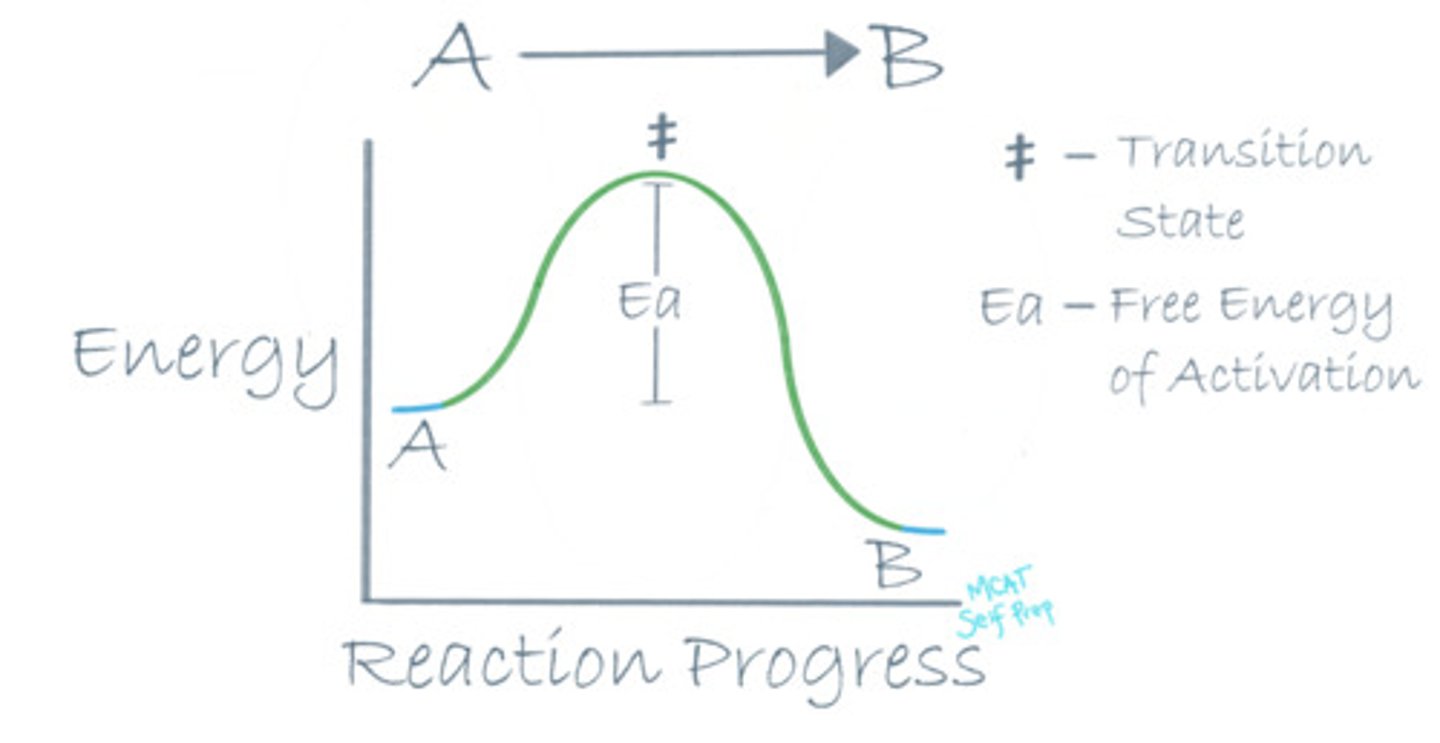
Where is the free energy of activation (Ea) in a reaction coordinate diagram?
The free energy of activation (Ea) in a reaction coordinate diagram is the difference between the energy level of the transition state (top of the graph) and the energy level of the reactants.
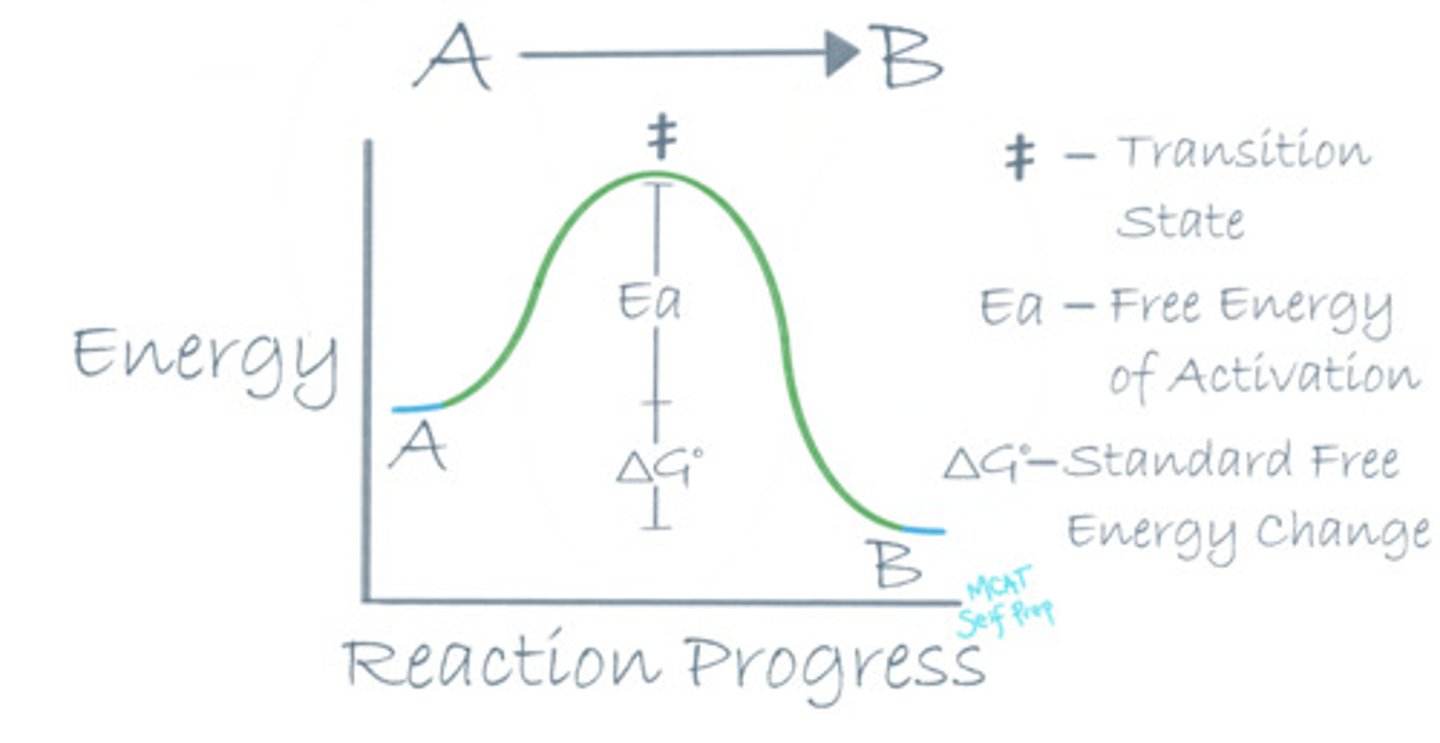
CRB Recall the Reaction Diagram in the previous card, and that enzymes catalyze Spontaneous reactions. Do they catalyze Endergonic or Exergonic reactions?
Because these reactions occur spontaneously, they must have a negative ΔG, meaning that they are Exergonic Reactions (energy is allowed to exit the substrates, becoming more stable).
Where is the standard free energy change (ΔG°) in a reaction coordinate diagram?
The standard free energy change (ΔG°) in a reaction coordinate diagram is the difference between the energy levels of the reactants and the products.
The Ea for a certain reaction is 32 kJ. That means it takes 32 kJ to do what?
The free energy of activation (Ea) is the amount of energy that a molecule needs to have input in order to break the reaction barrier to ultimately get to the transition state. Is the energy input required to start a reaction. In this case, it would take 32 kJ to start the reaction.
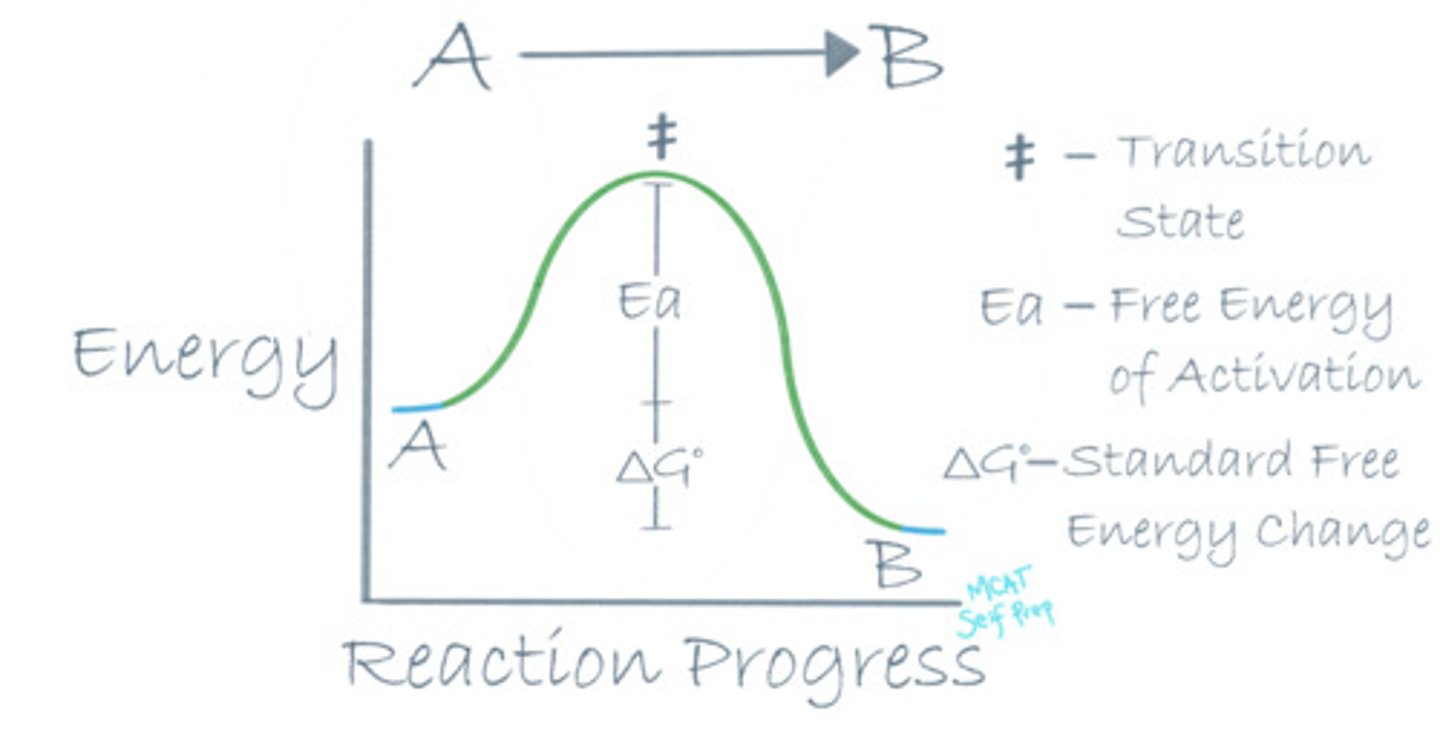
The ΔG° for a certain reaction is -14 kJ. This means that 14 kJ of energy will do what?
The standard free energy change (ΔG°) is the net change in energy level between our products and reactants and it is also the energy that is released into or taken from the environment once the reaction is over. In this case, 14 kJ are released from the reaction.
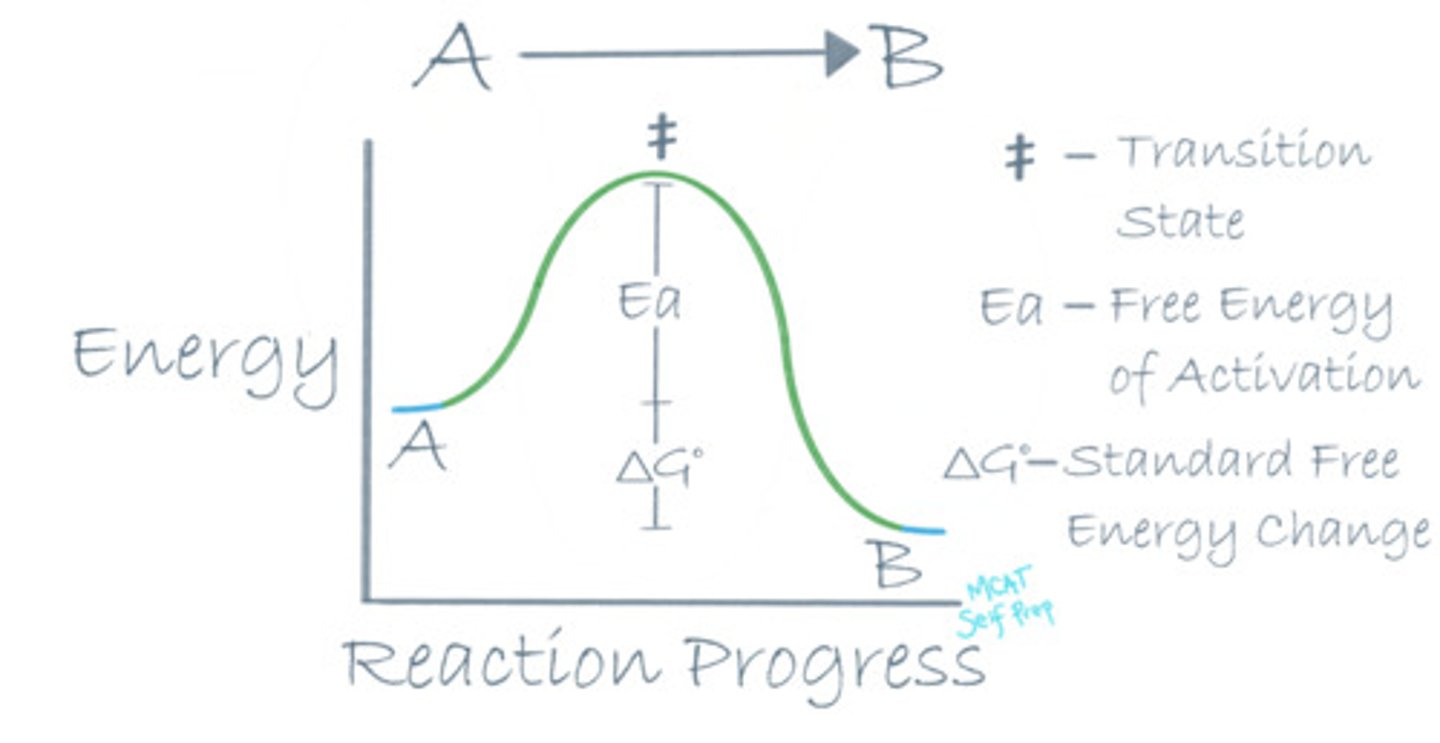
How can you tell that a reaction is a spontaneous reaction by looking at the reaction coordinate diagram?
A spontaneous reaction on a reaction coordinate diagram appears to have the products at a lower energy level than the reactants.
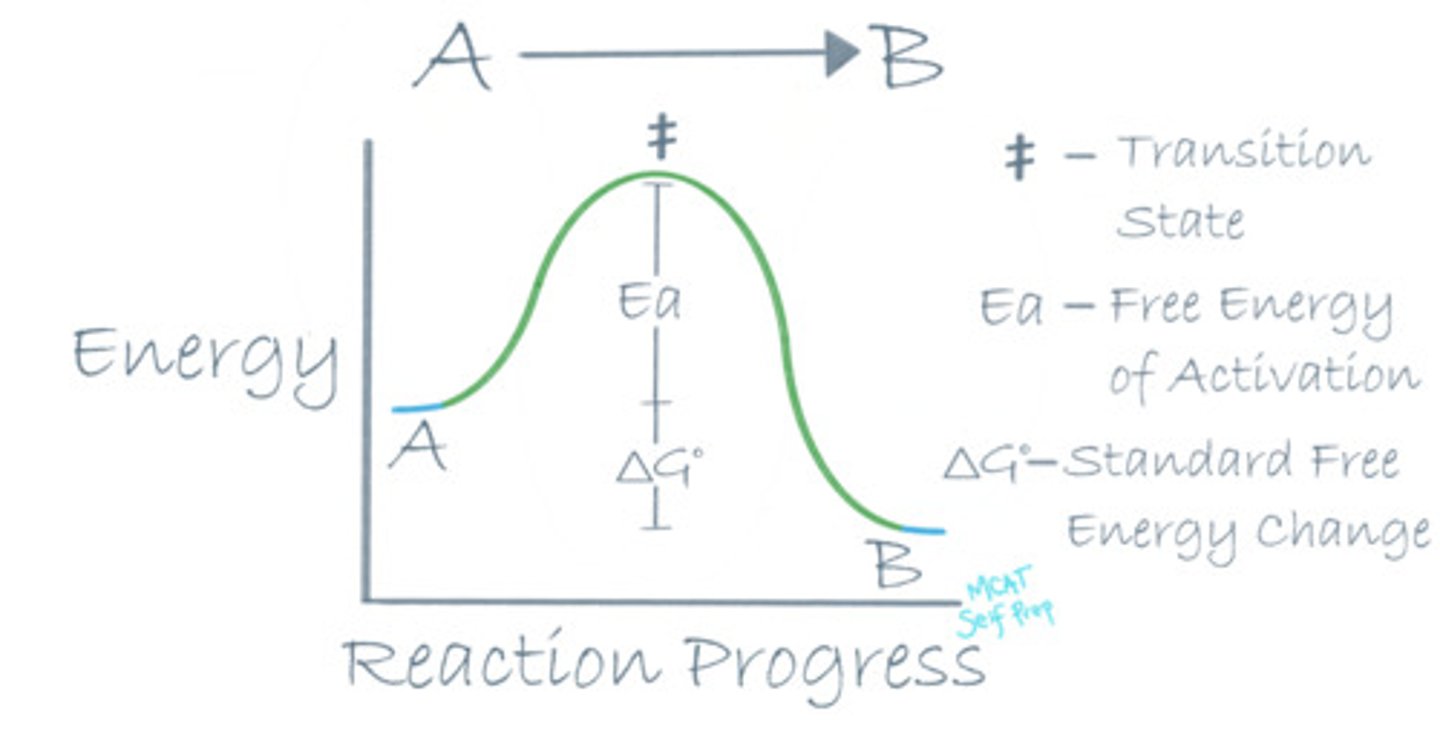
Which of the following gives you information about a reaction's speed?
I. ΔG°
II. Ea
III. Eproducts
(A) I Only
(B) II Only
(C) I and II Only
(D) II and III Only
(B) II Only
The free energy of activation (Ea) relates to the speed of a reaction. ΔG° does not!
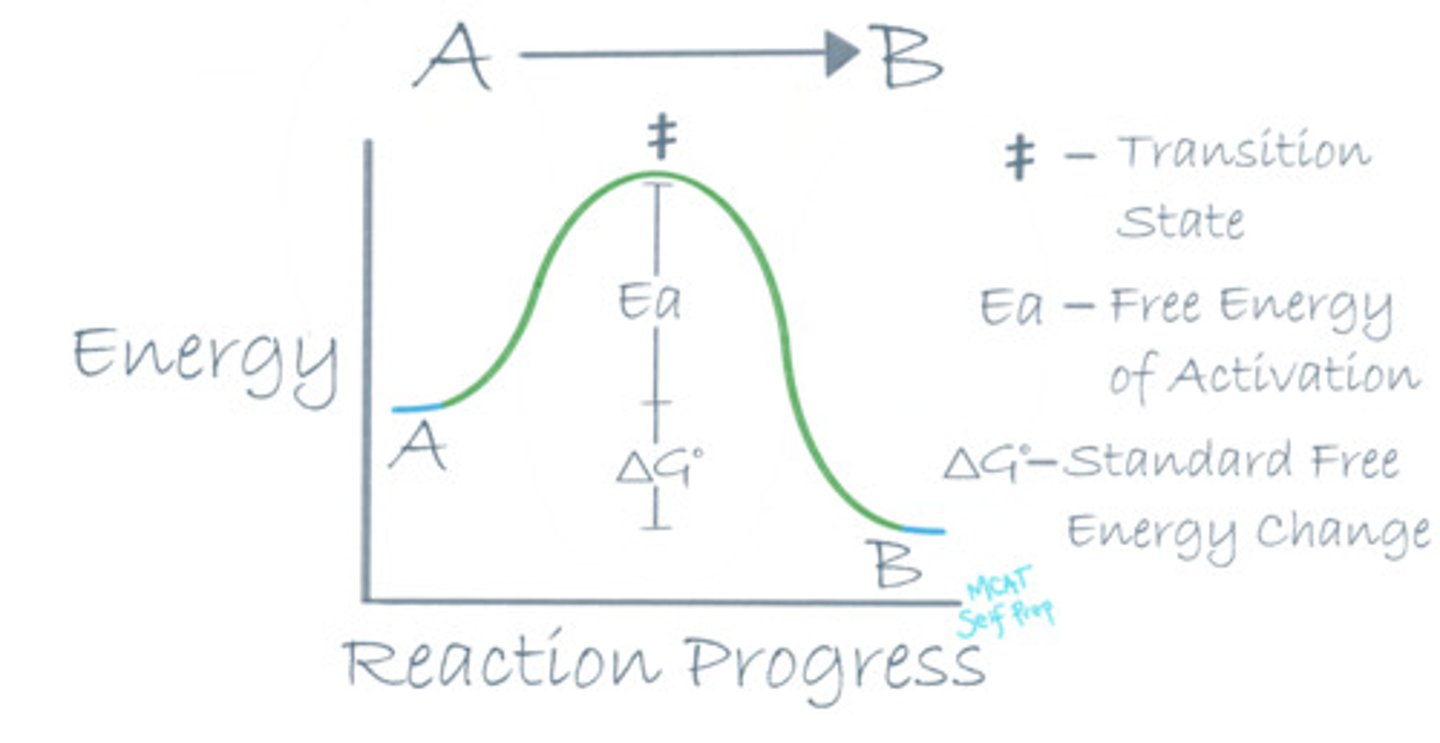
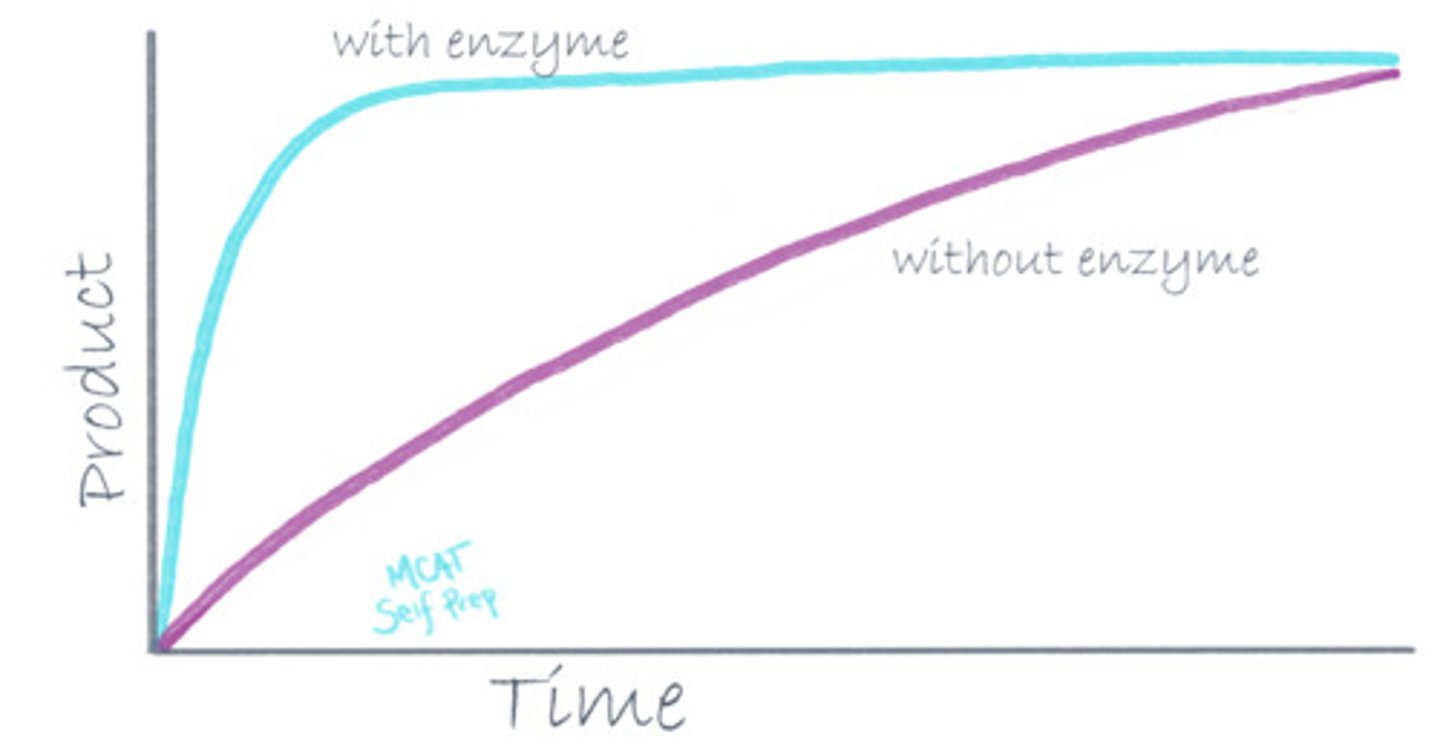
What is different about a catalyzed reaction compared to an uncatalyzed reaction in terms of the reaction coordinate diagram?
(A) Starting point of a reaction
(B) End point of a reaction
(C) Standard free energy of the reaction
(D) Free energy of activation
(D) Free energy of activation
An enzyme lowers the free energy of activation by stabilizing the transition state, making a catalyzed reaction go faster. An uncatalyzed reaction will have a much higher free energy of activation and have a less stable transition state.
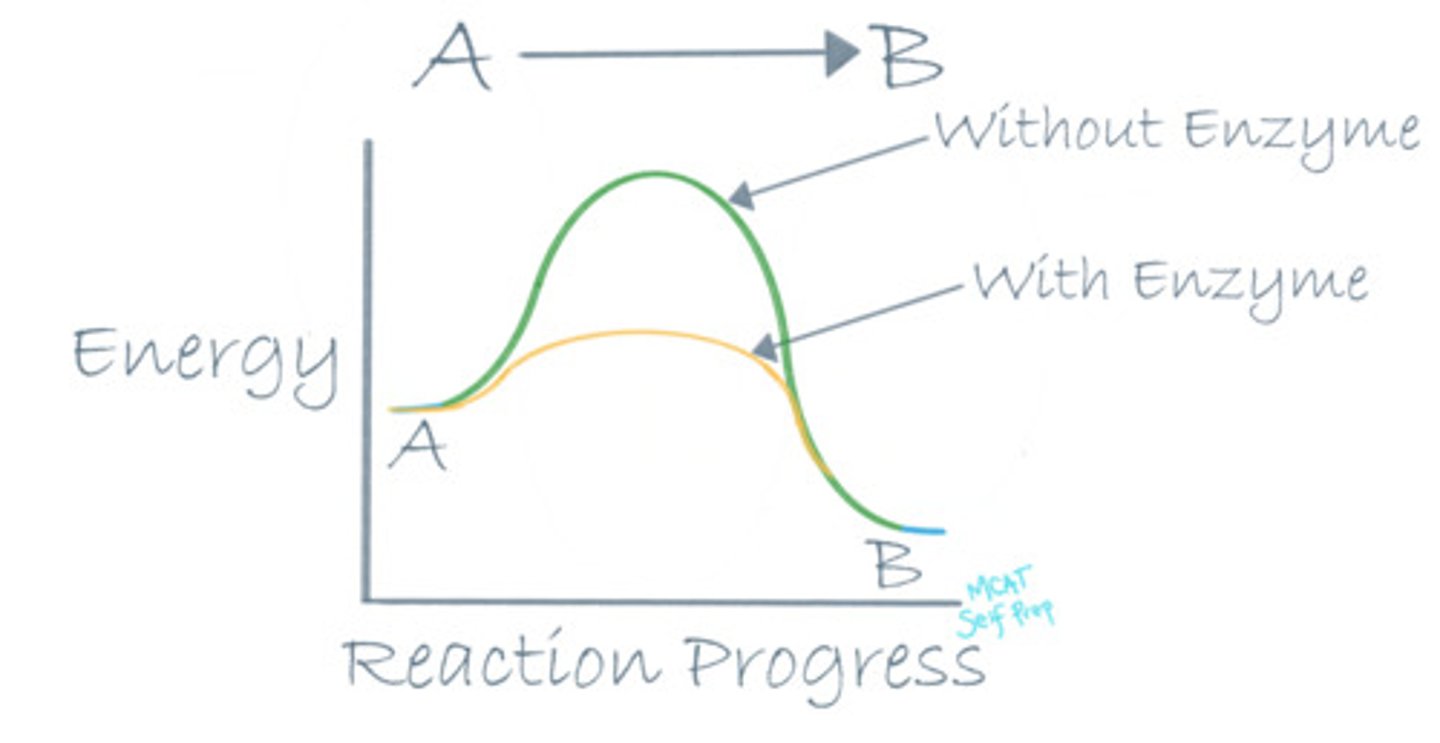
True or False? Enzymes are consumed during a reaction.
False. Enzymes are NOT consumed when they catalyze a reaction and the same enzyme can catalyze reactions over and over again.
Describe the relationship between enzymes and substrates.
A substrate is any molecule that an enzyme acts on. Substrates are the reactants that the enzyme will help turn into product.
What is the enzyme-substrate complex?
The enzyme-substrate complex consists of an enzyme and a substrate bound together.
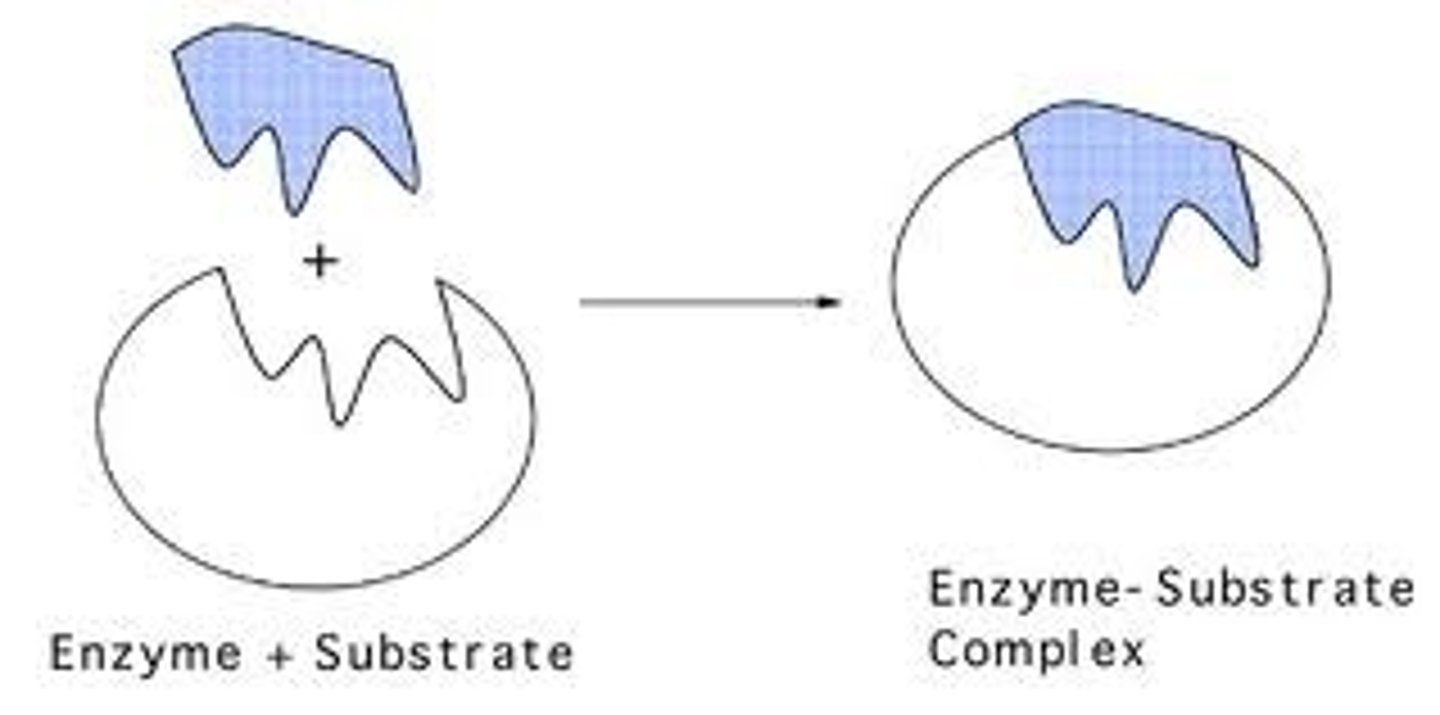
What is the induced fit model of enzymes?
The induced fit model of enzymes says that enzymes and substrates do not fit together like rigid puzzle pieces (as suggested in the "lock and key" model), but rather act like two pieces of clay that will both "mold together" and alter their shape/conformation to tightly bond.
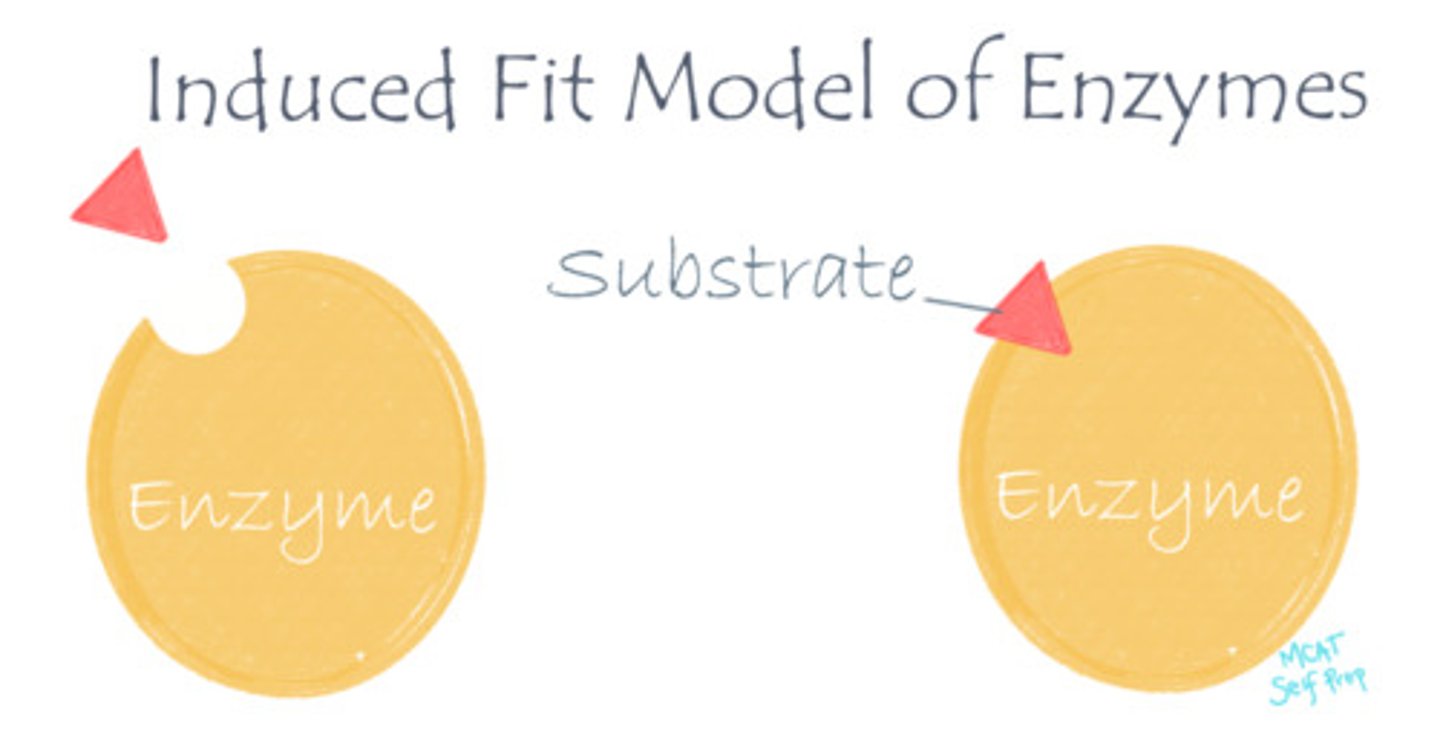
At which point of a reaction do the enzyme and substrate bind at their maximum strength?
(A) Just before the Transition State
(B) During the Transition State
(C) Just after the Transition State
(D) At the end of a reaction
(B) During the Transition State
The enzyme is most tightly bound to its substrate during the transition state (induced fit stage).
True or False? An enzyme can only fit one substrate in its active site at a time.
False. Although an active site will only catalyze a single reaction at a time, some active sites will bind to multiple substrates at the same time. For instance, during lactic acid fermentation, lactose dehydrogenase binds to two substrates, NADH and pyruvate, at the same time.
What occurs at the active sites vs. allosteric sites of an enzyme?
The active site is where the reaction takes place, while the allosteric site is where regulation takes place.
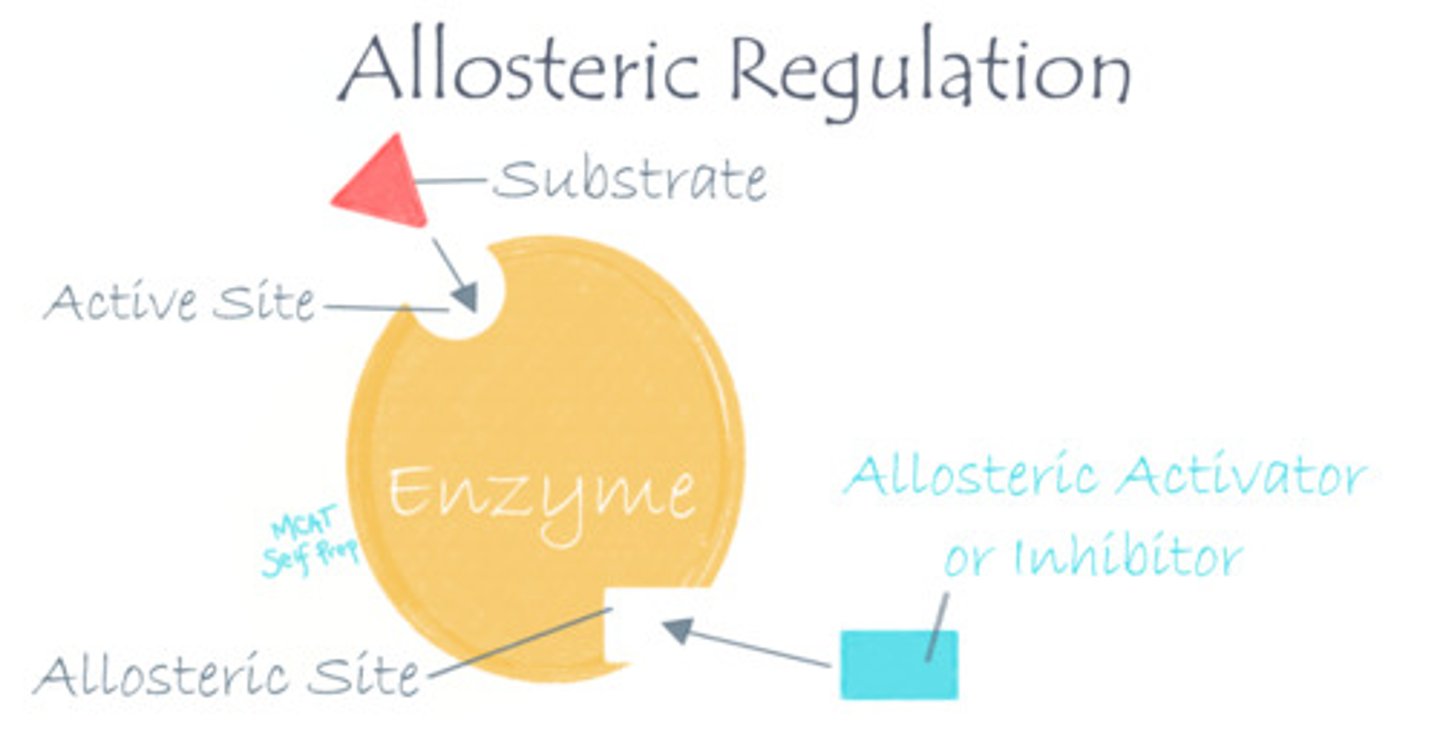
What kind of molecule would bind at an allosteric site?
I. Activators
II. Inhibitors
III. Substrates
(A) I Only
(B) II Only
(C) I and II Only
(D) I, II, and III
(C) I and II Only
Regulatory molecule, such as activators and inhibitors, will bind at an allosteric site. Their binding will change the enzyme's ability to catalyze reactions.
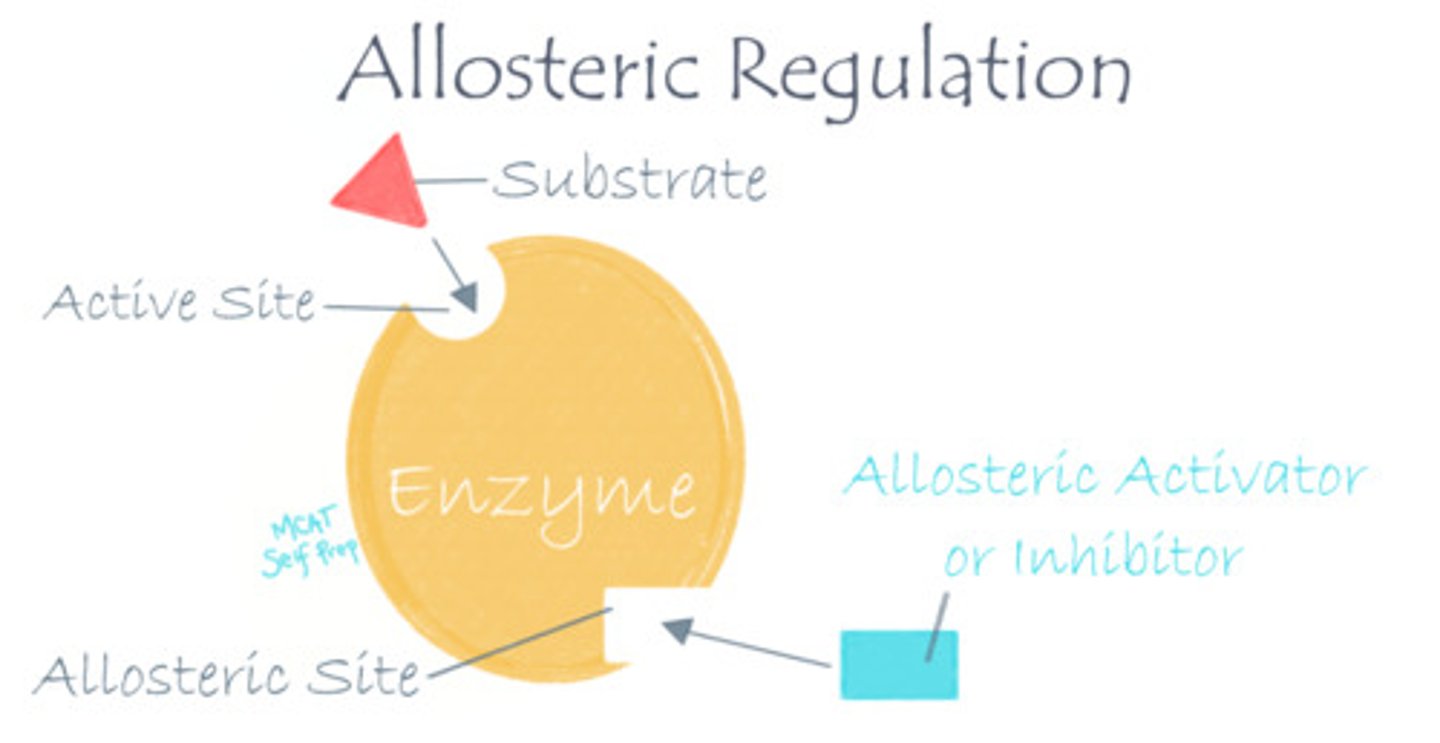
When an inhibitor binds to an allosteric site, how might the enzyme's active site change? What about when an activator binds?
When an inhibitor binds to an allosteric site, the enzyme's active site may change shape in a way that prevents it from binding to the substrate anymore.
When an activator binds to an allosteric site, the enzyme's active site may change shape in a way that allows it to better bind to the substrate.
Describe the mechanism of action for a transferase enzyme.
Transferase enzymes catalyze the reaction of moving a functional group from molecule A to molecule B.
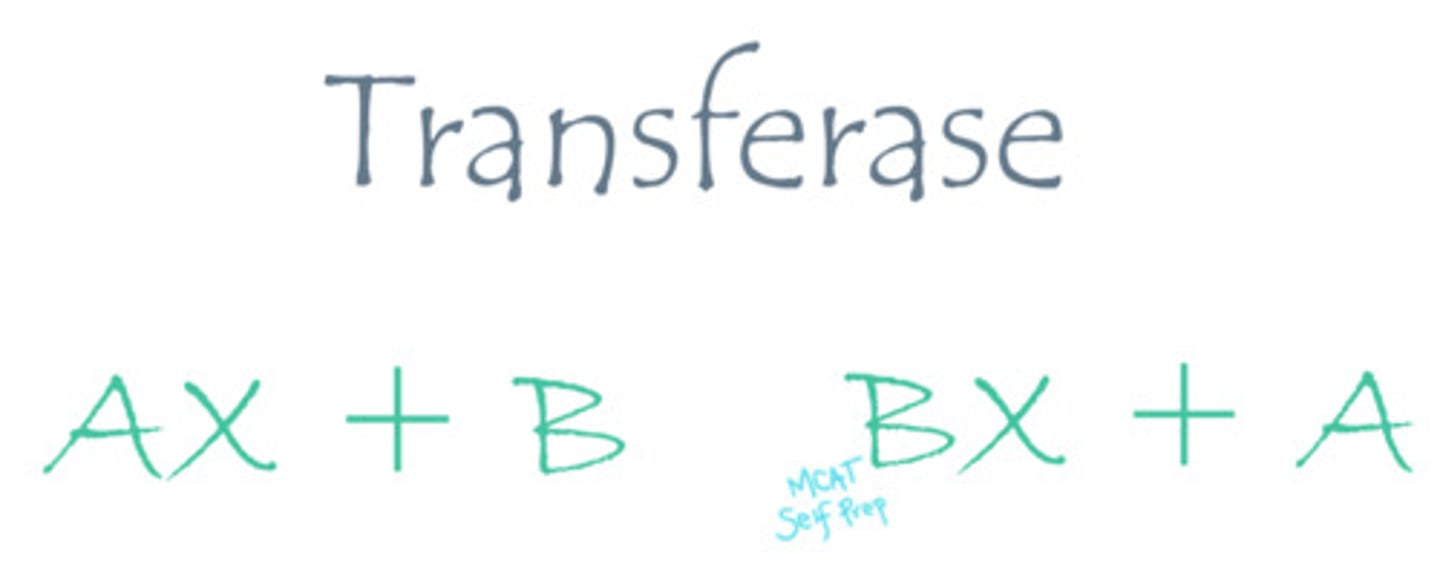
CRB A major factor in DNA transcription is the amount of methyl groups surrounding the gene of interest. There is an enzyme that uses S-adenosyl Methionine as a methyl donor and attaches that methyl group to the nucleotide of interest. Would this be considered a Transferase Enzyme?
Because this enzyme is moving a functional group (the methyl group) from one molecule to another, this would be considered a Transferase reaction. The enzyme described is actually called DNA Methyltransferase.
CRB Is a Kinase, which adds Phosphate groups to various cellular targets (oftentimes to enzymes), a Transferase Enzyme? Why or why not?
A Kinase is a Transferase Enzyme, because it transfers a Phosphate group from a donor molecule (oftentimes ATP) to a cellular target (like another enzyme).
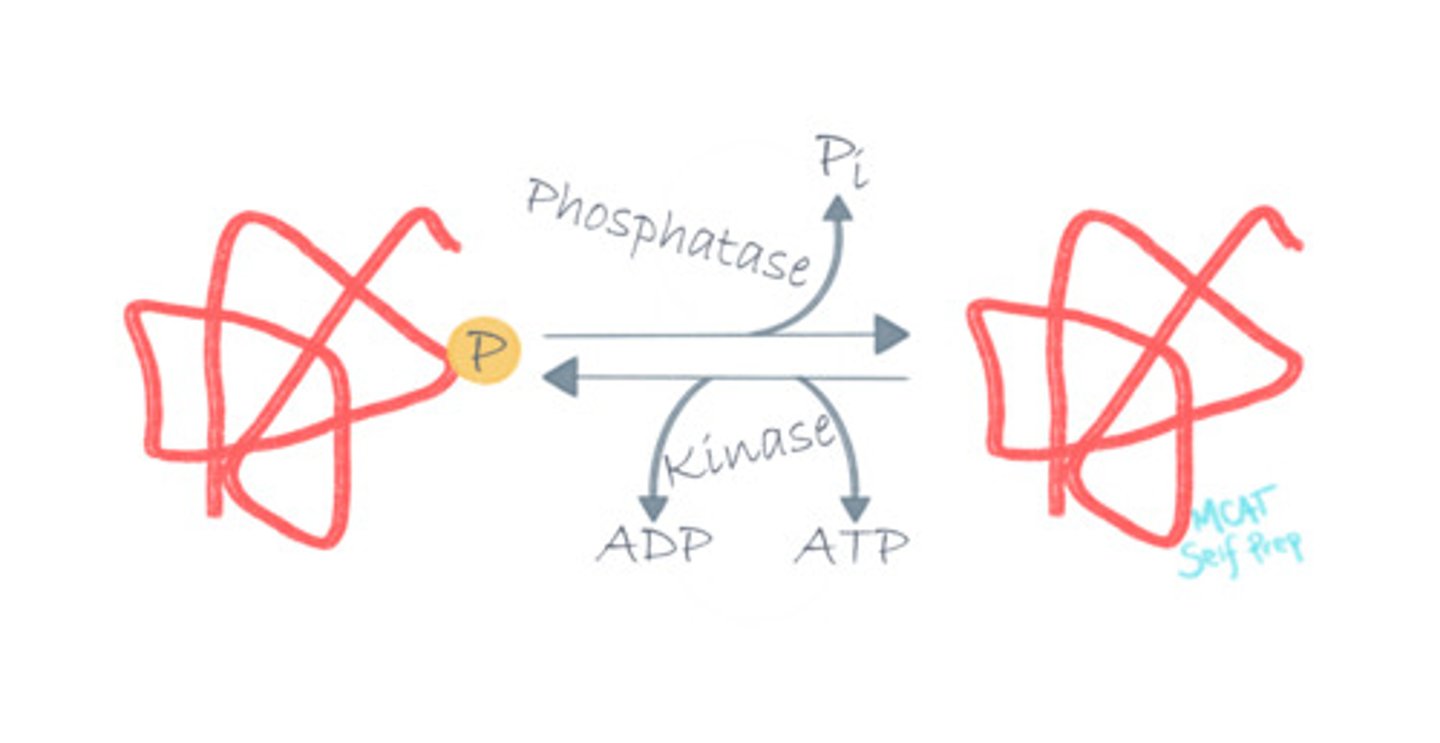
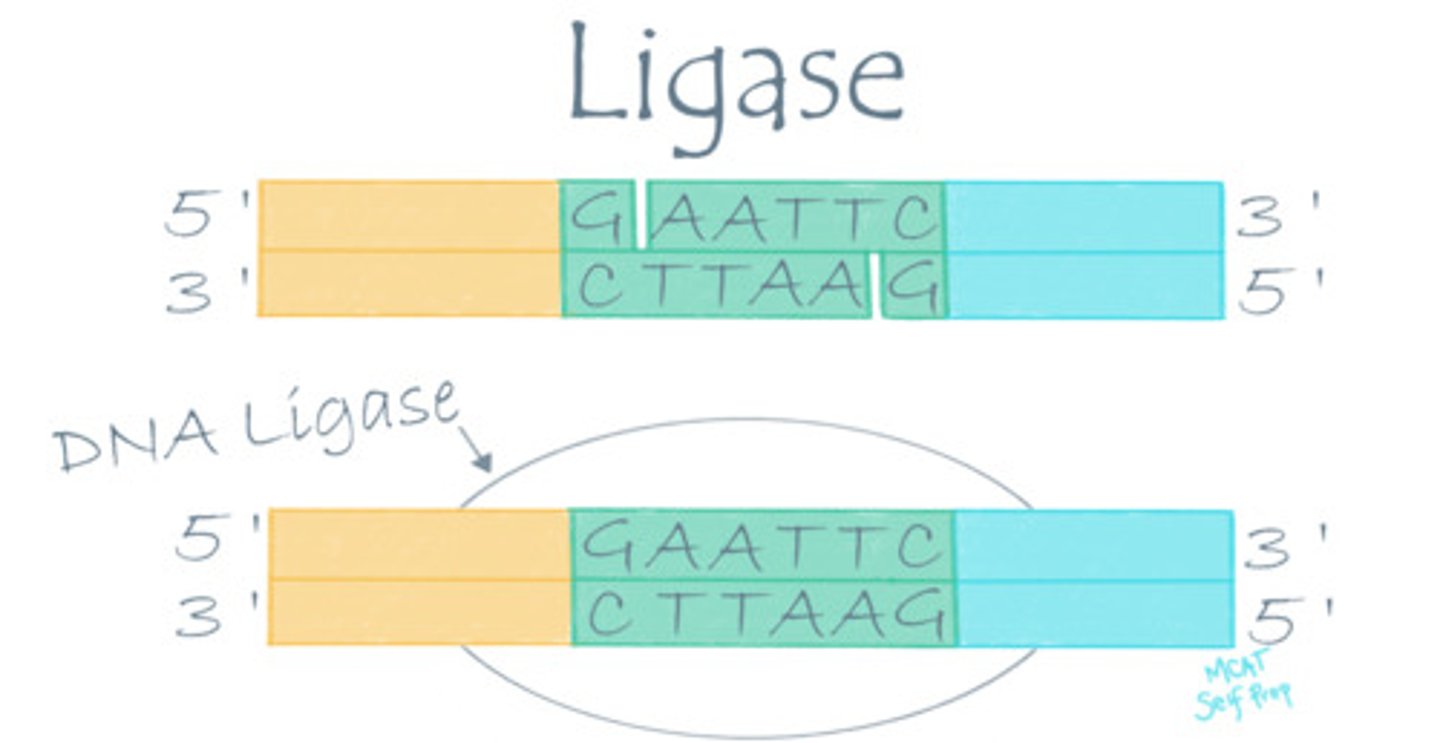
Describe the mechanism of action for a ligase enzyme.
Ligase enzymes catalyze reactions between molecule A and molecule B to form a complex molecule AB.
Describe the mechanism of action for an oxidoreductase enzyme? Oxidase enzyme? Reductase enzyme?
Oxidoreductase enzymes can catalyze both oxidation and reduction reactions.
Oxidase enzymes catalyze reactions that take electrons away from a molecule.
Reductase enzymes catalyze reactions that give electrons to a molecule.
CRB Fill in the blanks: In an Oxidoreductase enzyme's reactions, the electron donor is called the _____________, and the electron acceptor is called the ____________. This is more closely related to the ____________ definition of Acids and Bases.
(A) Reductant, Oxidant, Bronsted-Lowry
(B) Reductant, Oxidant, Lewis
(C) Oxidant, Reductant, Bronsted-Lowry
(D) Oxidant, Reductant, Lewis
(B) Reductant, Oxidant, Lewis
In an Oxidoreductase enzyme's reactions, the electron donor is called the Reductant, and the electron acceptor is called the Oxidant. This is more closely related to the Lewis definition of Acids and Bases.
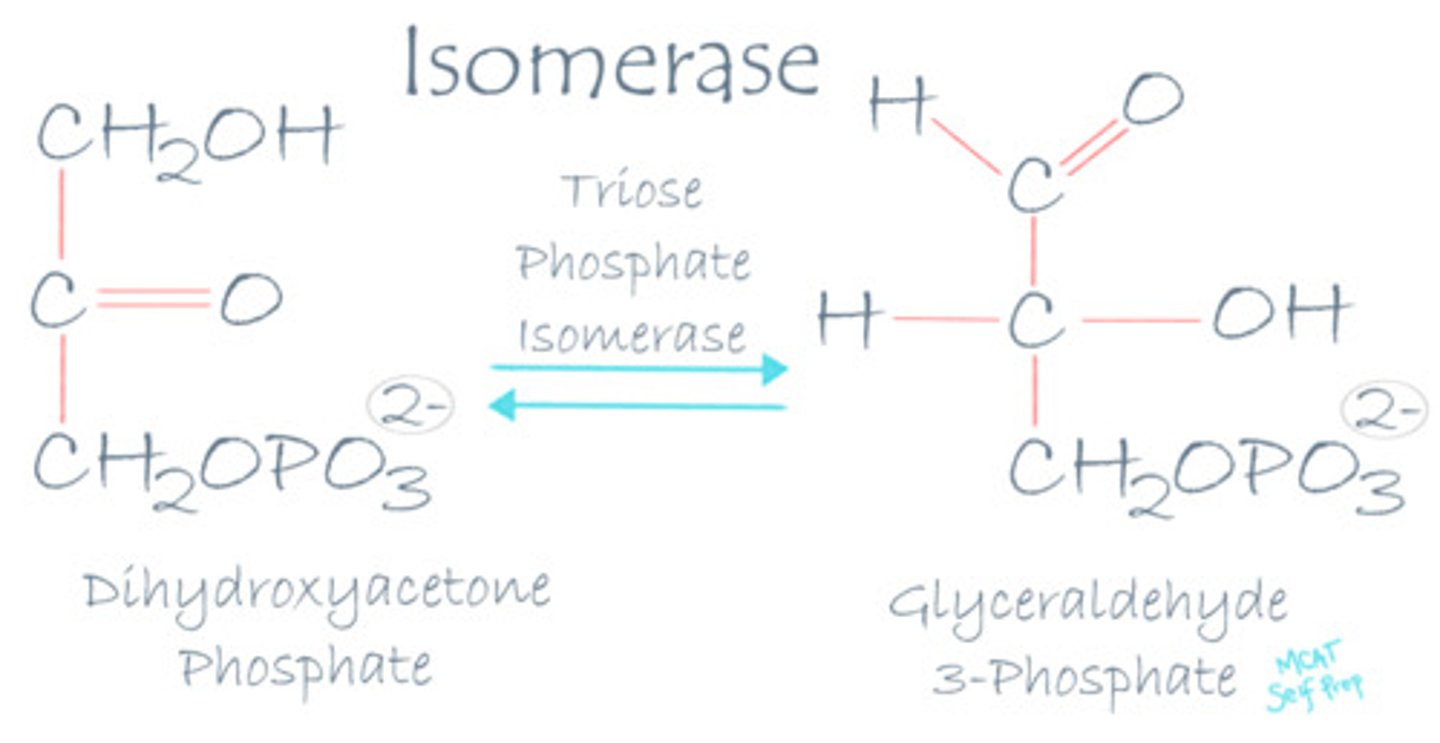
Describe the mechanism of action for an isomerase enzyme.
Isomerase enzymes are involved in reactions in which a molecule is converted into one of its isomers.
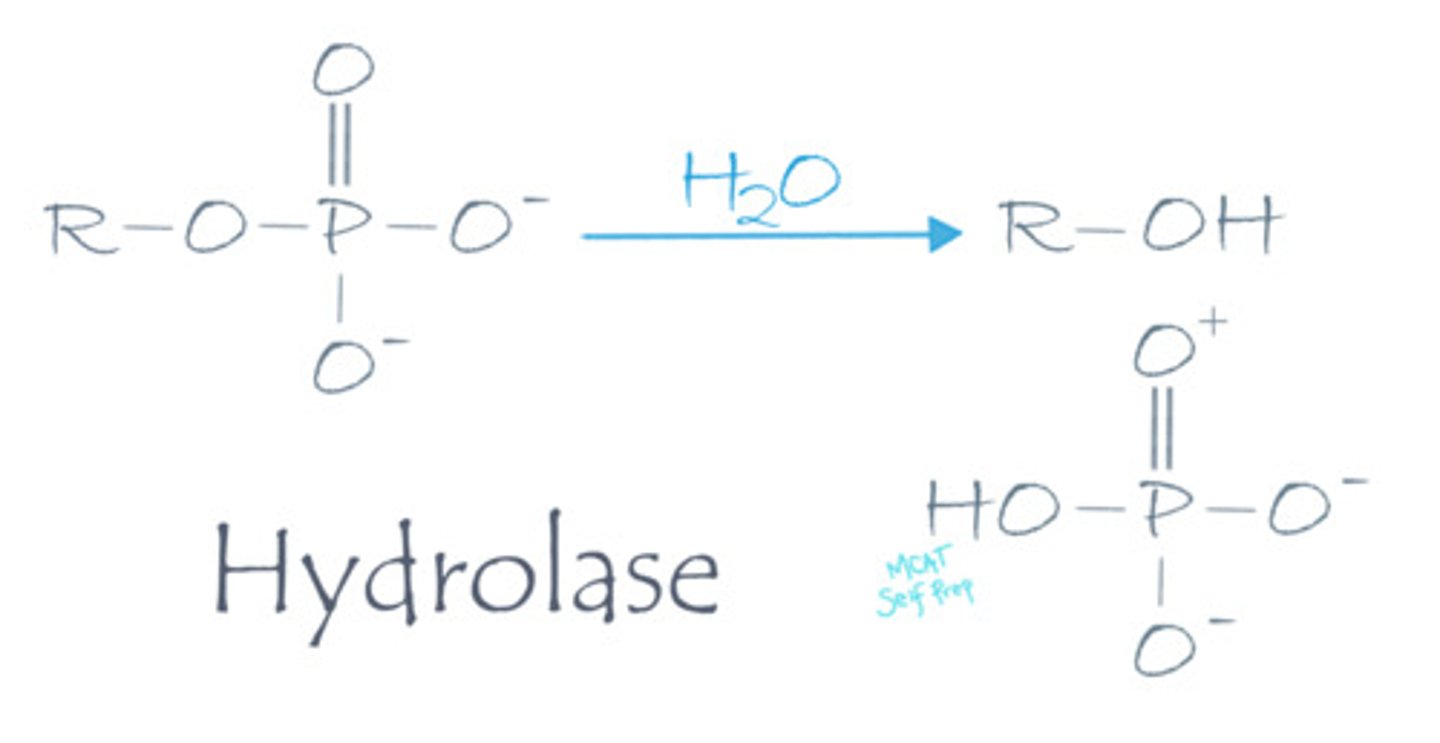
Describe the mechanism of action for a hydrolase enzyme.
Hydrolase enzymes cleave a molecule into two separate molecules using water as a substrate.
Describe the mechanism of action for a lyase enzyme.
Lyase enzymes catalyze the dissociation of molecules without the use of water or oxidation.
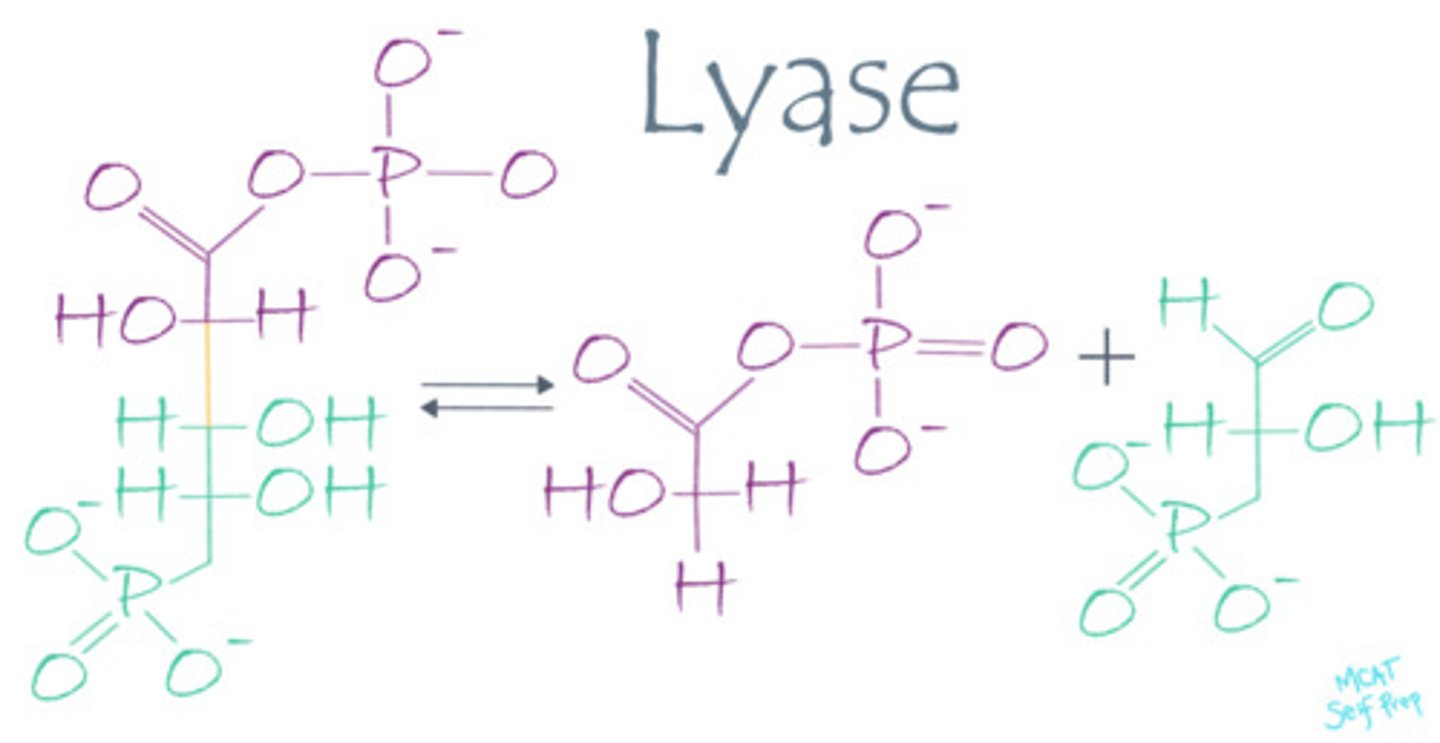
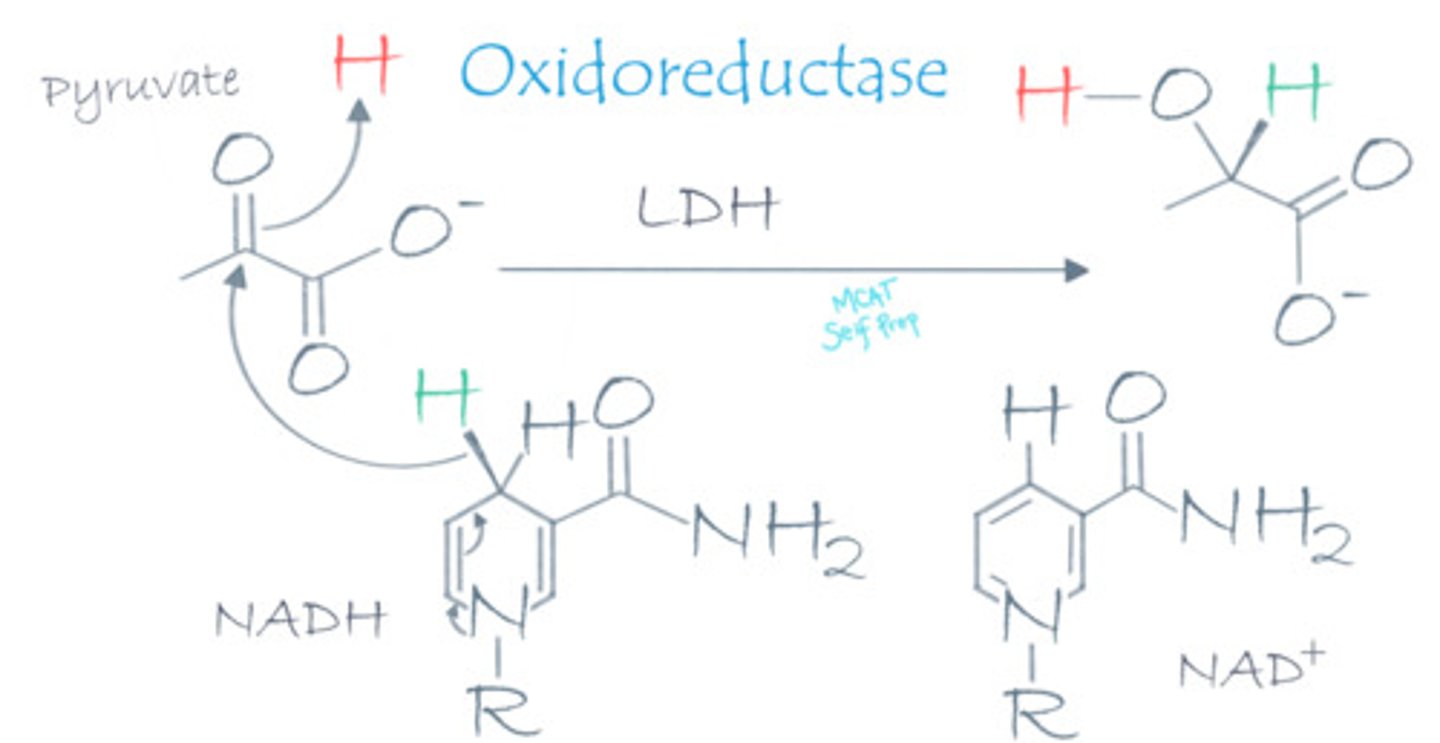
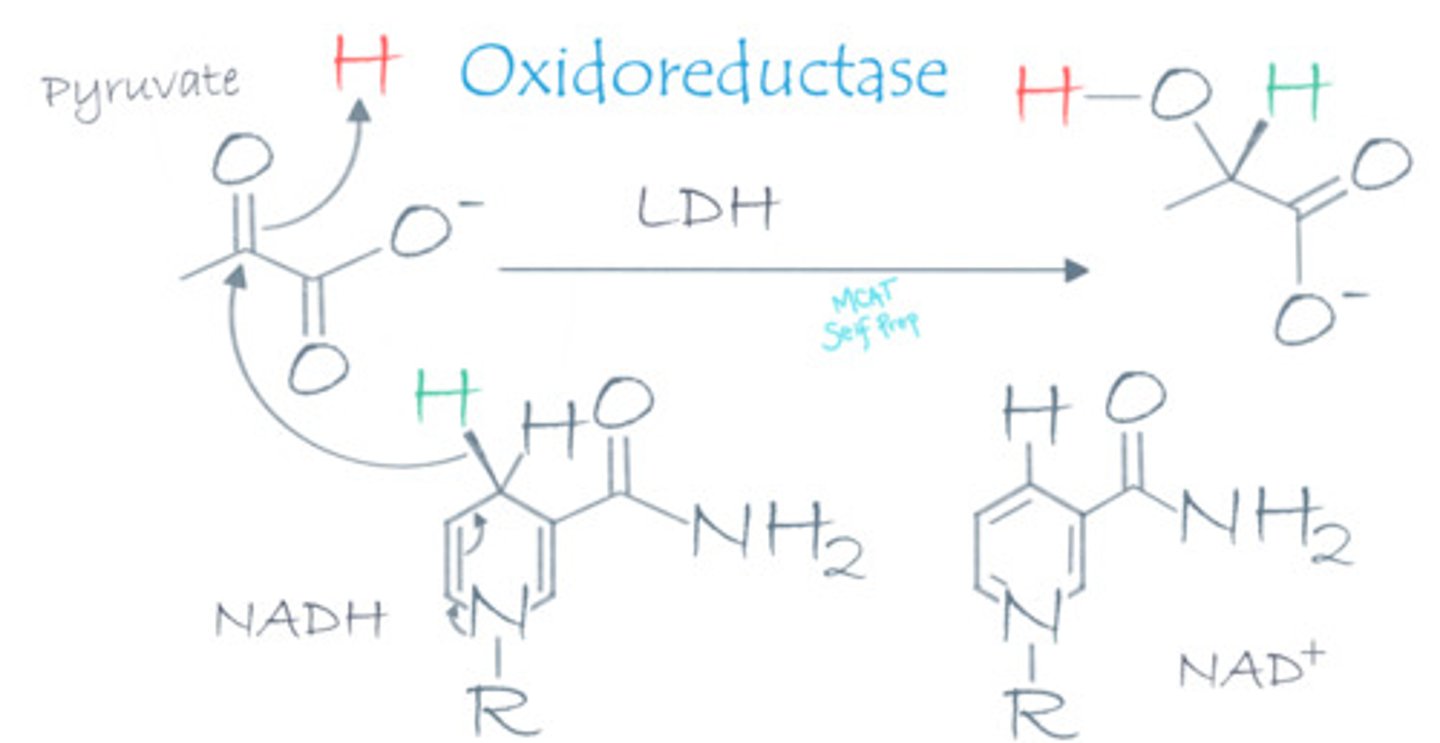
During lactic acid fermentation, lactate dehydrogenase catalyzes the transfer of electrons from NADH to pyruvate. Why is this enzyme a "dehydrogenase" when it is an oxidoreductase enzyme?
Lactate dehydrogenase is an oxidoreductase enzyme. "Dehydrogenase" simply refers to the removal of hydride (H-), which is equivalent to removing a proton and 2 electrons, from the NADH coenzyme.
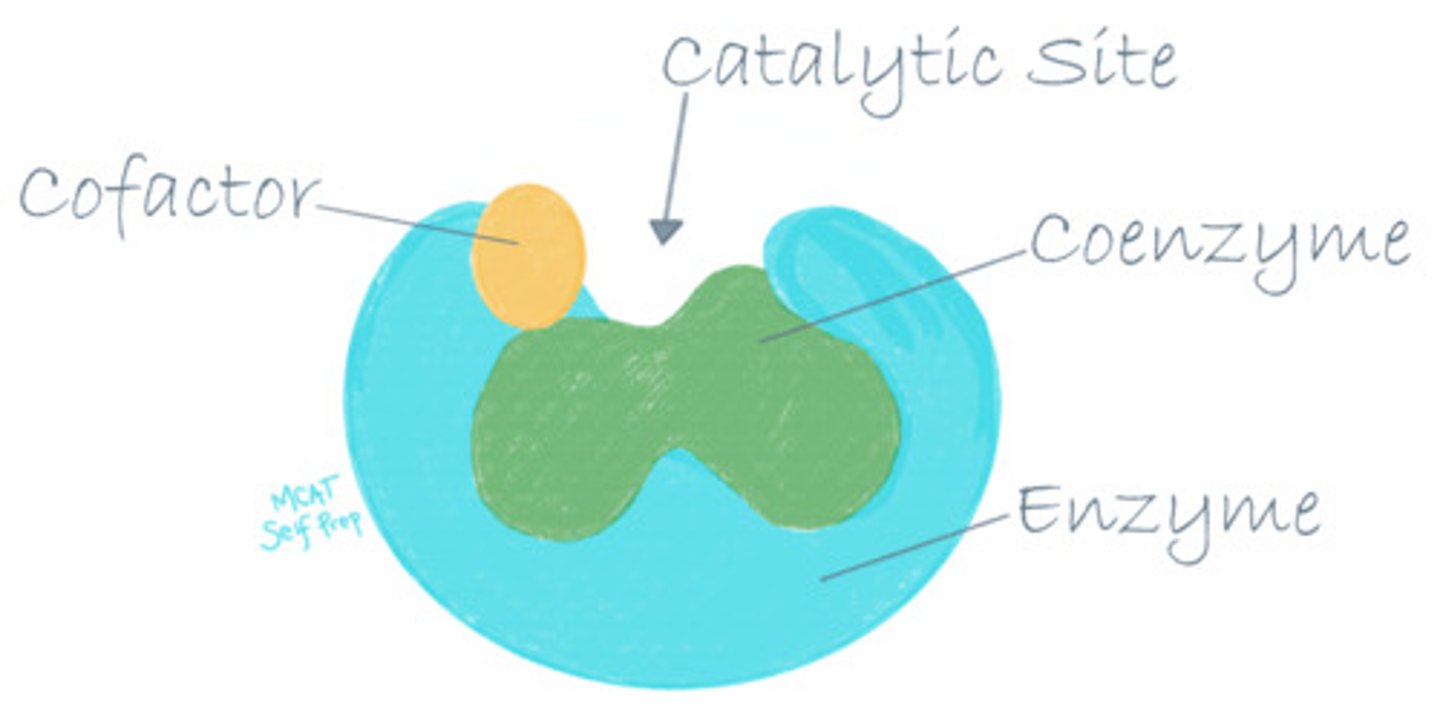
CRB Compare Coenzymes, Cofactors and Prosthetic Groups.
Coenzymes, Cofactors and Prosthetic Groups all are necessary for their enzymes to properly function.
Coenzymes are organic molecules, whereas cofactors are typically metal ions.
Prosthetic groups are very tightly bound Cofactors or Coenzymes, so that you almost always see the enzyme with its Prosthetic Group.
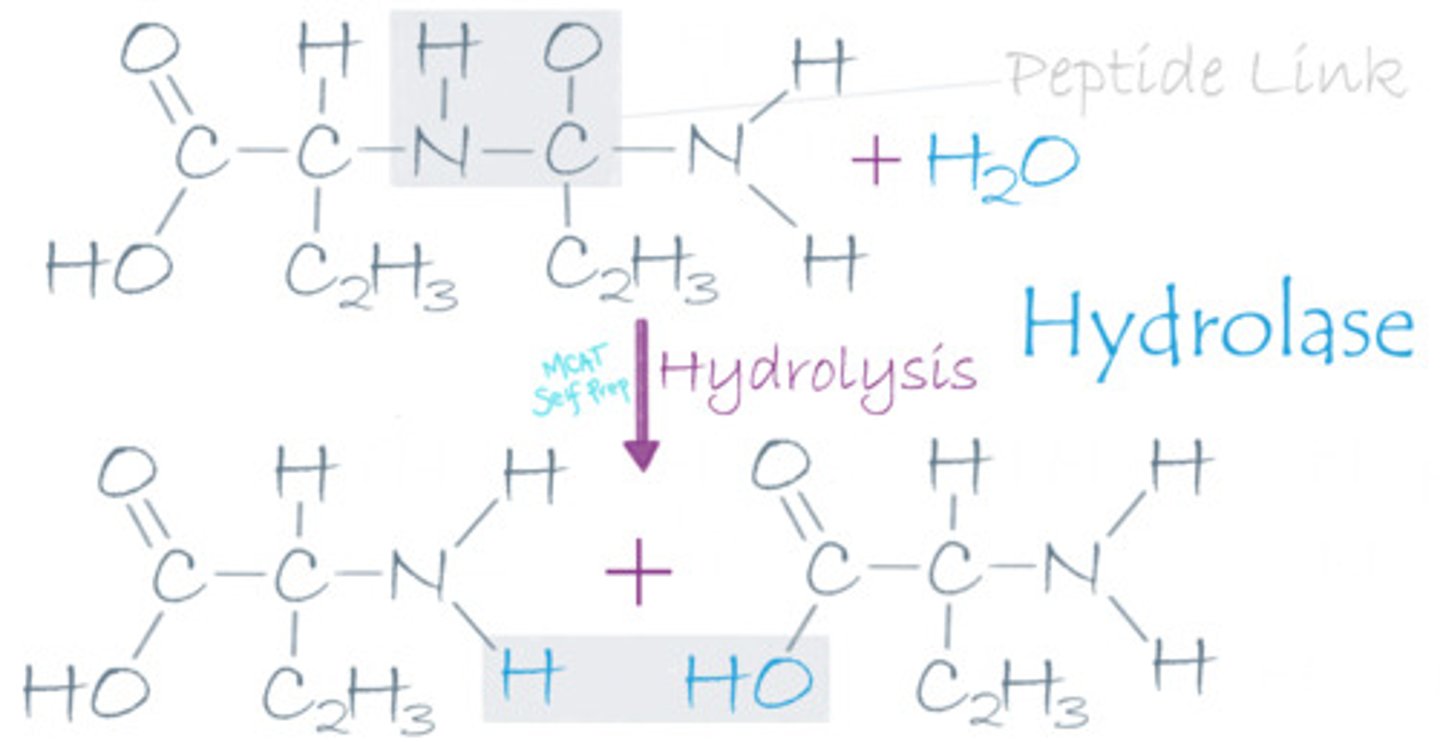
A lysine-alanine dipeptide reacts with water, releasing two free amino acids. What type of enzyme was involved in this reaction?
(A) Hydrolase
(B) Ligase
(C) Lyase
(D) Isomerase
(A) Hydrolase
A hydrolase enzyme was involved because it used water to break this peptide bond, producing two amino acids. Also note that a hydrolase enzyme that breaks peptide bonds can be called a protease.
Struggling with the one-letter abbreviations, three-letter abbreviations, structures, and the essential properties of Amino Acids? Learn how to conquer any Amino Acid MCAT question using Andrew's Amino Acid Mastery Course @ https://mcatselfprep.com/course/andrews-amino-acid-mastery-course/
CRB Based on the previous question, you know that a Protease is a type of Hydrolase. Which of the following is NOT a fellow Hydrolase?
(A) Lipidase
(B) Nuclease
(C) Aconitase
(D) Phosphatase
(C) Aconitase
An Aconitase is a Lyase enzyme.
Lipidases, Nucleases and Phosphatases are all Hydrolase enzymes.
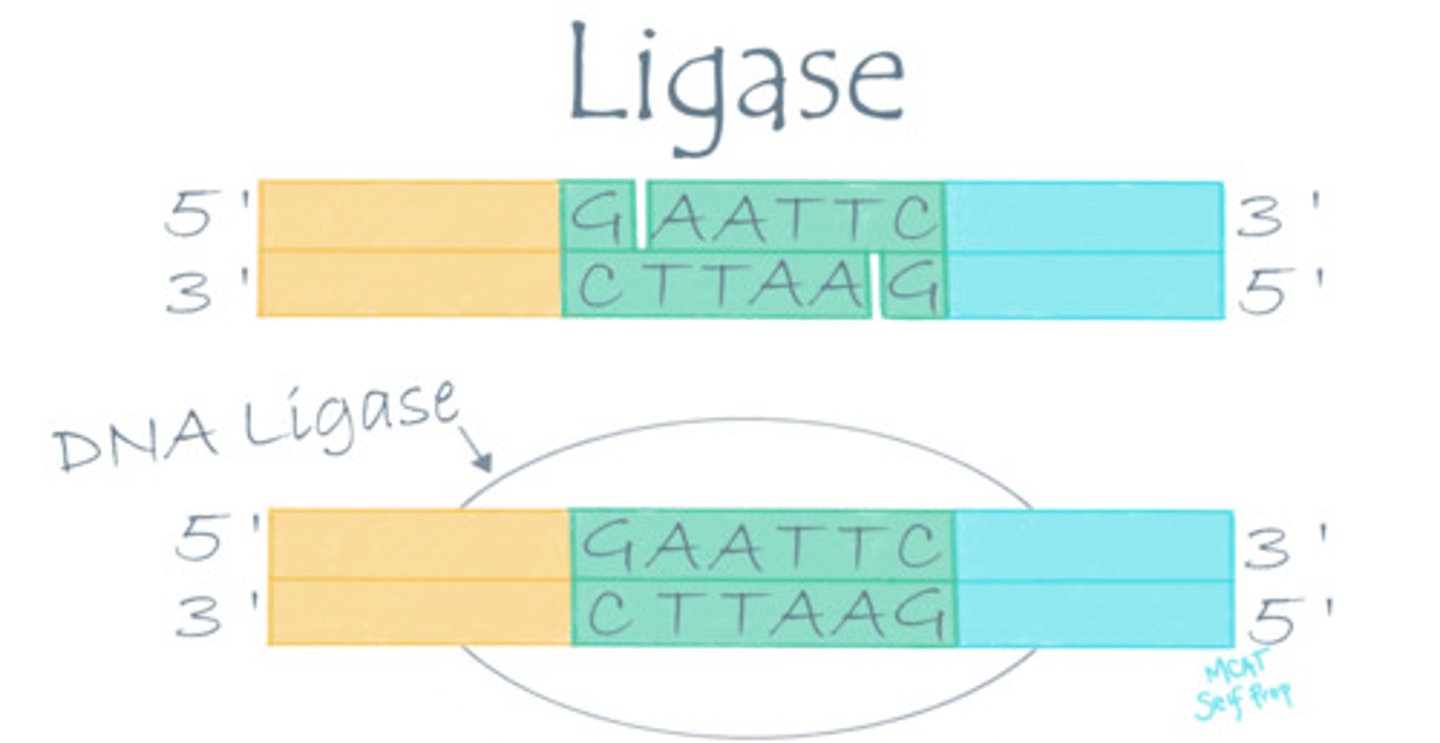
During DNA replication, two strands of DNA are joined together. What kind of enzyme would be involved in this reaction?
(A) Hydrolase
(B) Ligase
(C) Lyase
(D) Transferase
(B) Ligase
A ligase enzyme (DNA ligase) is involved in DNA replication because two separate DNA strands (molecules) are being joined to form a single strand.
During protein translation, amino acids bound to tRNA molecules are transferred to the growing polypeptide chain. What kind of enzyme would be involved in this reaction?
(A) Hydrolase
(B) Ligase
(C) Oxidoreductase
(D) Transferase
(D) Transferase
This transferase enzyme (peptidyl transferase) will take a functional group (amino acids) bound by one molecule (tRNA) and transfer them to another molecule (the growing polypeptide chain).
An enzyme catalyzes the breakdown of an arginosuccinate molecule into arginine and succinate (without the use of water or oxidation) during the urea cycle. What kind of enzyme is this?
(A) Hydrolase
(B) Ligase
(C) Lyase
(D) Oxidoreductase
(C) Lyase
A lyase enzyme (argininosuccinate lyase) is involved because it does not use water or oxidation to break a bond.
Struggling with the one-letter abbreviations, three-letter abbreviations, structures, and the essential properties of Amino Acids? Learn how to conquer any Amino Acid MCAT question using Andrew's Amino Acid Mastery Course @ https://mcatselfprep.com/course/andrews-amino-acid-mastery-course/
CRB Compare Lyases and Ligases.
Both Lyases and Ligases can catalyze synthesis reactions without Oxidation-Reduction occurring.
Lyases can also catalyze the splitting of these bonds without water or Oxidation-Reduction occurring, and typically work with small molecules. This 2nd reaction is what lyases are typically known for.
Ligases typically work with larger molecules and require ATP.
In one of the steps in glycolysis, glucose-6-phosphate is converted into fructose-6-phosphate. What type of enzyme would be involved in this reaction?
(A) Oxidoreductase
(B) Ligase
(C) Isomerase
(D) Transferase
(C) Isomerase
An isomerase enzyme (phosphoglucose isomerase) is involved in this reaction since glucose-6-phosphate in converted into its isomer, fructose-6-phosphate.
Struggling to memorize the metabolic pathways (such as glycolysis and beta-oxidation)? Learn them like the back of your hand using Andrew's Metabolic Pathways Mastery Course @ https://mcatselfprep.com/course/andrews-metabolic-pathways-mastery-course/
CRB True or false? An Isomerase can catalyze reactions to form Constitutional Isomers and converting between Stereoisomers.
True. An Isomerase can catalyze reactions to form Constitutional Isomers and converting between Stereoisomers.
In a lyase reaction, what structures are usually formed as a result of breaking a covalent bond?
I. Hydroxy Groups
II. Double Bonds
III. Ring Structures
(A) I Only
(B) II Only
(C) I and II Only
(D) II and III Only
(D) II and III Only
Lyase catalysis reactions must generate either a double bond between two atoms or a ring structure.
Compare co-enzyme vs. co-factor structure and function?
Co-enzymes are carbon-based (organic) carrier molecules that carry/hold small particles for an enzyme to make catalysis run more smoothly.
Co-factors are non-protein chemical compounds (typically metallic ions) that are directly involved in the enzyme's catalytic mechanism. They do not "carry" anything like co-enzymes do, but rather stabilize the enzyme or substrate.
A positively charged magnesium ion (Mg) is used to stabilize the negatively charged backbone of DNA, improving DNA Polymerase's binding to DNA during replication. Is Magnesium an example of a co-enzyme or a co-factor?
The magnesium ion (Mg) is an example of a co-factor because it is stabilizing the DNA and is directly involved in catalysis. Also, it is not carbon-based, so it cannot be a co-enzyme. Cations are very common co-factors!
During lactic acid fermentation, lactate dehydrogenase uses NADH to transfer electrons to the pyruvate molecule in order to convert pyruvate into lactic acid. NADH an example of a co-enzyme or a co-factor?
NADH is an example of a co-enzyme because it helps carry electrons for lactate dehydrogenase. It is also an organic molecule as are most co-enzymes.
Vitamins and minerals may act as:
I. Co-factors
II. Co-enzymes
III. Enzymes
(A) I Only
(B) III Only
(C) I and II Only
(D) II and III Only
(C) I and II Only
Vitamins and minerals are various co-enzymes and co-factors that your body may not be able to build from scratch and you may need to get them from your diet.
What is the difference between vitamins and minerals?
Vitamins are carbon-based (organic) co-enzymes, whereas minerals are inorganic or metallic co-factors. Minerals may also help structurally, such as calcium, which is an important component of bone and teeth.
True or False? Enzymes are dependent upon their local environment.
True. Enzymes are related to their local environment in that they function best under very specific environmental conditions, which can be either pH-dependent and/or temperature-dependent. Any change to the surrounding pH or temperature can lead to a loss of enzyme functionality.
What is alpha amylase responsible for?
Alpha amylase is responsible for breaking down complex carbohydrates like starches into small simple carbohydrates like individual sugars.
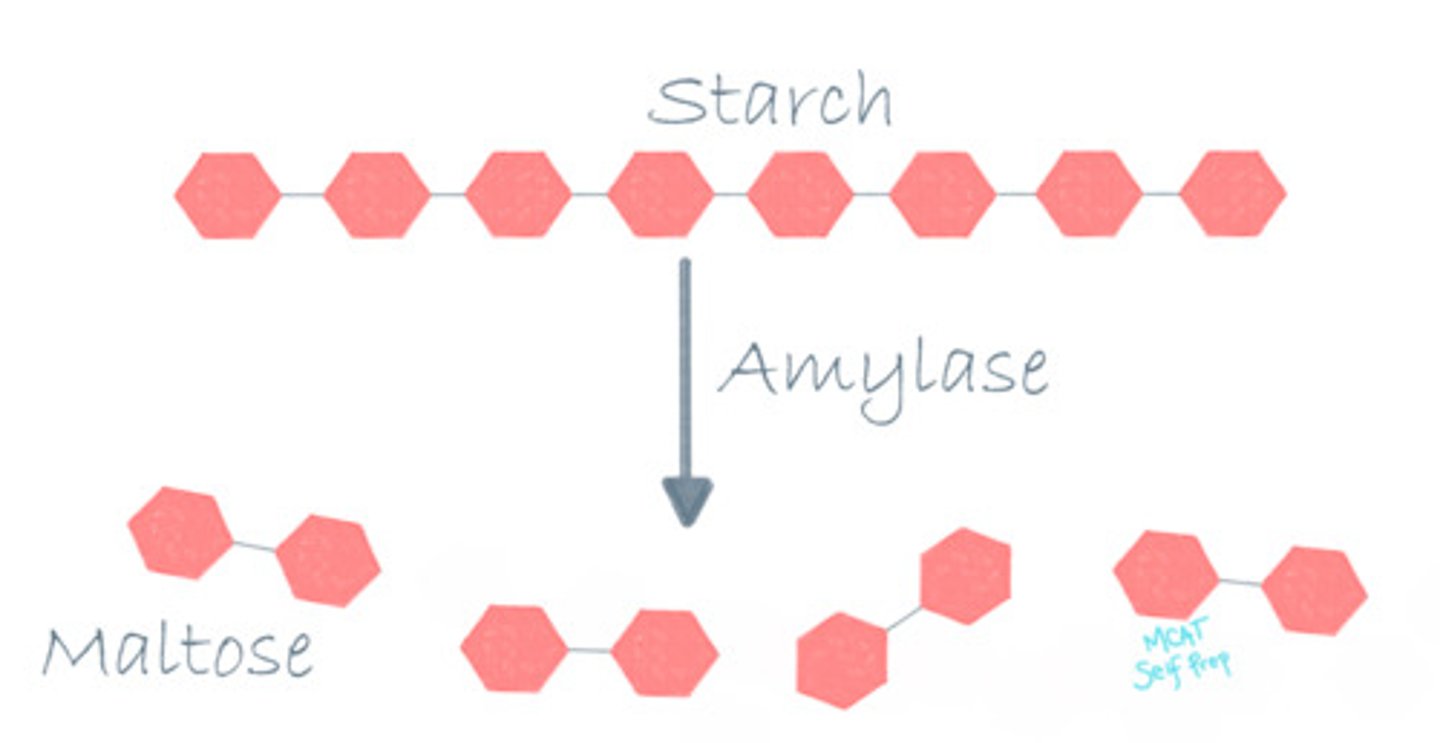
True or False? Alpha amylase works just as well in the mouth as it does in the stomach.
False. Alpha amylase will not be able to function and would denature in the stomach (pH = 2) because it is a pH-dependent enzyme and works best at a pH of 7, which is similar to the pH of the human mouth. Thus, alpha amylase will work best in the mouth and will fail to function in the stomach.
Using DNA Polymerase as an example, explain how pH changes can alter the function of a pH-dependent enzyme.
DNA Polymerase typically relies on an electrostatic interaction between the Mg2+ co-factor and an Aspartate residue to stabilize the highly-negative charge of DNA at normal pH. However, in a very acidic environment, the Aspartate will become protonated, eliminating the essential electrostatic interaction.
Explain how temperature changes can alter the structure and thus the function of an enzyme.
At higher temperatures, the protein geometry can be disrupted by interfering with quaternary, tertiary and secondary structures. This changed geometry can affect active sites, making enzymes less effective.