🎚️ Lecture 6: Regulation of the Activity of Transcription Factors
1/10
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
11 Terms
Lipid Hormone Regulation of Transcription Factors
Direct Regulation
Transcription factor activity can be directly regulated by lipid hormones
Hormone Signaling
A cell sends a signal to other cells by secreting a lipid-soluble hormone
Hormone Entry and Receptors
Lipid-soluble hormones diffuse through the plasma membrane
They bind a dedicated class of proteins
These are lipid-hormone receptors also called transcriptional activators
Cytoplasmic Receptors
Some receptors are located in the cytoplasm
Hormone binding forms a receptor–hormone complex
The complex moves to the nucleus if not already there
It becomes an active transcription factor
It binds specific response elements of genes
This regulates gene expression
Nuclear Receptors
Other receptors are located in the nucleus
Hormone binding changes their activity
Shared Features
Lipid-hormone receptors and transcription activators share significant homology
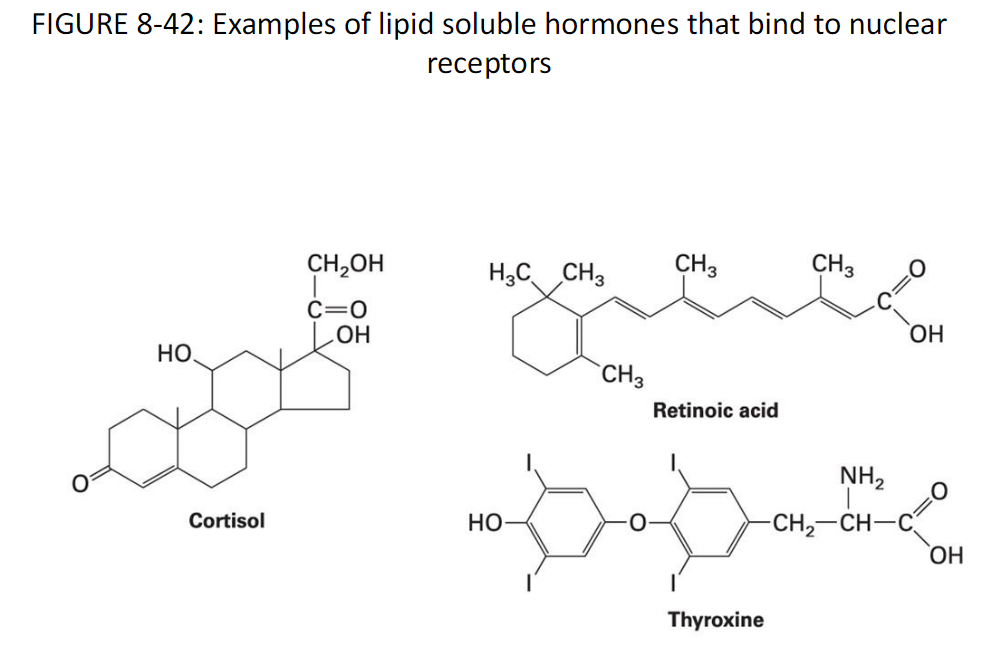
Lipid-Soluble Hormones and Nuclear Receptors
Figure 8-42 Examples
Examples of lipid-soluble hormones that bind to nuclear receptors
Retinoic Acid
Cortisol
Thyroxine
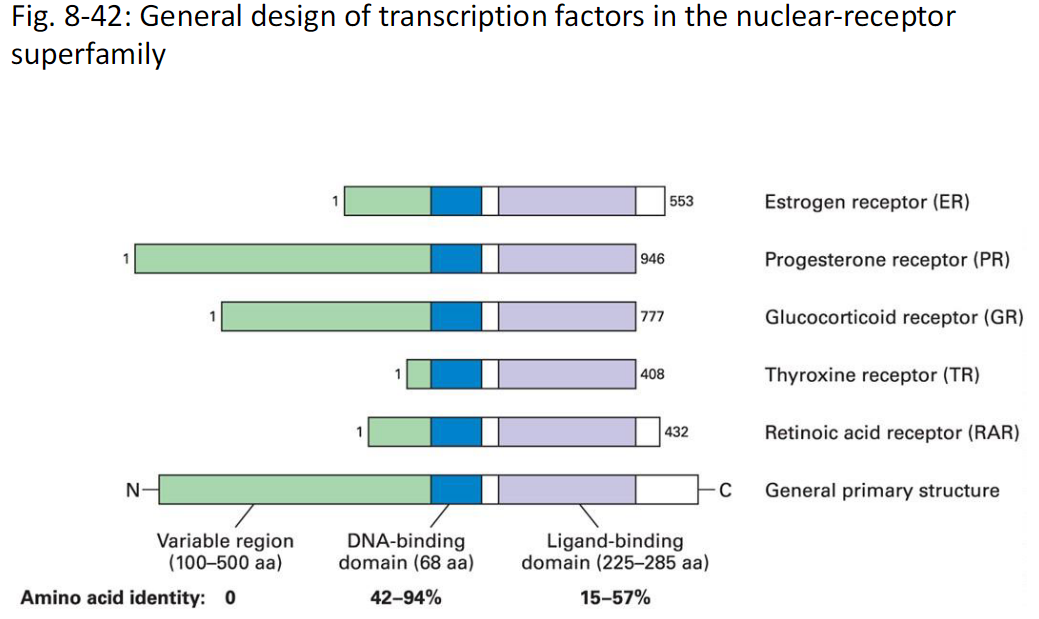
Nuclear Receptor Transcription Factors Structure
Figure 8-42
General design of transcription factors in the nuclear-receptor superfamily
Overall Structure
N terminus to C terminus
General primary structure is shared
Variable Region
Located at N terminus
Length is 100–500 amino acids
Low amino acid identity
Identity range 0
DNA-Binding Domain
Length is 68 amino acids
Responsible for binding DNA
Amino acid identity is high
Identity range 42–94%
Ligand-Binding Domain
Located toward C terminus
Length is 225–285 amino acids
Binds hormone ligand
Amino acid identity range 15–57%
Examples of Nuclear Receptors
Estrogen receptor ER
Progesterone receptor PR
Glucocorticoid receptor GR
Thyroxine receptor TR
Retinoic acid receptor RAR
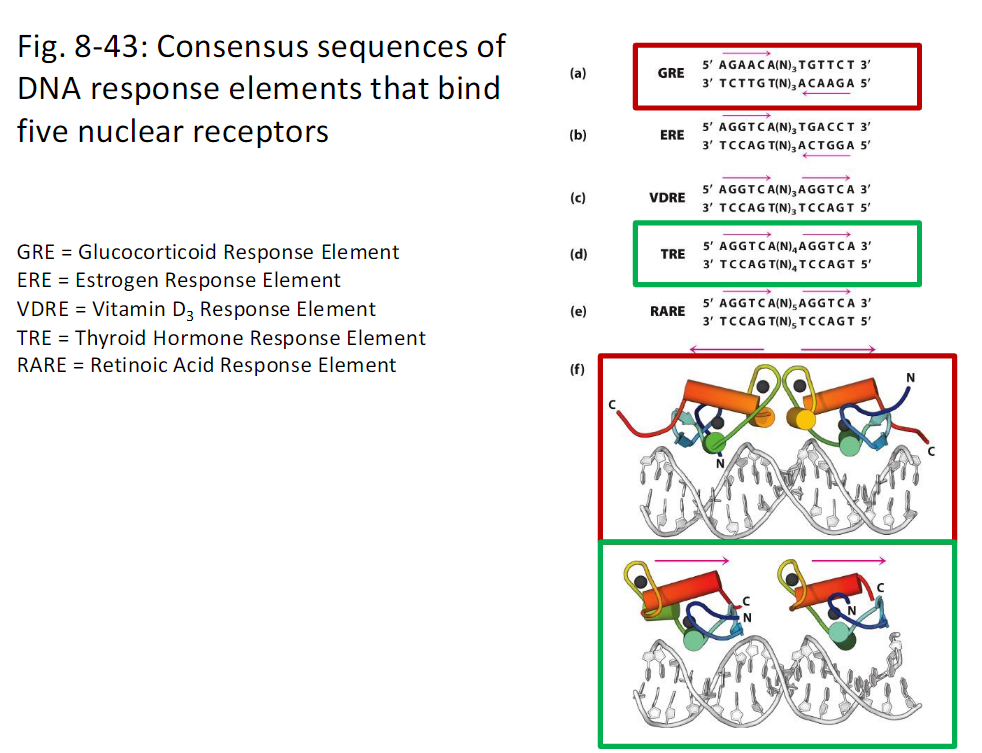
DNA Response Elements for Nuclear Receptors
Figure 8-43
Consensus sequences of DNA response elements that bind nuclear receptors
Glucocorticoid Response Element GRE
DNA response element that binds the glucocorticoid receptor
Estrogen Response Element ERE
DNA response element that binds the estrogen receptor
Vitamin D3 Response Element VDRE
DNA response element that binds the vitamin D3 receptor
Thyroid Hormone Response Element TRE
DNA response element that binds the thyroid hormone receptor
Retinoic Acid Response Element RARE
DNA response element that binds the retinoic acid receptor
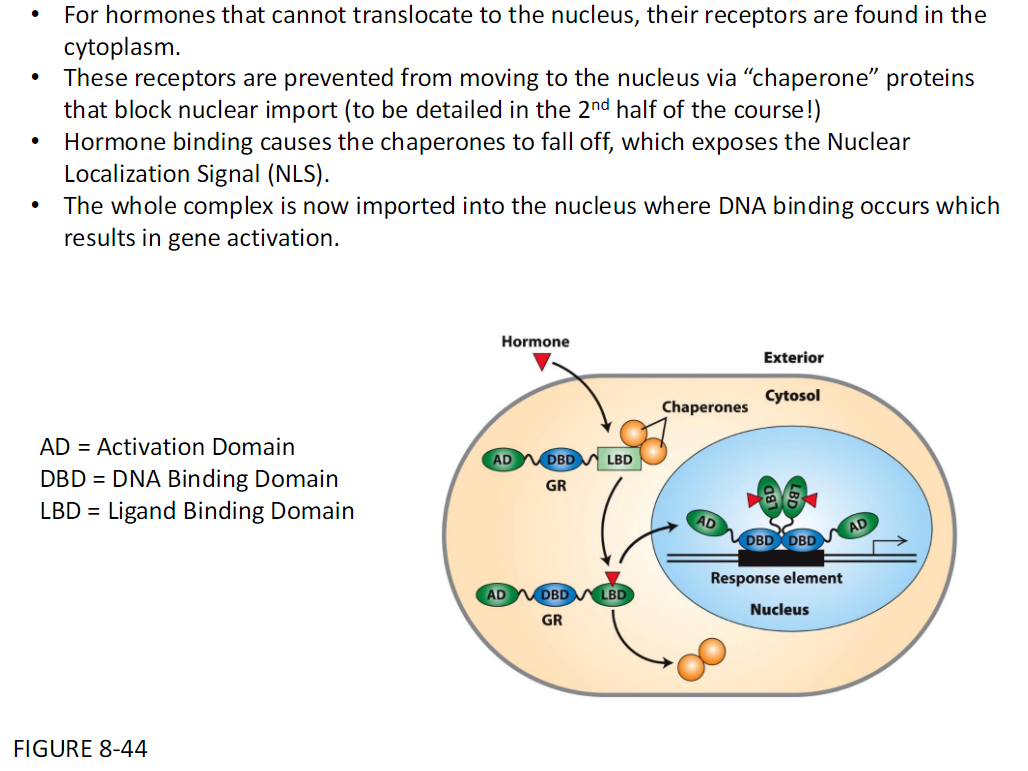
Cytoplasmic Hormone Receptors and Nuclear Translocation
Cytoplasmic Receptors
For hormones that cannot enter the nucleus, their receptors are in the cytoplasm
Chaperone Proteins
Receptors are kept out of the nucleus by chaperone proteins
Chaperones block nuclear import
Hormone Binding
Hormone binding makes chaperones detach
This exposes the Nuclear Localization Signal NLS
Nuclear Import and Activation
The receptor–hormone complex is imported into the nucleus
DNA binding occurs
This results in gene activation
Domains
AD = Activation Domain
DBD = DNA-Binding Domain
LBD = Ligand-Binding Domain
Eukaryotic Activators and Repressors
DNA Binding
Activators and repressors usually bind directly to DNA in promoters or enhancers
Gene Expression Regulation
From these positions they recruit multi-subunit co-activator or co-repressor complexes
These complexes modulate chromatin structure or interact with the Mediator and General Transcription Factors
Activators
Stimulate assembly of preinitiation complexes
Move nucleosomes away from promoters
Specificity
A cell must produce the specific set of activators required for a specific promoter or enhancer to express a particular gene
Repressors
Often work by building repressive chromatin structures
Mechanisms
Chromatin remodeling can lead to transcriptional repression or activation
Recall
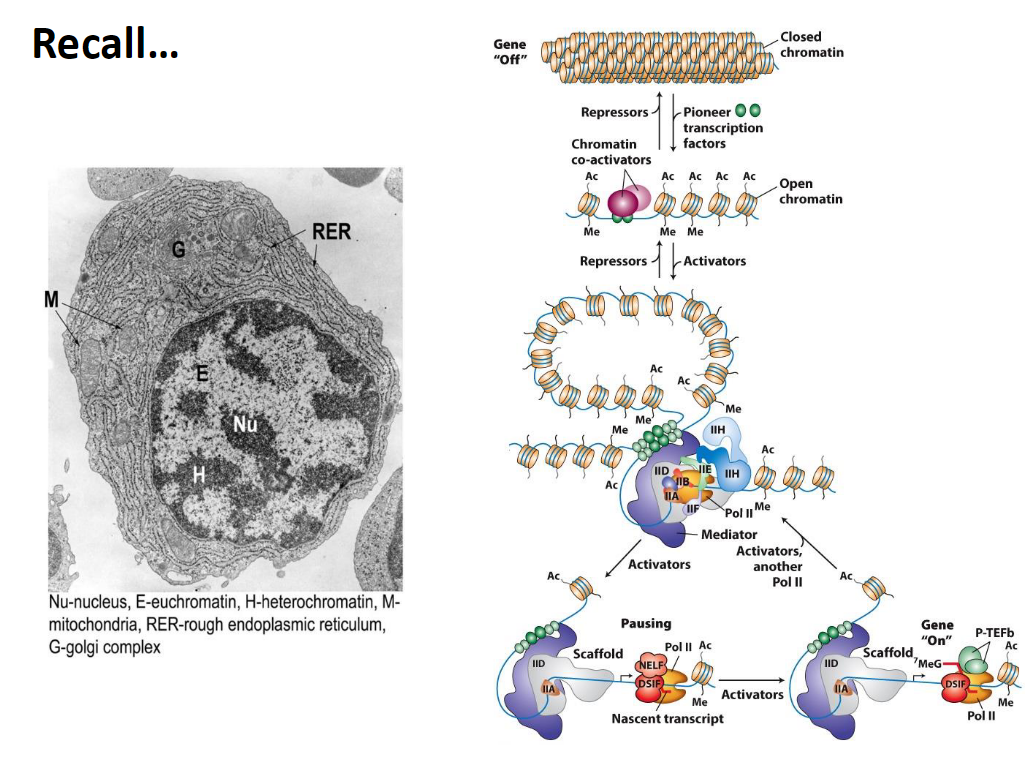
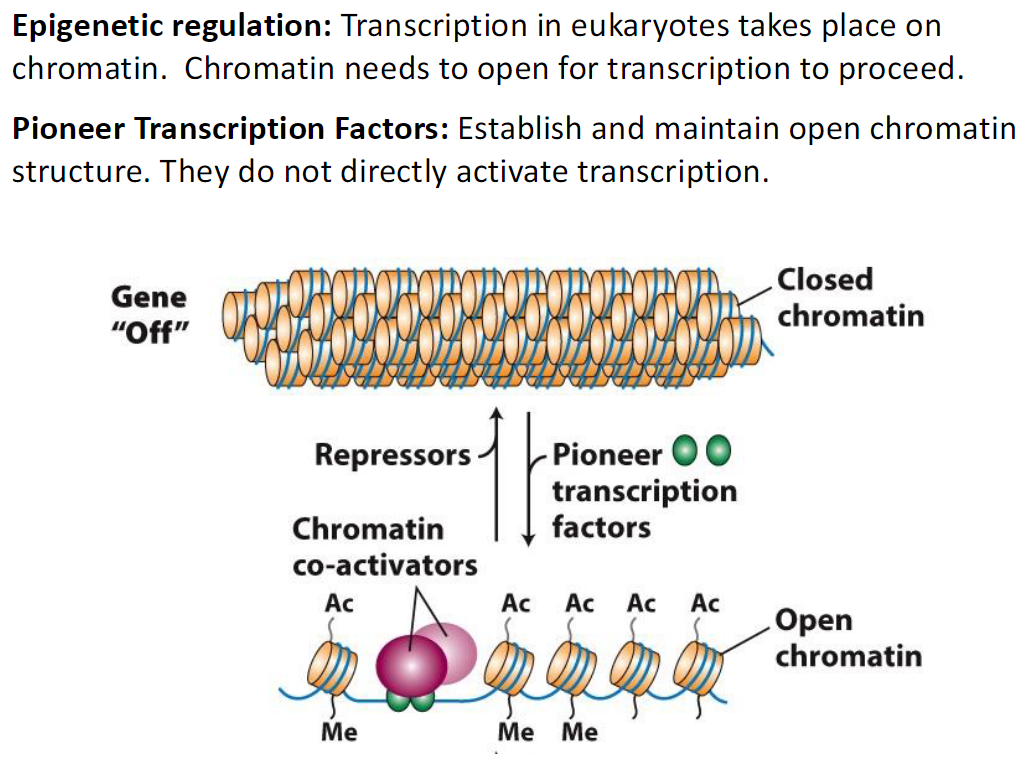
Epigenetic Regulation and Pioneer Transcription Factors
Epigenetic Regulation
Transcription in eukaryotes occurs on chromatin
Chromatin must open for transcription to proceed
Pioneer Transcription Factors
Establish and maintain open chromatin structure
Do not directly activate transcription
Gene Off
Chromatin is closed
Repressors and chromatin co-activators are present
Ac = acetylation, Me = methylation
Open Chromatin
Pioneer transcription factors help keep chromatin open
Allows transcription to occur
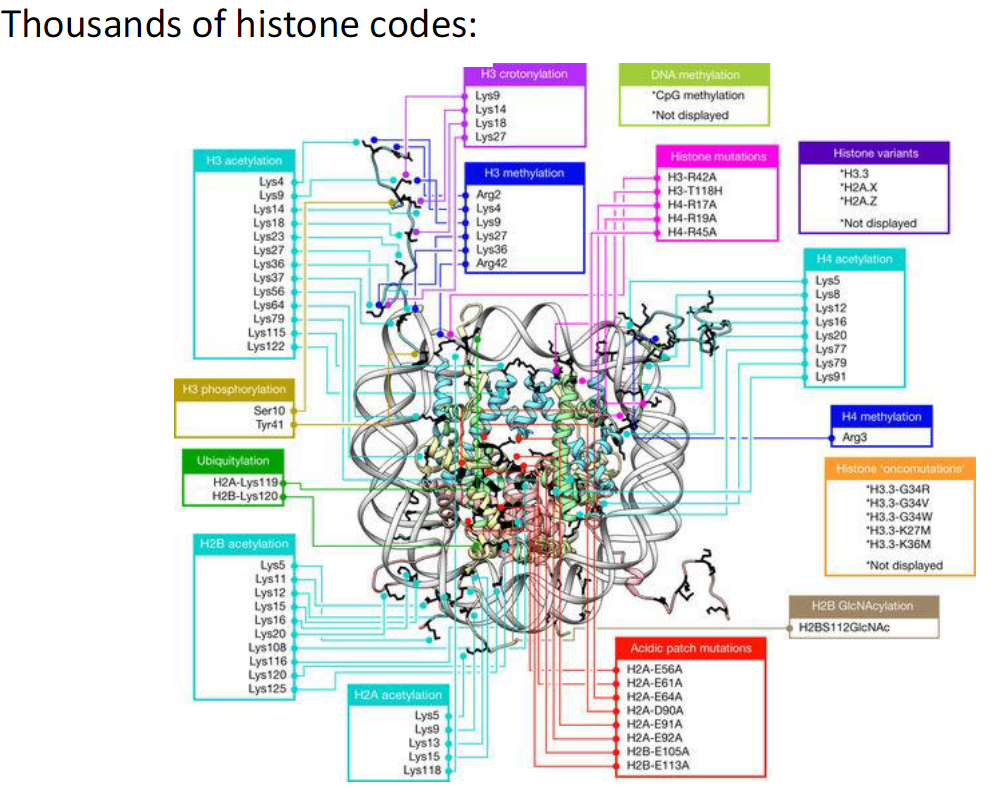
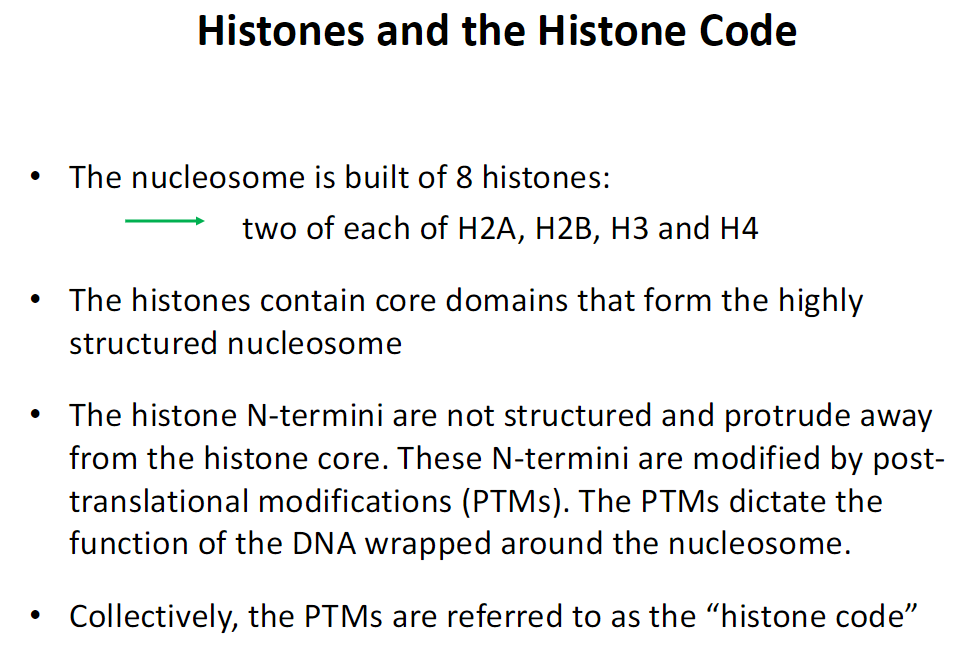
Histones and the Histone Code
Nucleosome Structure
The nucleosome is built of 8 histones
Two of each: H2A, H2B, H3, H4
Histone Core Domains
Core domains form the highly structured nucleosome
Histone N-Termini
N-termini are unstructured and protrude from the core
They are modified by post-translational modifications PTMs
Histone Code
PTMs dictate the function of the DNA wrapped around the nucleosome
Collectively, these modifications are called the “histone code”
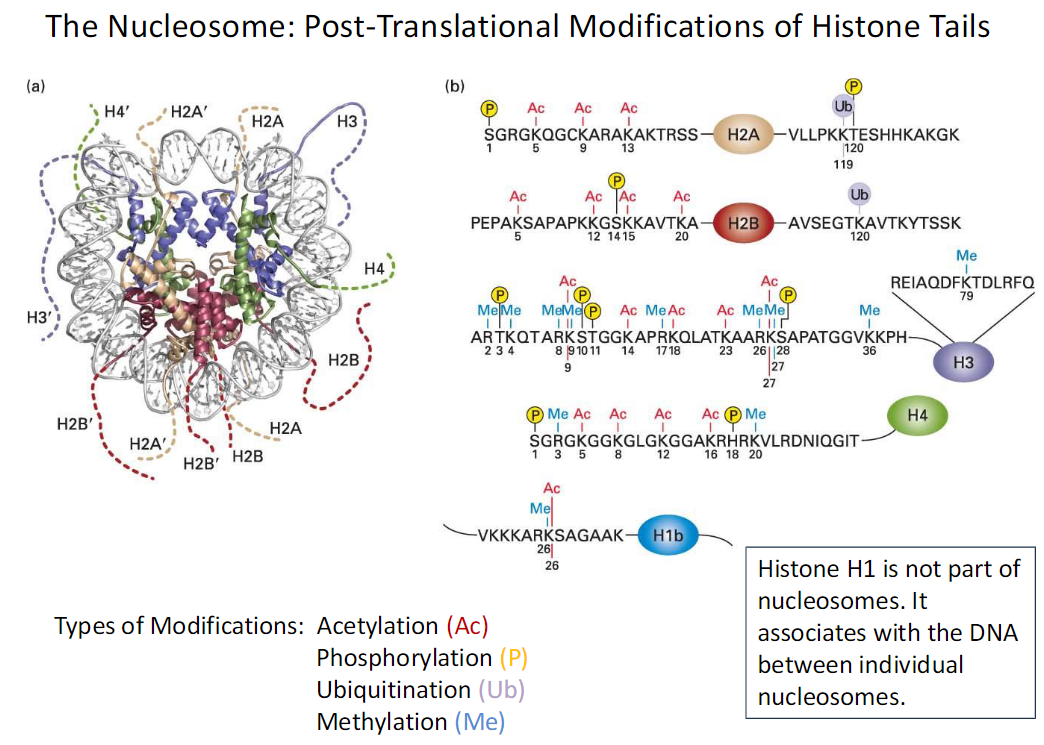
The Nucleosome: Histone Tail Modifications
Types of Modifications
Acetylation Ac
Phosphorylation P
Ubiquitination Ub
Methylation Me
Histone H1
Not part of the nucleosome
Associates with DNA between individual nucleosomes
Histone Codes
Diversity
There are thousands of possible histone codes
Different combinations of PTMs on histone tails create diverse regulatory signals