BIOL 326 E2
1/202
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
203 Terms
Chromosome characteristics
number, size, location of centromere, banding patterns
p
chromosome short arm
q
chromosome long arm
metacentric
centromere in middle
submetacentric
centromere slightly off center
acrocentric
centromere is significantly off center, but not at the end
telocentric
centromere at end; p may be nearly nonexistent
banding patterns
revealed when chromosomes are treated w/ stain
karyotype
photographic representation in which all of the chromosomes w/in a single cell are arranged in a standard fashion
chromosome numbering
according to size; Largest = smallest #
(sex chromosomes are just X and Y)
G banding
1. Chromosomes are treated w/ heat or proteolytic enzymes that partially digest chromosomal proteins
2. Chromosomes are exposed to Giemsa stain
3. Chromosomal regions that bind the Giemsa molecules heavily produce dark bands; in other regions, light bands are produced
dark G-bands
more tightly compacted regions
importance of banding
distinguishes chromosomes, detects changes in chromosome structure, assesses evolutionary relationships b/t species
euploid
organisms w/ a chromosome number that is an exact multiple of a chromosome set (ex/ fruit flies - 8 chromosomes, 2 sets = 4 chromosomes/set)
aneuploid
organisms w/ a chromosome number that is NOT an exact multiple of a set
trisomic
organism w/ three copies of a chromosome instead of 2 (2n+1)
monosomic
organism w/ one copy of chromosome instead of 2 (2n-1)
aneuploidy in humans
can cause miscarriage; ~50% of all spontaneous abortions are due to alterations in chromosome #
most common survivable trisomies
trisomy 13, 18, or 21 (down-syndrome), and sex chromosome abnormalities
trisomy 1
believed to either cause gametes to be inviable or lethal at such an early developmental stage that it prevents successful implantation of the embryo
trisomy X
X-chromosome inactivation (barr bodies) allows for the normal expression of most X-linked genes
meiotic nondisjunction
can occur during anaphase I or II
meiotic nondisjunction during A1
an entire bivalent migrates to one pole → 4 abnormal haploid cells
meiotic nondisjunction during A2
net result of 2 abnormal and 2 normal haploid cells
trisomic nondisjunction
if a gamete carrying an extra chromosome unites with a normal gamete
monosomic nondisjunction
if a gamete missing a chromosome is viable and participates in fertilization
complete nondisjunction
rare; all of the chromosomes undergo nondisjunction and migrate to one of the daughter cells
daughter cell w/ all chromosomes may complete meiosis and form two diploid cells, which give rise to diploid gametes
daughter cell w/ no chromosomes is nonviable
deletions and duplications
changes in the total amount of genetic material w/in a single chromosome
deletion
a segment of chromosomal material is missing causing the affected chromosome to be deficient in a significant amount of genetic material
deficiency
refers to the condition of a chromosome that is missing a region
duplication
occurs when a section of a chromosome is repeated w/in a chromosome
inversions and translocations
chromosomal rearrangements; total amount of material is unchanged
inversion
a change in the direction of the genetic material along a single chromosome
translocation
one segment of a chromosome becomes attached to a different chromosome/different part of the same chromosome
simple translocation
a single piece of chromosome is attached to another chromosome
reciprocal translocation
two different chromosomes exchange pieces, altering both of them
how deletions occur
a chromosome breaks in one or more places, and a fragment of the chromosome is lost OR
recombination occurs at incorrect locations b/t homologous chromosomes
terminal deletions
fragments w/ the centromere
non-terminal deletions
fragments w/out the centromere; lost bc they do not find their way into the nucleus after mitosis, resulting in degradation in cytosol
interstitial deletion
when two ends of a broken chromosome reconnect, and the central fragment is lost
deletions due to incorrect recombination
Result in one chromosome w/ a deletion and one w/ a duplication
how duplications occur
abnormal crossover events during meiosis; crossover may occur at misaligned sites on homologs
misalignment
occurs when a chromosome has repetitive sequences
repetitive sequences
two or more homologous segments that have identical or similar sequences
nonallelic homologous recombination
occurs at homologous sites (such as repetitive sequences), but the sites are not alleles of the same gene, resulting in one chromatid w/ a duplication and another w/ a deletion
deletion phenotypic consequences
usually produce detrimental effects on phenotype; larger deletions are more harmful bc more genes are missing
duplication phenotypic consequences
tend to be less phenotypically harmful than deletions; larger duplications are more harmful
gene family
two or more genes in a particular species that are similar to each other; occurs by duplication
copy number variation (CNV)
a type of structural variation in which a DNA segment (≥1000 bp) commonly exhibits copy number differences among members of the same species; occurs at the population level
how CNV occurs
may occur bc some members of a species carry a chromosome missing a gene or part of a gene OR
bc of nonallelic homologous recombination OR
bc of a duplication
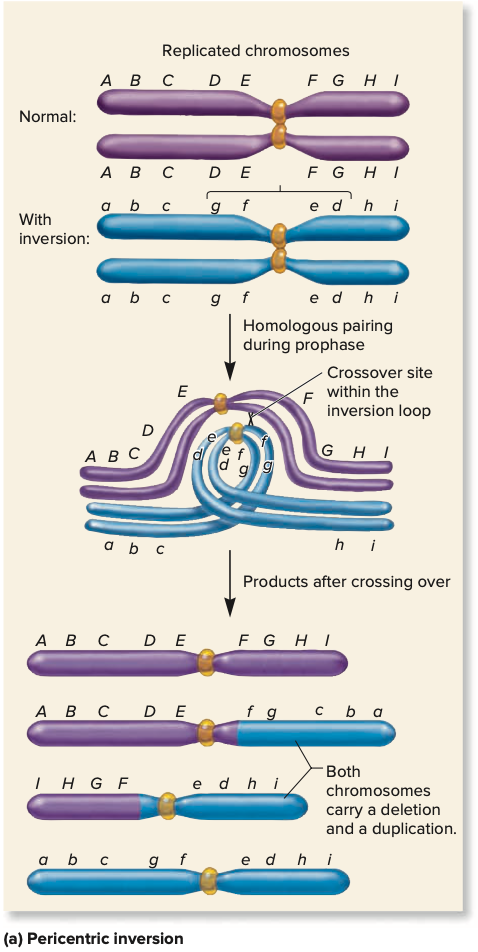
pericentric inversion
if the centromere lies w/in the inverted region of the chromosome
paracentric inversion
if the centromere lies outside the inverted region of the chromosome
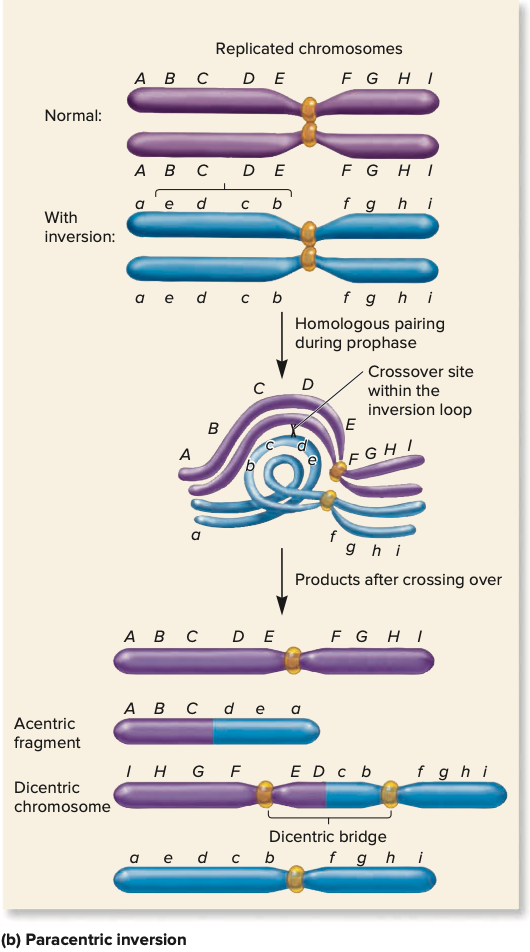
acentric fragment
piece of chromosome w/out any centromere - lost and degraded during cell division
dicentric chromosome
contains two centromeres, region connecting two centromeres is a dicentric bridge
balanced translocation
total amount of genetic material is unchanged; typically no phenotypic consequences
unbalanced translocation
significant portions of genetic material are duplicated/deleted; associated w/ phenotypic abnormalities and can be lethal
how balanced translocations occur
Mechanism 1:
1. Cells are exposed to agents that cause chromosomes to break
2. Broken ends lack telomeres and are therefore reactive
3. A reactive end readily binds to another reactive end (via DNA repair enzymes), so if multiple chromosomes are broken, reactive ends may join incorrectly to produce a reciprocal translocation
Mechanism 2: nonhomologous chromosomes may crossover and exchange pieces
how unbalanced translocations occur
occurs in offspring of parents with balanced translocation
nucleic acid
Term was derived from Friedrich Miescher's discovery of DNA in 1869
- He identified an unknown phosphorus-containing substance isolated from the nuclei of WBCs - he named it nuclein
- As the structure of DNA and RNA became better understood, it was determined they are acidic molecules bc they release H ions in solution and have a net negative charge at neutral pH
- Nuclein + acid = nucleic acid = DNA and RNA
genetic material criteria: information
genetic material must contain info necessary to construct an entire organism; blueprint for determining the inherited traits of an organism
genetic material criteria: transmission
during reproduction, the genetic material must be passed from parents to offspring
genetic material criteria: replication
genetic material must be copied to pass it from parent to offspring
genetic material criteria: variation
the genetic material must vary in ways that can account for phenotypic differences w/in each species
nucleotide components
at least one phosphate group, a pentose sugar, and a nitrogenous base
pentose sugars
deoxyribose (DNA) and ribose (RNA)
purines
double ring structures: adenine (A) and guanine (G)
pyrimidines
single ring structures: thymine (T), cytosine (C) (DNA), and uracil (U) (RNA)
phosphodiester linkage
attachment of phosphate to 3' carbon in one nucleotide and the 5' carbon in the next nucleotide
why DNA has directionality
all sugar molecules are oriented in the same direction
5' end
phosphate group
3' end
sugar hydroxyl (OH) group
DNA backbone
phosphates and sugar molecule; phosphate group connects two sugar molecules via phosphodiester linkage; negatively charged due to negative charge on each phosphate; bases project from the backbone
base pairing rules
A-T, G-C, A-U (RNA)
chargaff's rule
the amount of A in DNA = the amount of T & the amount of G = the amount of C
antiparallel arrangement
in a DNA double helix, one strand is 5' to 3' and the other is 3' to 5'
A-T
two hydrogen bonds
GC
three hydrogen bonds
helicase
breaks the h-bonds b/t DNA strands
topoisomerase II
alleviates positive supercoiling (overwounding)
single-stranded binding protein
keeps the parental strands apart
primase
synthesizes an RNA primer
DNA polymerase III
synthesizes a daughter strand of DNA
DNA polymerase II
proofreads the daughter strand of replicated DNA and corrects any base pairing errors
DNA polymerase I
removes the RNA primers and fills in w/ DNA
DNA ligase
covalently links the Okazaki fragments together
leading strand
continuous synthesis; one RNA primer is made at the origin, and then DNA pol III attaches nucleotides in 5' to 3' direction as it slides toward the opening of the replication fork
lagging strand
Discontinuous synthesis; RNA primers repeatedly initiate synthesis in 5' to 3' direction, away from the replication fork
okazaki fragment
segments of lagging strand; each contains a short RNA primer at the 5' end
completion of okazaki fragment synthesis
1. Removal of RNA primers by DNA pol I
2. Synthesis of DNA by DNA pol I in the region where the primers were removed
3. Covalent attachment of adjacent fragments of DNA by DNA ligase
replisome
Primosome + two DNA polymerase II
primosome
complex formed by DNA helicase and primase
termination of DNA replication in E. coli
1. there is a pair of termination sequences on the opposite side of the chromosome from oriC (origin of replication) known as ter sequences
2. The protein, termination utilization substance (Tus) binds to the ter sequences and stops the movement of the replication forks
3. One of the ter sequences, T1, prevents the advancement of the fork moving left to right
4. Another ter sequence, T2, prevents the advancement of the fork moving right to left
5. DNA replication ends when oppositely advancing forks meet, usually at T1 or T2
dNTP
nucleotide about to be attached to the growing strand; contains three phosphate groups attached at the 5' carbon atom of deoxyribose
how nucleotides are connected
1. dNTP first enters the catalytic site of DNA polymerase and binds to the template strand
2. Next, the 3' OH group in the nucleotide on the end of the growing strand reacts with the PO42- group on the dNTP
- Highly exergonic reaction resulting in a covalent bond b/t 3' and 5' end
- Formation of the bond causes the newly made strand to grow in the 5' to 3' direction
how proofreading occurs
occurs by the removal of nucleotides in the 3' to 5' direction at the 3' exonuclease site; after the mismatched nucleotide is removed, DNA polymerase resumes DNA synthesis in the 5' to 3' direction
telomeres
highly repetitive telomeric sequences w/in the DNA and the specific proteins that are bound to those sequences
telomerase
recognizes the sequences at the the end of eukaryotic chromosomes and synthesizes the telomeric sequences
introns
non coding intervening sequences
exons
regions of an RNA molecule that remain after splicing has removed the introns
origins of replication
chromosomal sites necessary to initiate replication; eukaryotic chromosomes contain many origins