BIOL 2500 - Topic 10 (part 4)
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
Restriction enzymes allow us to get…
DNA fragments
What do we do with DNA fragments
1.) We can amplify them using PCR
2.) Then we can insert them into bacterial DNA, specifically vectors, resulting in the formation of recombinant vectors, otherwise known as molecular cloning
Molecular cloning
1.) The technique where you insert a restriction fragment into a vector
2.) The vector is then inserted into an organism, usually bacteria, which amplifies the recombinant DNA molecule within the cell, thereby making DNA clones
What do you do once you have the DNA clones?
You can isolate the DNA products (i.e. the clones) for DNA sequencing and other downstream methodologies
Vectors
The carrier fragment of DNA that contains all the necessary genes to replicate the DNA in the cell
How do you get the vector inside the bacterium?
Electroporation, where you electrocute the bacteria to make their cell membrane permeable, allowing the vector to enter the cell
Dideoxynucleotide DNA sequencing
Aka Sanger method DNA sequencing, an in vitro DNA synthesis technique that determines the nucleotide sequence of a DNA fragment
Where do you get the DNA fragment for Sanger DNA sequencing
From vector cloning or PCRs
The Sanger method DNA sequencing process closely resembles…
PCR
The Sanger method DNA sequencing process requirements
1.) Genomic DNA (amplified to high concentrations, most likely via PCR)
2.) The reaction mixture (primers, taq, buffer, dNTPs)
3.) ddNTPs
Sanger method DNA sequencing reactions
It is done using four different synthesis reactions, one for each different dNTP/ddNTP
Use of Sanger DNA sequencing back then
It was the technique used to sequence the first genome
ddNTPs
Dideoxynucleotides, which are dNTPs but they lack two oxygens, such that they have a 2’ and 3’-H rather than a 2’ and 3’-OH
Role of the ddNTPs
1.) Since they lack the 3’-OH, they cannot form phosphodiester bonds
2.) Therefore, they terminate the growing DNA strand, which produces various fragments that are 1 nucleotide longer/shorter than the other
Sanger DNA sequencing is sequencing by ____________
Termination
Concentrations of dNTPs vs. ddNTPs
1.) There are high concentrations of dNTPs and low concentrations of ddNTPs
2.) Therefore, more dNTPs will bind to the growing strands
3.) But when a ddNTP does bind, it results in the production of fragments of varying lengths
How does sanger DNA sequencing determine the sequence
It determines the DNA sequence by looking at the termination products, specifically based on its position in the gel electrophoresis, as the ddNTPs are usually radioactivally or fluorescently labelled
Sanger DNA sequencing reaction on the gel electrophoresis
Bottom of the gel: the 5’ end of the synthesized complementary strand, where the smallest fragments are
Top of the gel: the 3’ end of the synthesized complementary strand, where the longest fragments are
Which end are the primers located in the gel electrophoresis of the Sanger sequencing method
The 5’ bottom end of the gel
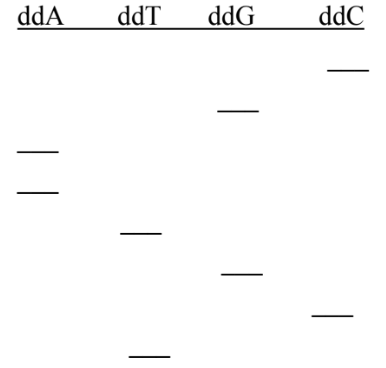
What is the template strands sequence
Complementary strand: 3’ CGAATGCT 5’
Template strand: 5’ GCTTACGA 3’