lecture 24, 25 & 26
1/96
Earn XP
Description and Tags
Genetic Engineering and Genomics
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
97 Terms
What is biotechnology?
Using living things or parts of them to create useful products or solve problems (biology + technology).
Give an example of ancient biotechnology.
Using yeast to brew beer.
Give an example of modern biotechnology.
Engineering bacteria to produce insulin for diabetics.
What is genetic engineering?
A specific part of biotechnology where we directly change an organism’s genes for practical purposes.
Give an example of genetic engineering in plants.
Making a plant produce a vaccine naturally.
How can genetic engineering be described in simple terms?
Rewriting the code of life to give new abilities or traits.
What is the main focus of genetic engineering?
Altering the DNA of organisms to obtain desired traits.
: What is a plasmid?
A small, circular piece of DNA used as a delivery vehicle for genes.
What is a vector in genetic engineering?
A delivery tool, often a plasmid or virus, used to insert genes into an organism
How do scientists insert a gene into a plasmid?
By cutting the plasmid and the gene with restriction enzymes and joining them to form recombinant DNA.
What happens after a plasmid carrying a gene is inserted into bacteria?
The bacteria replicate, producing many copies of the gene and its protein product.
What is gel electrophoresis?
A technique that separates DNA fragments by size; smaller fragments move faster through a gel.
How do Restriction Fragment Length Polymorphisms (RFLPs) work?
DNA is cut with restriction enzymes, and fragments are sorted by size to compare DNA samples.
What are Short Tandem Repeats (STRs)?
Tiny, repeated DNA sequences used to create unique genetic fingerprints.
Why are STRs important in forensics?
They help identify individuals based on unique DNA patterns.
What is PCR (Polymerase Chain Reaction)?
A molecular copy machine that makes millions of copies of a DNA segment.
What are the three steps of PCR?
Denaturation (separate strands), Annealing (primers attach), Extension (build new DNA strands).
What is genomics?
The study of an organism’s entire set of genes (its genome).
What is bioinformatics?
Using computers to store, analyze, and interpret large amounts of biological data.
What is gene expression?
How active a gene is; how much of its product is being made.
Name methods to measure gene expression.
Real-Time PCR, Northern Blotting, RNA-Seq.
What is functional genomics?
Studying what genes do and how they interact inside cells.
How do scientists test gene function?
By turning genes on or off, or increasing their expression, and observing the effects.
: Name medical applications of biotechnology.
Gene therapy to fix or modify genes in patients.
Name agricultural applications.
Creating GMOs like pest-resistant or larger crops.
Name environmental applications.
Bioremediation using engineered organisms to clean pollution.
What is proteomics?
The large-scale study of all proteins in an organism.
Why is proteomics important?
It shows what genes actually do after producing proteins.
Name some ethical concerns in biotechnology.
Safety of GMOs, gene therapy, human gene editing, access to treatments.
Name social impacts of biotechnology.
Effects on environment, health, agriculture, and biodiversity.
What is the first step in genetic engineering?
Find a gene of interest.
What is the next step after finding the gene?
Insert it into a plasmid, often with a marker gene.
How is recombinant DNA introduced into bacteria?
Using vectors such as plasmids, viruses, or other delivery methods.
What is done after bacteria receive recombinant DNA?
They are grown into clones, producing many copies of the gene and its protein.
How are proteins harvested from genetically engineered bacteria?
By isolating the specific proteins produced by the introduced gene.
What are restriction enzymes?
Molecular scissors that cut DNA at specific sequences.
What type of ends does EcoRI produce?
Sticky ends (overhangs).
What type of ends does HindIII produce?
Blunt ends (straight cuts).
What type of ends does BamHI produce?
Sticky ends.
What is gene splicing?
Joining pieces of DNA from different sources to make recombinant DNA.
Which enzyme seals DNA fragments together?
DNA ligase.
What is recombinant DNA?
DNA created by combining pieces from different sources.
How are genes identified after cloning?
Using nucleic acid hybridization with labeled DNA probes.
What is a Bacterial Artificial Chromosome (BAC)?
A vector that carries ~300 kb of DNA.
What is a Yeast Artificial Chromosome (YAC)?
A vector that carries up to ~1,000 kb of DNA.
What is microinjection?
Using a tiny needle to inject DNA directly into a cell’s nucleus.
What is electroporation?
Using electric shocks to create holes in cell membranes for DNA entry.
What is a gene gun?
A device that shoots DNA-coated particles into cells.
What is a transgenic organism?
An organism with genes from another species.
Give an example of a transgenic animal.
salmon engineered with growth hormone genes to grow faster.
What is Sanger sequencing?
A method to read the exact order of nucleotides in DNA using chain-terminating molecules.
What is automated sequencing?
A faster, machine-based version of Sanger sequencing using fluorescent labels.
What is massively parallel sequencing?
Sequencing millions of DNA fragments at once, reducing time and cost.
What are Open Reading Frames (ORFs)?
DNA sections containing instructions for proteins, between start and stop signals.
What are Single Nucleotide Polymorphisms (SNPs)?
Small DNA variations at a single nucleotide, important for studying genetic differences.
What are transposable elements (jumping genes)?
DNA sequences that can move around in the genome, contributing to diversity.
How much of the human genome can transposable elements make up?
Up to 44%.
What are multigene families?
Groups of related genes performing similar functions (e.g., globin genes).
What is transcriptomics?
The study of all mRNA molecules in a cell to see which genes are active.
How does the transcriptome relate to the proteome?
The transcriptome determines which proteins (proteome) are produced.
Why is proteomics more complicated than genomics?
Proteins can vary, be modified, and interact in complex networks.
Name three fields where biotechnology is applied.
Medicine, agriculture, and environmental science.
Give a medical biotechnology example.
Producing insulin via engineered bacteria.
Give an agricultural example.
Crops resistant to pests or herbicides.
Give an environmental example.
Bioremediation using engineered microorganisms.
What ethical questions arise with biotechnology?
Safety of GMOs, gene editing in humans, equitable access to therapies.
How can biotechnology affect society?
Impacts on health, environment, agriculture, and biodiversity.
What are vectors in genetic engineering?
Vehicles (plasmids, viruses) used to deliver DNA into cells.
What is the purpose of PCR in biotechnology?
To make millions of copies of a DNA segment quickly for analysis.
Why is recombinant DNA important?
It allows scientists to combine DNA from different sources to create new traits or products.
practice quiz #13
For a particular microarray assay (DNA chip), cDNA has been made from the mRNAs of a dozen patients' breast tumor biopsies. Which of the following types of evidence will researchers be looking for in order to determine if the cells are cancerous?
A. a particular gene that is amplified in all or most of the patient samples
B. a pattern of fluorescence that indicates which cells are over proliferating
C. a pattern shared among some or all of the samples that indicates gene expression differing from control samples
D. a group of cDNAs that match those in non-breast cancer control samples from the same population
E. A and B
Correct Answer: C
Reason: Microarrays are used to compare gene expression patterns between different samples. Cancerous cells typically have a different pattern of gene activity (some genes overexpressed, others underexpressed) compared to healthy cells. Researchers look for these differential expression patterns to identify cancer markers.
The specific function of the Cas9 protein in the CRISPR system is to
A. cut double stranded DNA of a targeted gene
B. guide the CRISPR system to a targeted gene
C. stimulate telomerase to extend chromosome length
D. repair DNA damage of a targeted gene
E. A and B
Correct Answer: A
Reason: Cas9 is a nuclease enzyme that acts as molecular scissors, specifically cutting double-stranded DNA at a target sequence identified by a guide RNA within the CRISPR system. The guide function is performed by the guide RNA, not Cas9 itself.
Because eukaryotic genes contain introns, their transcripts cannot be translated by bacteria, which lack RNA-splicing machinery. But if you want to engineer a bacterium to produce a eukaryotic protein, you can synthesize a gene without introns. A good way to do this is to __.
A. alter the bacteria so that they can splice RNA
B. use a phage to insert the desired gene into a bacterium
C. use a nucleic acid probe to find a gene without introns
D. us a restriction enzyme to remove introns from the gene
E. work backwards from mRNA using reverse transcriptase to make a version of cDNA
Correct Answer: E
Reason: Eukaryotic mRNA has already had introns removed through splicing. Using reverse transcriptase, a cDNA (complementary DNA) copy can be made from this mRNA, which will contain only the exon sequences and thus no introns. This intron-free cDNA can then be expressed in bacteria.
Suppose ENZ-1 and ENZ-2 are two different restriction enzymes. If various pieces of DNA are cut with either of these enzymes, which of the following cut DNAs would join together most easily to form recombinant molecules?
A. human DNA cut with ENZ-1 and gorilla DNA cut with ENZ-2
B. human DNA cut with ENZ-1 and human DNA cut with ENZ-2
C. human DNA cut with ENZ-2 and bacterial DNA cut with ENZ-2
D. bacterial DNA cut with ENZ-1 and gorilla DNA cut with ENZ-2
E. all of the above
Correct Answer: C
Reason: Restriction enzymes cut DNA at specific recognition sequences, often creating "sticky ends" (overhangs). DNA fragments cut with the same restriction enzyme will have complementary sticky ends, allowing them to anneal and be joined by DNA ligase, regardless of the species origin (as long as the recognition sequence exists). Therefore, DNA cut with ENZ-2 from any source (human or bacterial) will have compatible ends with other DNA cut with ENZ-2.
The last step in shotgun sequencing with the dideoxy chain termination method is to __.
A. randomly select DNA primers and hybridize these to random positions of chromosomes in preparation for sequencing.
B. clone small DNA fragments into plasmids and sequence each fragment.
C. cut genomic DNA into small fragments at random sites.
D. locate specific genes by FISH
E. use computer software to detect overlap in small fragments to order the overall sequence
Correct Answer: E
Reason: Shotgun sequencing involves first cutting the genome into many small, overlapping fragments (C), cloning them (B), and then sequencing each fragment (using methods like dideoxy chain termination, or Sanger sequencing, as mentioned in the note). The final crucial step (E) is to use sophisticated computer software to analyze the millions of short sequence reads, identify overlapping regions, and assemble them into the correct order to reconstruct the entire genome sequence
Gene duplication has played a critical role in evolution because it __.
A. produces redundant copies of existing genes, which are then free to mutate and adopt related functions
B. almost always introduces immediate benefits for the organism
C. increases the likelihood of viral infection in cells
D. increases the number of pseudogenes in the genome
E. increases the amount of DNA in the genome
Correct Answer: A
Reason: When a gene is duplicated, one copy can continue performing the original function, while the redundant second copy is 'freed' from selective pressure and can accumulate mutations. These mutations might lead to a new, related function without harming the organism, providing raw material for evolutionary innovation.
A mutated form of hemoglobin, so-called hemoglobin Lepore, exists in the human population. Hemoglobin Lepore has a deleted series of amino acids. If this mutated form was caused by unequal crossing over, what would be an expected consequence?
A. There should also be persons whose hemoglobin contains two copies of the series of amino acids that is deleted in hemoglobin Lepore.
B. The deleted region must be located in a different area of the individual's genome.
C. The deleted gene must have been lost through a translocation
D. Each of the genes in the hemoglobin multigene family must show the same deletion.
E. It would result in aneuploidy.
Correct Answer: A
Reason: Unequal crossing over occurs when homologous chromosomes misalign during meiosis, leading to an exchange that results in one chromatid losing a segment (deletion) and the other chromatid gaining an extra copy of that segment (duplication). Thus, if one individual has a deletion (like hemoglobin Lepore), there must also be individuals with a corresponding duplication somewhere in the population.
What characteristic of short tandem repeats (STRs) also called microsatellites makes it useful for DNA fingerprinting?
A. The number of repeats varies widely from person to person or animal to animal.
B. The sequence of DNA that is repeated varies significantly from individual to individual.
C. The sequence variation is acted upon differently by natural selection in different environments.
D. Every racial and ethnic group has inherited different short tandem repeats.
E. All of the above
Correct Answer: A
Reason: As stated in the note under "Short Tandem Repeats (STRs)", "everyone has slightly diffenret numbers at different part of their geno" and "scientist look at avriations to create a unqiue genetic fingerprint". It is the number of times a specific sequence is repeated at a given locus that varies considerably between individuals, making STRs highly polymorphic and ideal for individual identification.
Which of the following terms is LEAST related to the rest?
A. RT-PCR
B. CRISPR-Cas9
C. Northern Blotting
D. FISH with RNA probes
E. Illumina sequencing
Correct Answer: B
Reason: RT-PCR, Northern Blotting, and FISH with RNA probes (often used to detect specific RNA molecules in cells) are all techniques primarily used to analyze or measure gene expression (i.e., the presence and quantity of RNA transcripts). Illumina sequencing is a high-throughput method for determining DNA (or RNA after reverse transcription) sequences. CRISPR-Cas9, however, is a gene-editing tool used to change the DNA sequence (e.g., cut or modify genes), not primarily to measure or sequence existing genetic material.
Human proteomics presents a particular challenge because
A. the number of proteins in humans probably far exceeds the number of genes.
B. a cell's proteins differ with cell type.
C. proteins are extremely varied in structure and chemical properties.
D. A and B only
E. A, B, and C
Correct Answer: E
Reason: As indicated by the "Proteomics" section, proteins are the workers encoded by genes. The complexity arises because a single gene can produce multiple protein variants (isoforms), proteins undergo post-translational modifications, and the set of proteins expressed varies greatly depending on cell type, developmental stage, and environmental conditions. Additionally, proteins have diverse structures and chemical properties, making their analysis challenging. All these factors contribute to the immense complexity of the human proteome, far exceeding the number of genes.
Imagine that you've sequenced the genome of a human pathogenic bacterium. In the early stages of analysis, you discover a stretch of DNA that has a significantly different GC content (the proportion of bases that are G and C) than the rest of the genome. Further examination of this region, using a computer program that searches for ORFs, shows there are roughly one dozen protein-coding regions. These are not found in the genome of a previously sequenced and related bacterium. These sequences do, however, predict protein products strikingly similar to those of another bacterial pathogen that is not closely related to the organism you're studying. You immediately suspect:
A. convergent evolution
B. that the sequence you've analyzed could not encode these proteins
C. a high rate of mutation
D. a low rate of mutation
E. lateral gene transfer
Correct Answer: E
Reason: The distinct GC content, presence of new genes not in related species, and similarity to genes in unrelated pathogens are classic hallmarks of lateral (or horizontal) gene transfer. This process allows bacteria to acquire genetic material, often virulence factors or antibiotic resistance genes, from other species, even distantly related ones.
What percent of the DNA in the human genome are regions coding for protein?
A. 0.5%
B. 1.5%
C. 15.0%
D. 20.0%
E. 44.0%
Correct Answer: B
Reason: Only a very small fraction of the human genome (approximately 1.5%1.5%) consists of protein-coding exons. A larger portion is transcribed into RNA (including non-coding RNAs), but the actual protein-coding sequences are quite limited. The vast majority of the genome is non-coding, including introns, regulatory sequences, and repetitive DNA.
rela quiz #13
Question 1
A paleontologist has recovered a bit of tissue from the 400-year-old preserved skin of an extinct dodo (a bird). The researcher would like to compare DNA from the sample with DNA from living birds. Which of the following would be most useful for increasing the amount of dodo DNA available for testing?
Answer Choices:
A. PCR
B. gel electrophoresis
C. electroporation
D. Southern blots
E. RFLP analysis
Correct Answer: ✔ A. PCR
Explanation:
PCR amplifies tiny amounts of DNA, producing millions of copies — ideal for ancient or degraded DNA.
Question 2
When a typical restriction enzyme cuts a DNA molecule, the cuts are staggered so that the DNA fragments have single-stranded ends. This is important in recombinant DNA work because __________.
Answer Choices:
A. the single-stranded ends serve as starting points for DNA replication
B. it enables researchers to use the fragments as introns
C. the fragments will bond to other fragments with complementary single-stranded ends
D. it allows a cell to recognize fragments produced by the enzyme
E. only single-stranded DNA segments can code for proteins
Correct Answer: ✔ C. the fragments will bond to other fragments with complementary single-stranded ends
Explanation:
Staggered cuts produce sticky ends, which base-pair with matching sequences, allowing insertion into plasmids.
Question 3
From the figure below, which of the following organisms has the lowest gene density with respect to genome size?
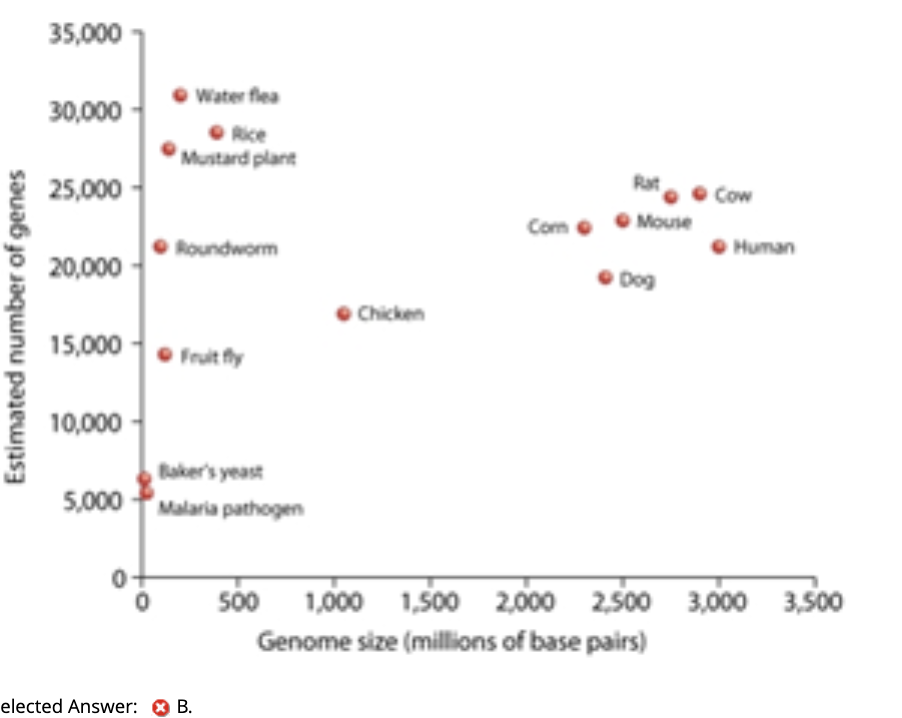
Answer Choices:
A. water flea
B. malaria pathogen
C. human
D. Baker’s yeast
E. chicken
Correct Answer: ✔ C. human
Explanation:
Humans have a huge genome with much noncoding DNA, meaning fewer genes per million base pairs → lowest gene density.
Question 4
Why might the cricket genome have eleven times as many base pairs as that of Drosophila melanogaster?
Answer Choices:
A. Crickets must have more noncoding DNA.
B. Crickets must make many more proteins.
C. Drosophila are more complex organisms.
D. Crickets have higher gene density.
E. A and B
Correct Answer: ✔ A. Crickets must have more noncoding DNA.
Explanation:
Genome size differences usually reflect differences in repetitive/noncoding DNA, not gene number.
Question 5
Chromosomal rearrangements may be important in evolution because __________.
Answer Choices:
A. chromosome rearrangements lead to gene duplication, thus generating a "spare" copy of the gene that is free to evolve and acquire a new function
B. chromosome rearrangements may cause genetic diseases
C. chromosomal rearrangements caused the extinction of dinosaurs
D. chromosome rearrangements cause new genes to evolve
E. offspring who inherit two differently arranged sets of chromosomes instantly become a new species
Correct Answer: ✔ A. chromosome rearrangements lead to gene duplication
Explanation:
Gene duplication provides an extra copy that can mutate and evolve new functions — a major evolutionary mechanism.
Question 6
You wanted to visualize the pattern of gene expression viewing the whole organism of a Drosophila larva. The preferred technique in this situation would be
Answer Choices:
A. CRISPR-Cas9
B. FISH
C. RNA interference
D. RT-PCR
E. DNA microarrays
Correct Answer: ✔ B. FISH
Explanation:
FISH allows fluorescent visualization of where mRNA transcripts appear inside intact tissues/whole organisms.
Question 7
The human genome consists of approximately 21,000 genes, which is only slightly more than the genome of the nematode Canorhabditis. elegans. How can two organisms with similar numbers of genes be so different in overall complexity?
Answer Choices:
A.Extensive RNA splicing, which generates more than one polypeptide from a single gene, occurs in humans but is not typically seen in nematodes.
B. RNAs such as miRNAs that play regulatory roles in humans are not typically present in nematodes.
C. Human genes typically contain multiple exons, which can be spliced together in different ways, leading to the production of many more proteins than the number of proteins in the nematode.
D. Post-translational modifications of proteins are more common in humans than in nematodes.
E. All of the above
Correct Answer: ✔ E. All of the above
Explanation:
Humans have:
more alternative splicing
regulatory RNAs
more protein modification
→ greater complexity from similar gene numbers.
Question 8
Which arrangement of the following four enzymes represents the order in which they would be used in a typical gene-cloning experiment resulting in the insertion of a cDNA into a bacterial plasmid? Begin with the gene's mRNA transcript.
Answer Choices:
A. Restriction enzyme → reverse transcriptase → DNA polymerase → DNA ligase
B. Reverse transcriptase → restriction enzyme → DNA polymerase → DNA ligase
C. Reverse transcriptase → DNA polymerase → restriction enzyme → DNA ligase
D. Restriction enzyme → DNA ligase → reverse transcriptase → DNA polymerase
E. Reverse transcriptase → DNA ligase → DNA polymerase → restriction enzyme
Correct Answer: ✔ C. Reverse transcriptase → DNA polymerase → restriction enzyme → DNA ligase
Explanation:
Reverse transcriptase converts mRNA → cDNA
DNA pol makes it double-stranded
Restriction enzymes cut vectors + cDNA
Ligase seals them together
Question 9
The Alu family is a family of repetitive DNA elements 300bp long scattered through the human genome comprising almost 10% of the genomic DNA. Alu elements can replicate independently of the genomic DNA. Alu is first transcribed into mRNA by RNA polymerase III. The mRNA is then converted by reverse transcriptase to cDNA, which is inserted into the genome. Based on this description an Alu element must be a |
Answer Choices:
A. SINE
B. retrotransposon and SINE
C. LINE
D. retrotransposon and LINE
E. retrotransposon
Correct Answer: ✔ B. retrotransposon and SINE
Explanation:
Alu elements are:
short → SINE
use reverse transcriptase → retrotransposons
Question 10
If the CRISPR-Cas9 system was used in conjunction with multiple guide RNA molecules (multiplexing), what would be the most likely outcome?
Answer Choices:
A. disruption of a gene
B. disruption of more than one gene
C. repair of a gene
D. deletion of a chromosome
E. disruption and repair of one gene
Correct Answer: ✔ B. disruption of more than one gene
Explanation:
Multiple gRNAs target multiple sites, so several genes can be edited at once.
Question 11
The class activity related to “target practice” with tennis balls demonstrated the
Answer Choices:
A. process of lateral gene transfer
B. technique of inserting foreign DNA into plants to produce GMOs
C. procedure for creating a knockout mouse
D. movement of transposons to targeted locations in the DNA
E. evolutionary advantage of repetitive DNA in the human genome
Correct Answer: ✔ E. evolutionary advantage of repetitive DNA in the human genome
Explanation:
The activity modeled how repeated hits increase probability — mimicking repetitive DNA increasing chances of successful binding/interaction.
Question 12
Rank the following in order from the smallest to largest unit of genetic information: 1. codon, 2. ORF, 3. SNP, 4. EST, 5. STR
Answer Choices:
A. 3,1,5,4,2
B. 1,3,2,4,5
C. 4,3,1,2,5
D. 5,1,2,3,4
E. 1,4,3,2,5
Correct Answer: ✔ A. 3,1,5,4,2
Explanation:
Order small → large:
SNP → codon → STR → EST → ORF
Question 13
Like DNA microarrays, Northern blots analyze the levels of mRNA. However, Northern blots are almost always prepared with a probe tagged with one label, whereas DNA microarrays almost always use two groups of probes, each one tagged with a different fluorescent label. What advantage is conferred by using two labels for the probes of microarrays?
Answer Choices:
A. The two labels allow comparison of gene expression between two samples (i.e. control vs. treatment group)
B. The two labels increase the sensitivity of the microarray
C.Probing the array with two labels is like doing two experiments at once
D. The two labels allow the analysis of the expression of more genes
E. The two labels allow simultaneous assay of levels of DNA and RNA
Correct Answer: ✔ A. The two labels allow comparison of gene expression between two samples
Explanation:
One color = control
One color = treatment
→ Direct comparison on the same chip.