Topic 13 DNA replication
1/7
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
8 Terms
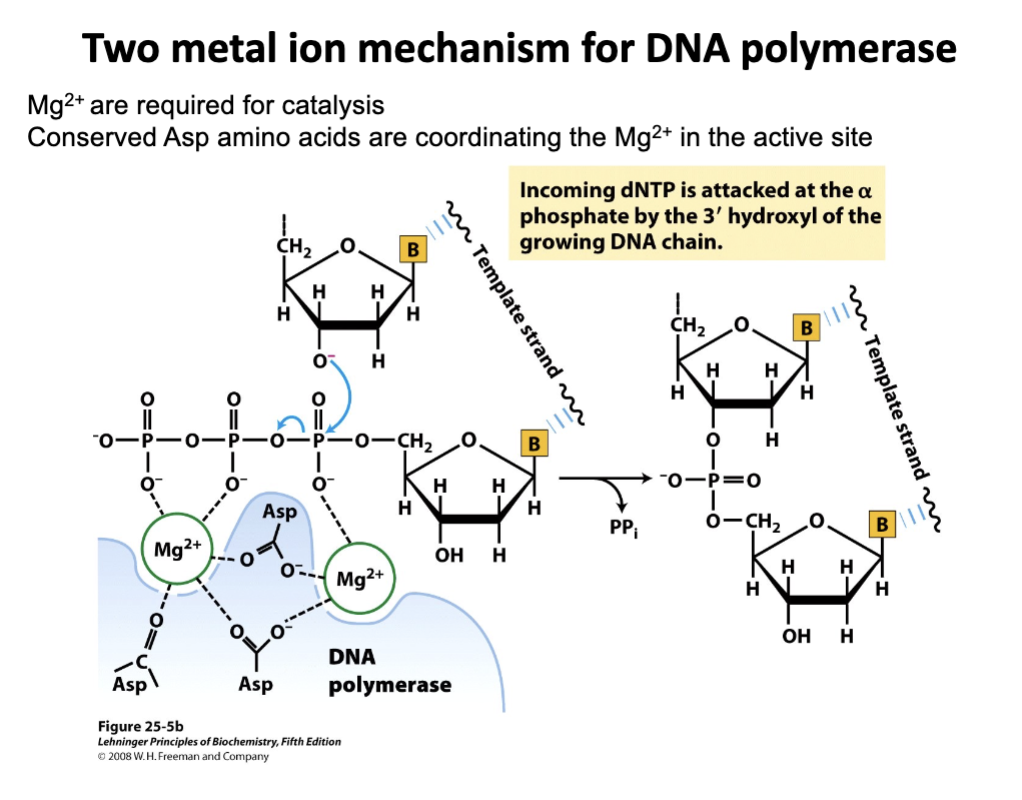
(learning objective) Explain why DNA polymerase requires metal ions for function
DNA polymerase needs metal ions like Mg2+ to stabilize negative charges and catalyze phosphodiester bond formation during DNA synthesis
(learning objective) Describe how DNA polymerase achieves specificity
Fingers, thumb, and palm model of DNA polymerase
Palm
Contains the active site that monitors base pairing and catalyzes bond formation when correct geometry is achieved
Fingers
Guide incoming dNTPs into the active site and close around correctly paired bases, enhancing specificity
Thumb
Holds the DNA in place, maintaining the correct positioning of the primer and template strand
Summary
Shape selectivity helps promote specificity of DNA replication. A conformational change occurs when the correct dNTP (deoxynucleoside triphosphate) is bound permitting the chemistry to occur
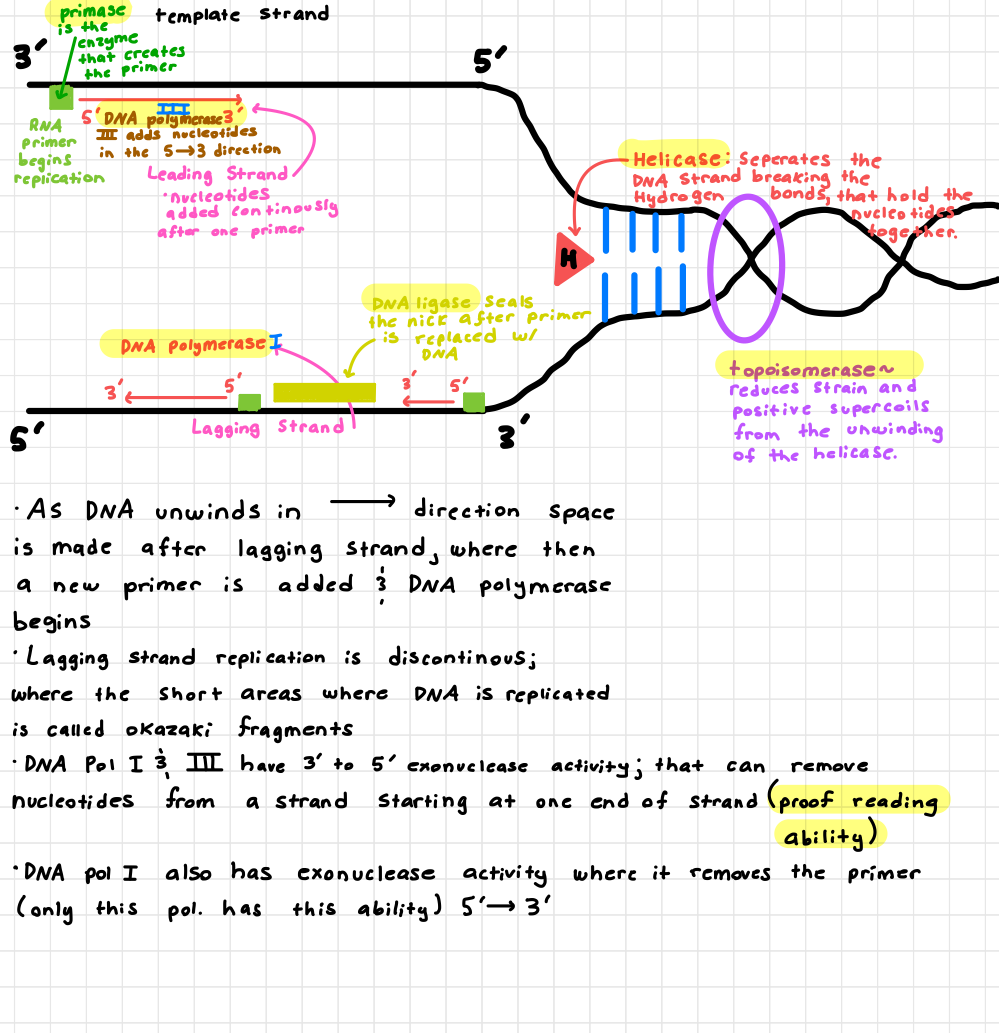
(learning objective) Describe how the components (helicase, topoisomerase, polymerase, primase, DNA ligase) of the replication machinery work together to copy DNA
Helicase
unwinds the DNA helix
Topoisomerase
relieves tension ahead of the fork
Primase
Makes RNA primers to start synthesis
DNA polymerase
adds new nucleotides to the growing strand
DNA ligase
seals gaps between fragments (especially lagging strand)
Together the coordinate to accurately and efficiently copy DNA
(learning objective) Describe how proofreading works
DNA polymerase proofreads by switching to its own exonuclease active site where it allows it to bind then cleaves nucleic acid if wrong to remove mismatched bases
Backtracks after removal and gets another try at added the correct dNTP
(learning objective) What happens at the replication fork
DNA is unwound by the helicase, and two new strands are synthesized
The leading strand is made continuously
Lagging strand is made in fragments (Okazaki fragments)
(learning objective) How is the leading and lagging strand synthesis different. What is the challenge in the lagging strand synthesis?
Leading strand is synthesized continuously toward the replication fork
Lagging strand is synthesized discontinuously away from the fork in short Okazaki fragments
Challenge
DNA polymerase can only add in the 5’ to 3’ direction, so lagging strand must keep restarting with new primers, making it more complex and requiring more enzymes.
(learning objective) Describe the Trombone model
The trombone model explains how the leading and the lagging strands are synthesized simultaneously
The lagging strand loops out like a trombone slide allowing synthesis in the same direction as the fork
each time an Okazaki fragment is finished, the loop resets with a new primer and clamp
this keeps both polymerases moving together despite opposite strand orientation
(learning objective) Describe key differences between prokaryotic and eukaryotic replication
Prokaryotic replication has one origin, uses DNA pol III, and is faster
Eukaryotic replication has many origins, uses different DNA pol and must also replication linear chromosomes with help from telomerase