2. Protein Targeting - Endoplasmic Reticulum
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
68 Terms
secretion to the ___ or ___ is the default situation for lipids and proteins synthesized in the ER
plasma membrane or outside world
proteins destined for the cytoplasm have no ___
targeting sequences- this is the default pathway
larger proteins destined for the nucleus have an ___
NLS and are taken up fully folded by nuclear pores (gated transport)
proteins targeted for the ER also have ___
signal sequences and are translocated across the ER membrane into a topologically distinct compartment (transmembrane transport)
ER functions (4)
site of protein synthesis for the endomembrane system
site of lipid synthesis
Ca++ storage
enzyme storage in some cell types (detoxifying enzymes in liver cells)
ER functions: site of protein synthesis for the endomembrane system
the secretory and endocytic pathways
ER, Golgi, lysosomes, endosomes, secretory vesicles
transmembrane and GPI-linked proteins
soluble proteins in the lumen of the endomembrane system
the ER is an ___, ___ structure
extensive, dynamic
the ER is a network (reticulum) of membrane tubules and sheets that stretches ___
from the nucleus to the outer periphery of the cell
the ER is constantly ___
changing shape, extending and retracting
during cell division, the entire ER network ___
breaks down into vesicles that partition into daughter cells
how many types of ER membranes can be seen in the electron microscope
2 types!! rough and smooth
appearance of rough ER under EM
studded with ribosomes, tends to be near the nucleus but is still extensive
appearance of smooth ER under EM
does not have ribosomes and tends to be in the more peripheral regions of the cell
RER ribosomes are bound to the membrane because of ___
nascent chains (new proteins) they are making
the proteins are being cotranslationally inserted into the membrane
what does a ribosome do again
translate mRNAs to make proteins
the two types of ER membranes can be separated by ___
gradient centrifugation
gradient centrifugation of the ER membranes
break cell mechanically
microsomes form; vesicles containing the membrane components that the ER had
Rough microsomes - dense due to ribosomes
Smooth microsomes - lighter due to lack of ribosomes (duh)
what is a polyribosome
A polyribosome, or polysome, is a complex of multiple ribosomes translating a single mRNA strand simultaneously
Free and Bound Polyribosomes
protein translation is often ___
mRNAs are often ___
once a ribosome has moved away from the start point on the mRNA, ___
many ribosomes can attach to a single mRNA in this way - it is now a ___
if an ER targeting signal is present in the protein, the ribosome and ___
free and bound polysomes appear identical except for ___
RER is efficient because the mRNA tends to remain near the ___
slow
long
another ribosome can attach
polyribosome or polysome
most cellular proteins are made on polysome
nascent chain dock to the membrane - Bound polysome
localization and the peptide being made
the membrane where there are abundant ribosomes being recycled
A signal sequence directs proteins to the ER
the ER signal sequence: __
N-terminal signal sequence for ___ membrane proteins
internal sequences for ___ membrane proteins
signal sequence is __ for ER membrane targeting
leads to ___ across ER membrane
N-terminal start transfer sequences are usually ___
the protein folds in the ___, with the help of ___
protein is now either ___ or ___
a stretch of hydrophobic amino acids
soluble and some membrane proteins
many membrane proteins
necessary and sufficient
co-translational translocation
cleaved by signal peptidase, so the sequence is not found in the mature protein
lumen, chaperones and other molecules
soluble in the lumen or associated with the ER membrane
proteins destined for the ER are transported by a ___ mechanism
cotranslational transmembrane
how is the ER targeting signal recognized?
signal sequence emerges from ribosome
associates with the Signal Recognition Particle (SRP)
the ___ recognizes the signal sequence
signal recognition particle (SRP)
SRP recognizes the ___
SRP is found ___ (where?)
discovered as a factor that allowed ___
SRP is a ___ particle; RNP = ___ + ___
binds to both the ___ and ___
binding of the SRP to the nascent peptide chain halts ___ when the peptide chain is ___ long
signal sequence
free in the cytoplasm
attachment of pure ribosomes to ER membranes
ribonucleoproteinh; RNA + 6 proteins
signal sequence and ribosome
translation; 70-100 amino acids long (long enough to protrude from the ribosome)
cotranslational translocation of proteins into the ER (no shot youre remembering this)
signal sequence emerges from ribosomes
associates with the SRP
SRP binds to signal sequence and ribosome
causes translation to pause
SRP associates with SRP-receptor in membrane and the ribosome docks onto translocon channel
this tight dock is what protects proteins from protease in our assays
translocon pore through the membrane opens
normally closed otherwise small particles would leak out and cytoplasm/ER compartments would be in equilibrium
signal sequence and adjacent nascent chains inserted pore
both SRP and SRP-receptor release from ribosome and translocon
translation resumes through the membrane pore and protein enterss lumen
SRP/SRP-receptor binding, ribosome docking, and SRP/SRP receptor release is somehow regulated by GTP binding/hydrolysis by BOTH SRP and SRP-receptor
N-terminal sequence is cleaved by signal peptidase
protein synthesis is completed with ribosome still docked
translocation channel closes and ribosome unlocks
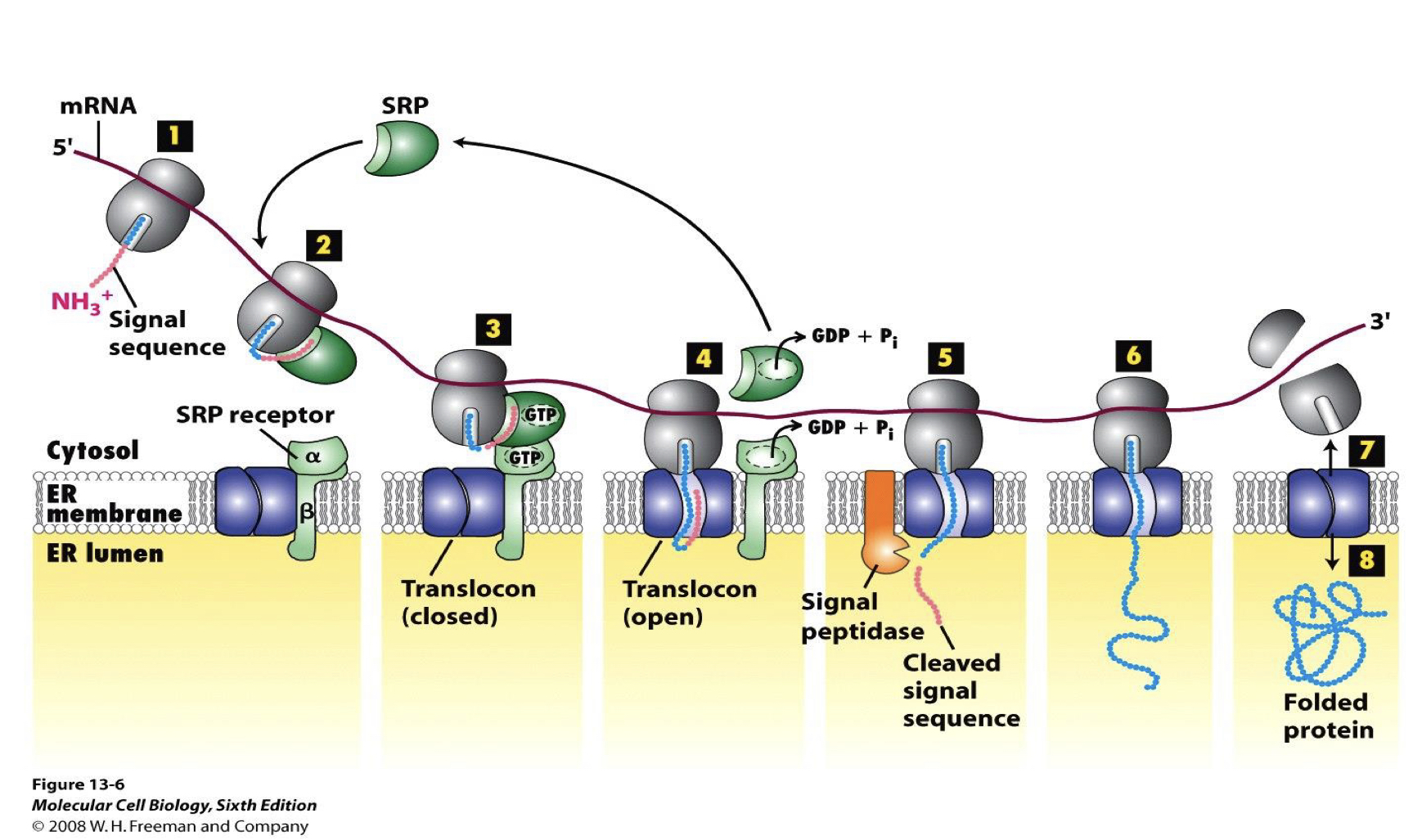
how do you demonstrate co-translational transport?
use microsomes and protease protection experiments to ask whether proteins are ever exposed
demonstrating co-translational transport using microsomes and protease protection experiments
you can mix mRNA encoding an ER-targeted protein with ____ and synthesize a protein (__)
if you added a protease to the tube during or after the synthesis, you find that your protein ___
if you repeat the experiment using microsomes, you find that your protein ___
if you add detergent during or after synthesis, ___
Conclusion:
the proteins are ___ during their synthesis - transport into the lumen of the membranes occurs ___ synthesis, aka: ___
ribosomes, ATP, tRNAs, and AAs (in vitro)
would be degraded (run a gel to determine size)
is not degraded, even if the protease is present during synthesis
in the presence of protease, the proteins are of course degraded
Conclusion:
protected; during; cotranslation translocation
a) mRNA + ribo + (ATP, tRNA, AAs etc) =
b) mRNA + ribo + (etc) + SRP =
c) mRNA + ribo + (etc) + SRP + SRP receptor =
d) mRNA + ribo + (etc) + sMicro + SRP + SRP receptor =
e) mRNA + ribo + (etc) + sMicro =
Full Protein, Protease Sensitive
a) +, +
b) 70-100 AAs, -*
c) +, +
d) +**, -
e) +,+
*70-100 amino acids protected by SRP and the ribosome itself
**slightly shorter due to cleaving og signal sequence by peptidase in sMicro membrane
Recap: sequential reactions and cotranslational translocation into the ER
Translation begins, exposing ___
Free SRP in cytoplasm binds ___
As a results of binding, SRP causes ___
SRP now binds ___ on the cytoplasmic side of the ER membrane, ribosome docks onto ___
__ and __ inserted into translocon
SRP/SRP-receptor dissociate and ___, ready to be used again
as a result of SRP release, translation ___; with signal peptide bound in translocon, ___ translocates alongside
_____
signal peptide
signal and ribosome
translation to stop
SRP-receptor; translocon
are released from ribosome
now resumes; polypeptide chain
N-terminal signal peptide cleaved
the N-terminal signal sequence is cleaved off by ___
signal peptidase
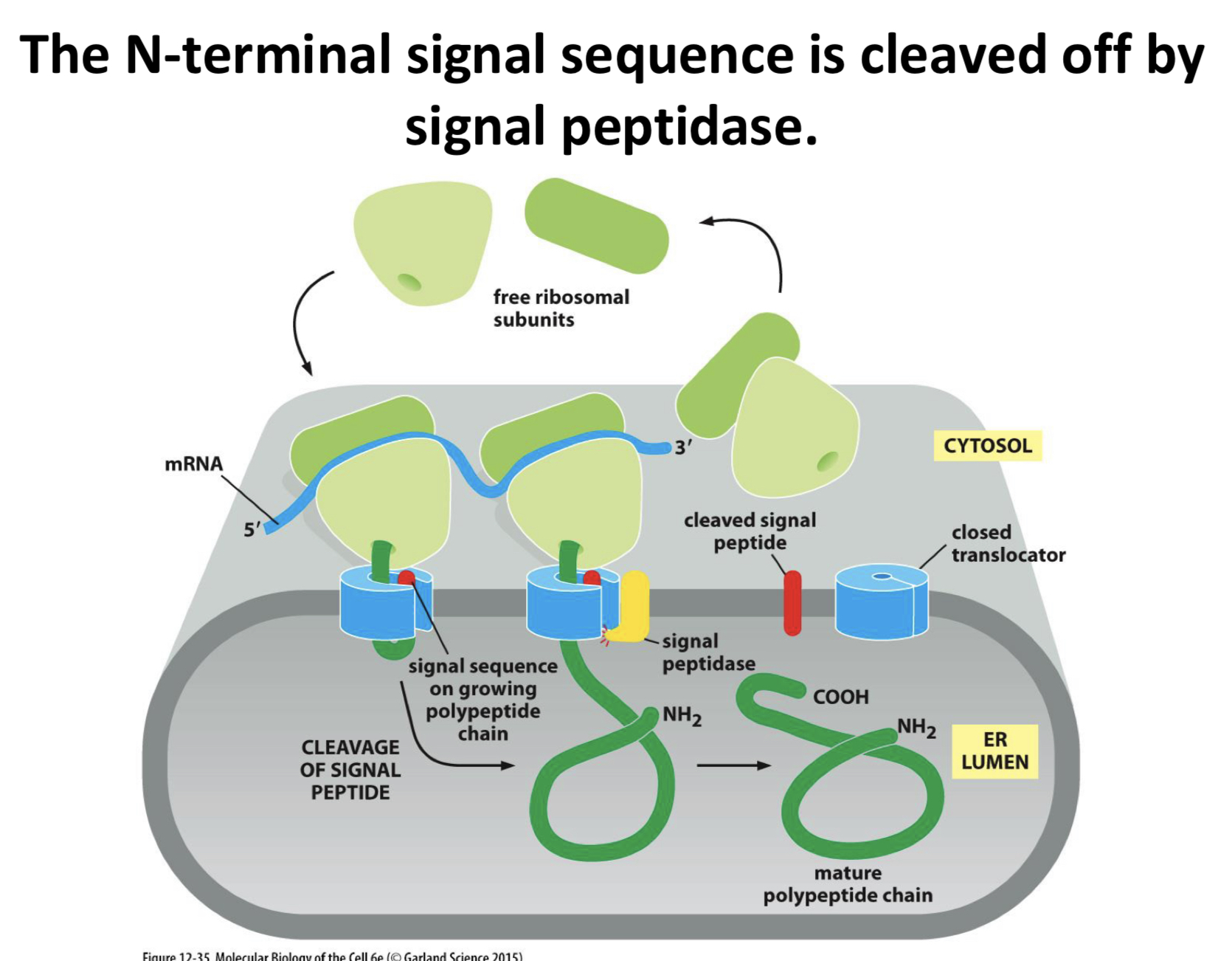
what is signal peptidase
a protease that recognizes a specific sequence and is embedded in the ER membrane and associated with the translocon complex
the cleaving of the N-terminal signal sequences causes what
releases soluble proteins into the lumen of the ER
proteins made in this way can eventually be secreted by the cell, or if they have appropriate additional signals, reside in the ER, Golgi, lysosomes, etc
if the signal sequences was not removed, what happens
the protein would stay stuck in the membrane, held by the signal sequence
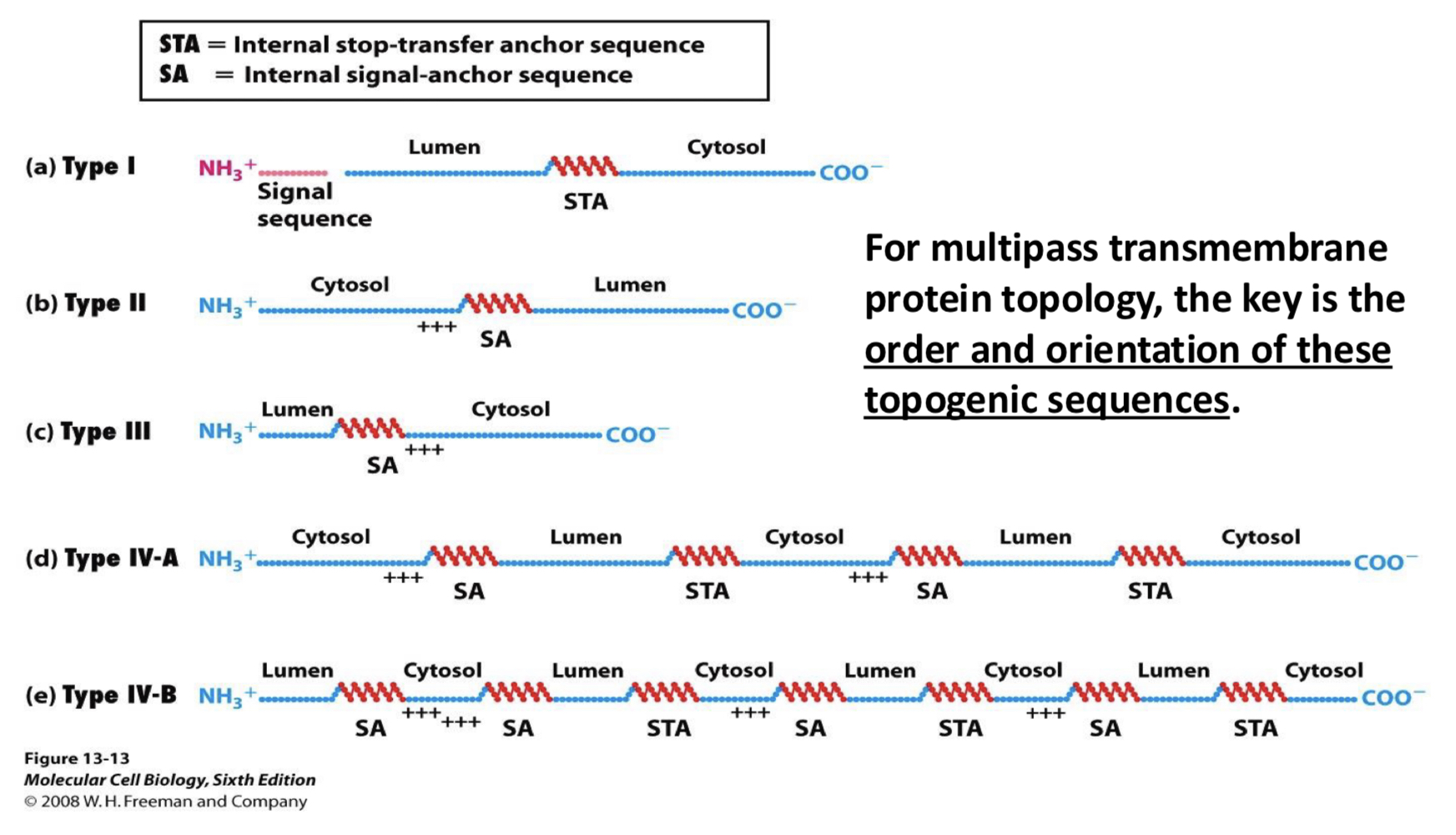
what are the different types of transmembrane proteins
Type I: single pass; N-terminal inside cell, C-terminal outside (signal sequence cleaved)
Type II: single pass; C-terminal inside cell, N-terminal outside
Type III: single pass; N-terminal inside cell, C-terminal outside
Type IV: multipass; N-terminal inside cell, C-terminal outside
GPI-linked proteins
one way to make single pass transmembrane proteins: ___ (Type I)
encode a stope transfer sequence
describe a stop transfer sequence
hydrophobic stretch of amino acids that is both a signal and the transmembrane domain
Stop-transfer Sequence and Type I Single Pass Protein
N-terminal sequence directs peptide to membrane
transfer begins
stop transfer sequence is made; ___ stops
hydrophobic stop transfer sequence moves laterally out of translocon and ___
protein continues to be made ___ (where?)
___/___ is removed
finished protein has N-terminal domain in ___ and C-terminal domain in ___
transfer to lumen
becomes anchored in the membrane
on the outside; “squeezes out” between ribosome and translocon seal
N-terminal signal/start transfer
in ER lumen; in cytoplasm
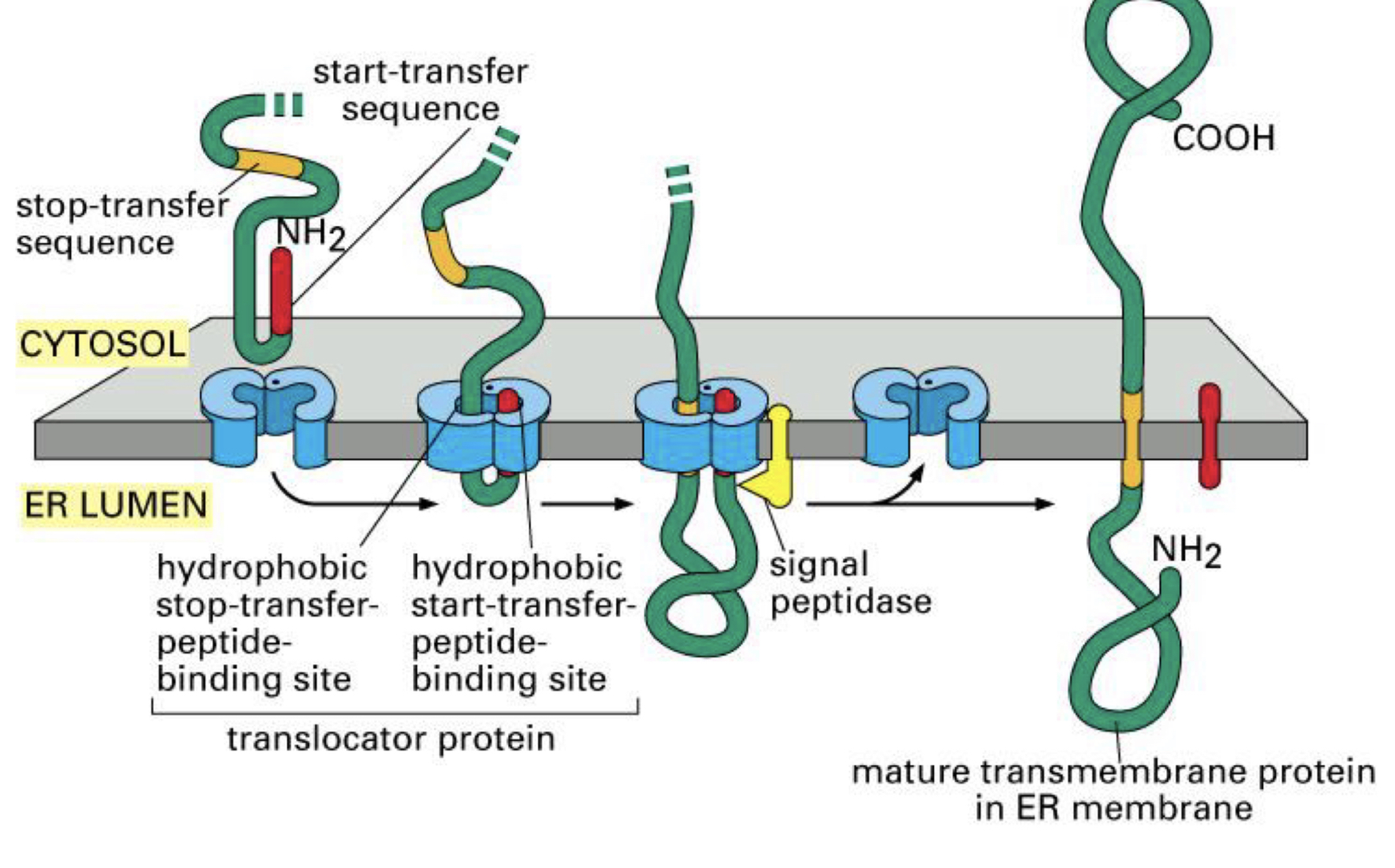
what would happen if you removed the stop transfer sequence from a Type I transmembrane protein?
it would go into the lumen and act more like a secreted protein
what would happen if you added a stop transfer sequence to a soluble ER protien?
it would act like a Type I transmembrane protein
another way to make a single pass transmembrane protein (Type 2 & 3): ___
the internal start transfer sequence
Start-transfer Sequence and Type 2 & 3 Single Pass Protein
no ___
an internal start transfer sequence aka. signal anchor sequence
sequence is both the ___ and ___
association with the ___ is delayed as the signal is further away from the N-terminus of the nascent chain
sometimes the___ ends up inside the lumen and sometimes in the cytoplasm; topology depends on ___
these signals are not ____, they are ___
N-terminal signal sequence
signal and the transmembrane anchor
SRP and ER membrane
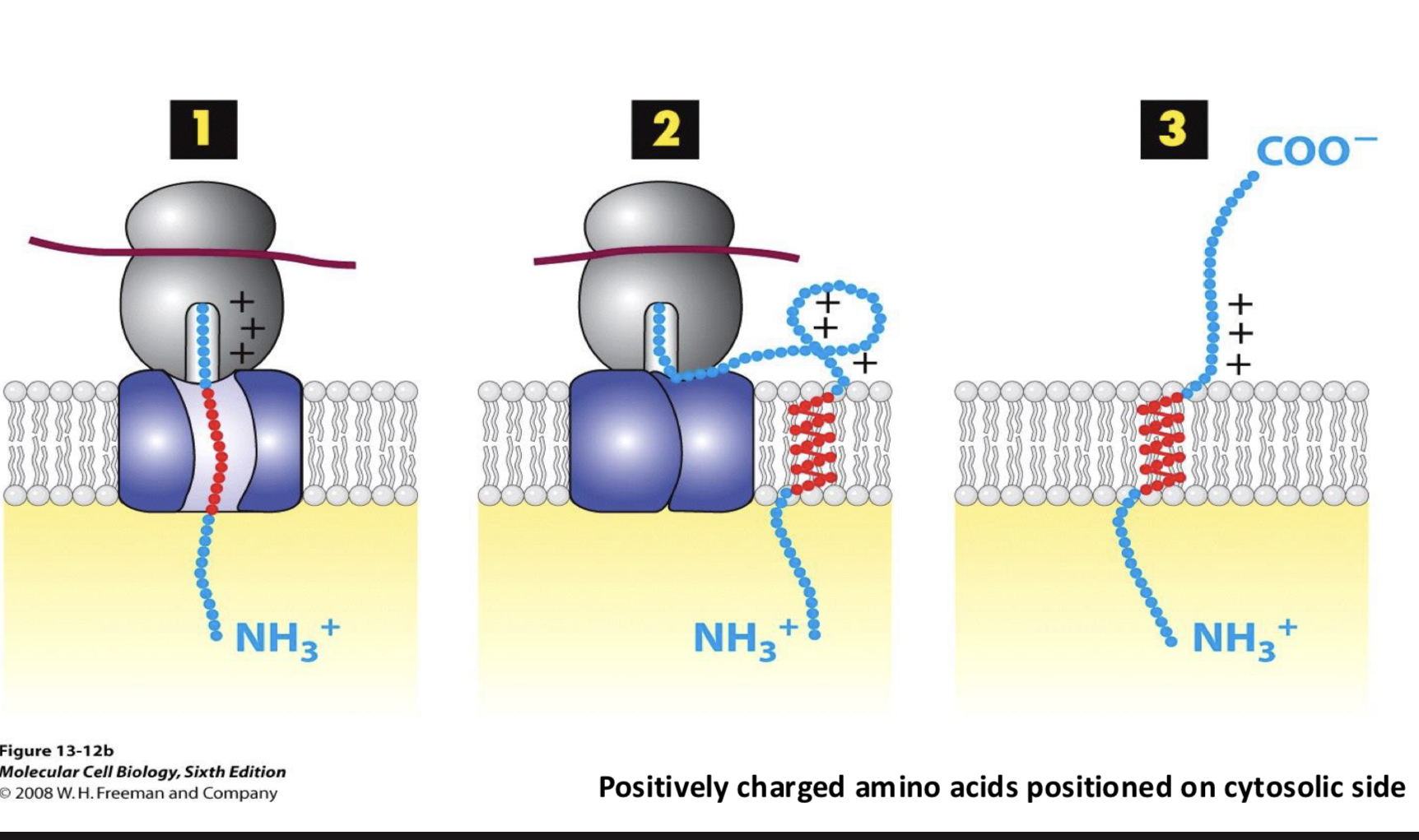
N-terminus; the orientation of a stretch of positively charged amino acids near the TM domain.
these + amino acids will be positioned on the cytoplasmic side of the membrane by the translocon
cleaved off, TM domains
Type II transmembrane protein has ___
positively charged amino acids residues at the N-terminal end of its signal sequence
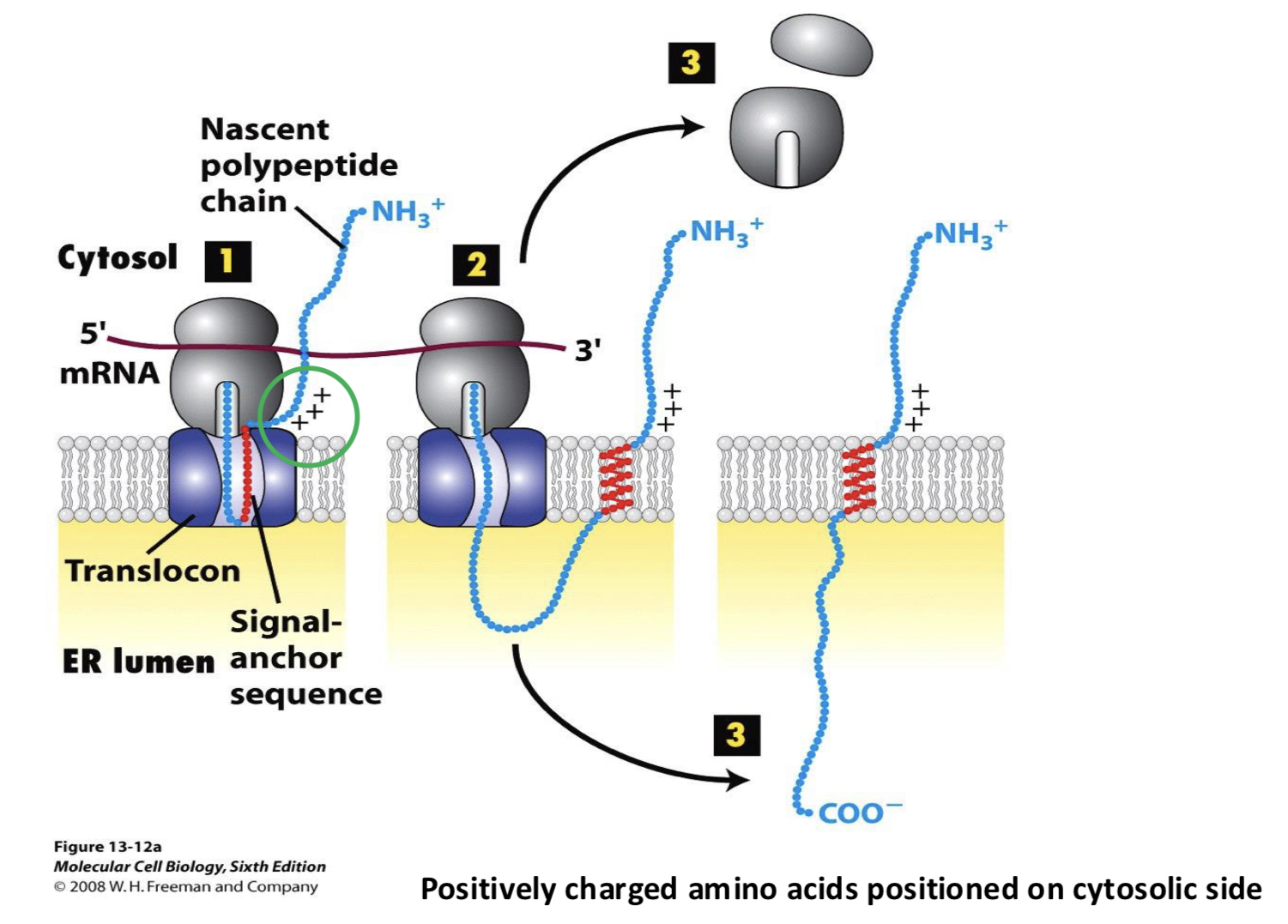
Type III transmembrane protein has ___
positively charged amino acid residues at the C-terminal end of its signal anchor sequence
Hydrophobic alpha helices as signals for protein transfer
N-terminal signal sequences, internal start transfer (signal anchor) sequences, and interna. stop sequences are all ___
N-term signal sequence will ___
internal signals are the ___
recognized by SRP, allowing ___
transmembrane domains are ___ while the protein ___
for multipass membrane protein topology (Type IV i think), the key is ___
hydrophobic (usually alpha helix)
be cleaved, others are not
transmembrane domains
ribosome docking and arrangement of signals in translocon
released laterally from the translocon into the ER membrane; continues to be made, either into the lumen or on the cytoplasmic side
order and orientation of these topogenic sequences
for multipass transmembrane protein transport (Type IV i think), the key is ___
the order and orientation of these topogenic sequences
in some eukaryotes such as yeast, some proteins are ___
translocated post-translationally
in post-translational translocation, how is the polypeptide chain “pulled” through the translocon and into the ER lumen
ADP clamps down on lumen side, random motion of the chain only allows it go inward
“thermal ratchet mechanism”
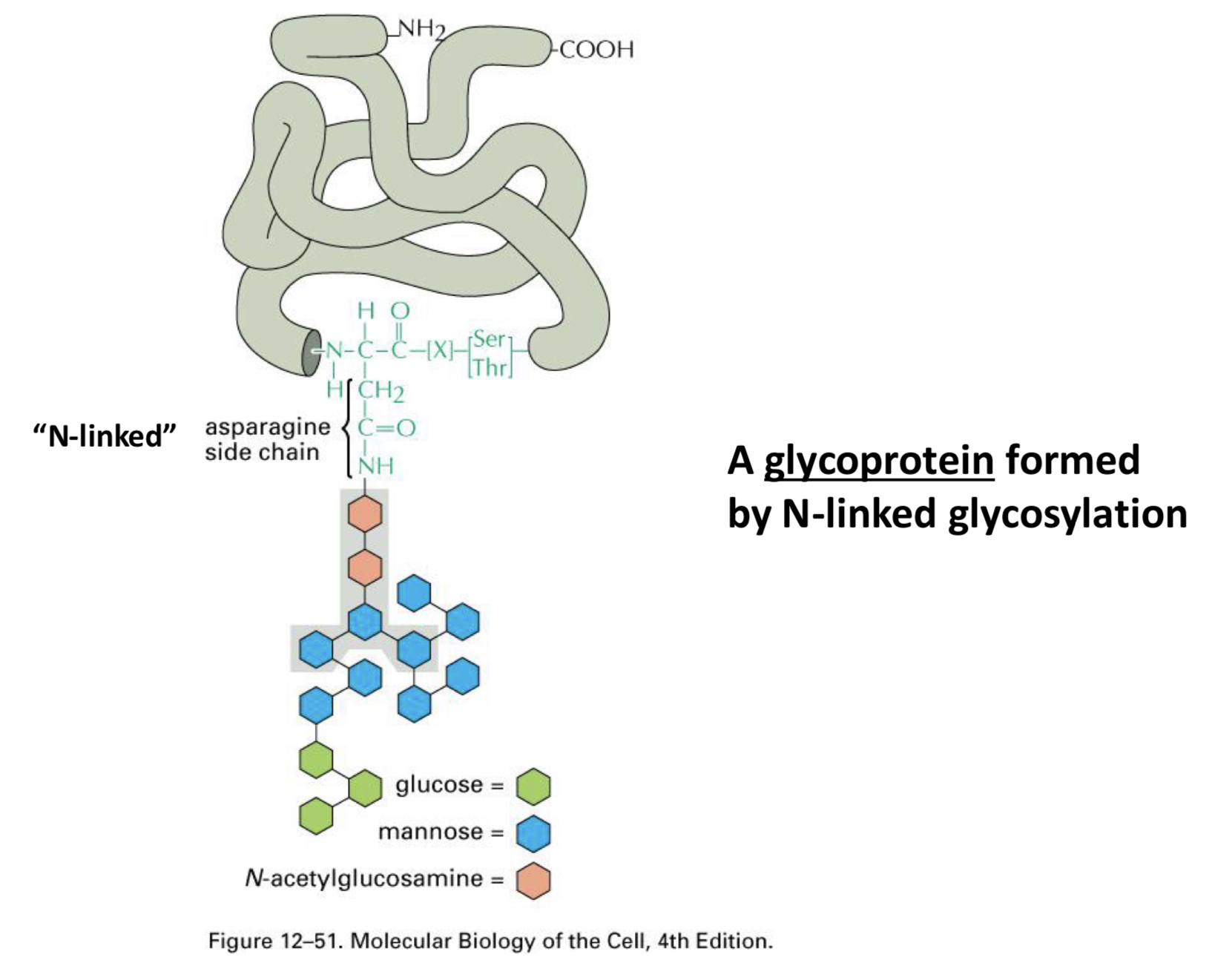
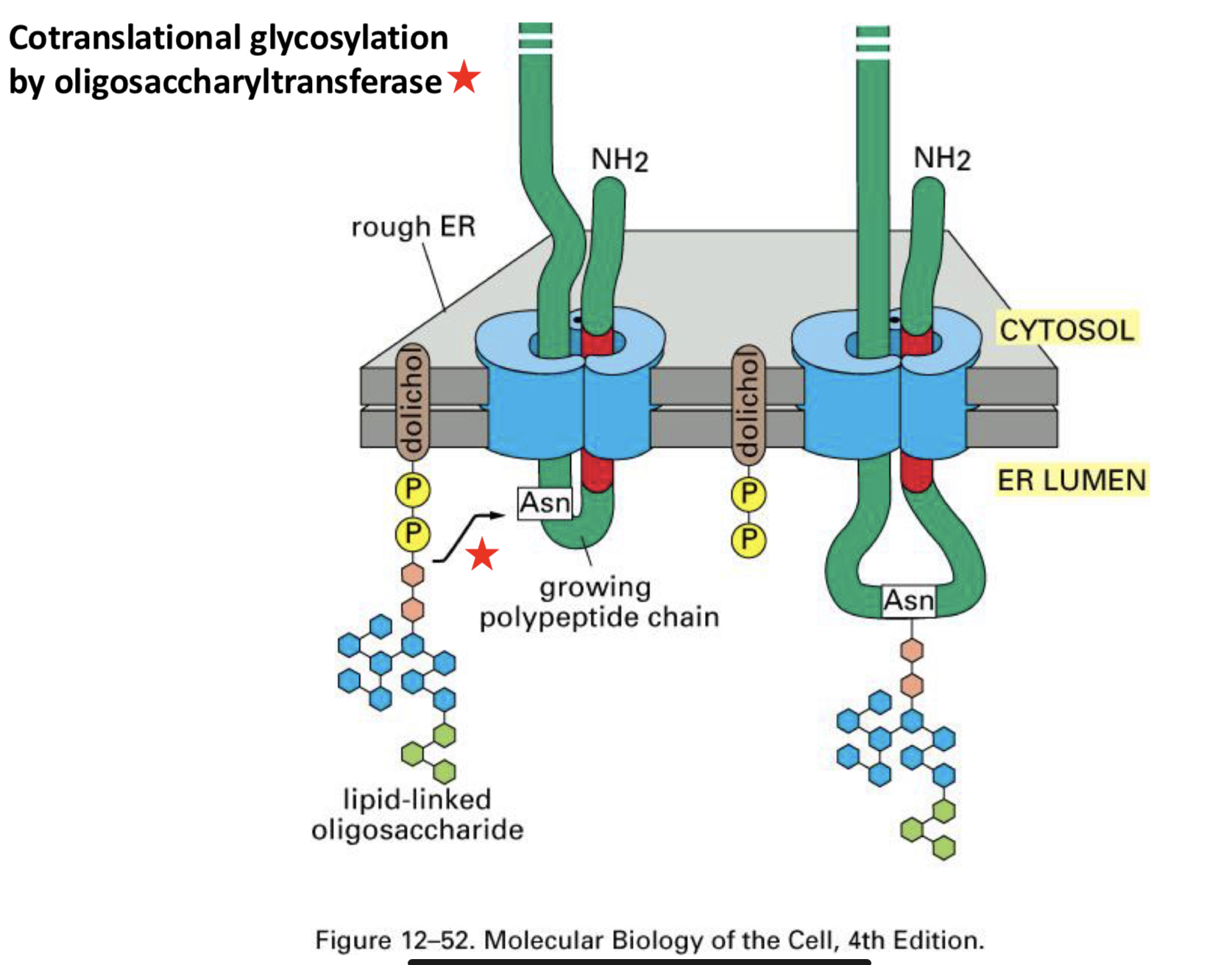
Glycosylation of Proteins
most proteins imported into the ER become ___
critical for quality control of ___ in the ER
further processing of these sugars in Golgi for ___
Oligosaccharyltransferase is an ___ associated with the ___
the sugars are added ___
N-linked Glycosylation; the signal for glycosylation is ___
the oligosaccharide is ___ in the ER and added ___
multiple oligosaccharides are often ___
this only happens in the ___
glycosylated
folding
proper protein function
integral membrane protein; translocon complex
cotranslationally during import
the amino acid sequence Asn-X-Ser and addition is to a nitrogen in Asn
preformed; as a block
added to a protein, each Asn-X-Ser will get its own oligosaccharide
lumen of the ER; there are no oligosaccharides of this sort on the cytoplasmic proteins
most proteins imported into the ER become ___
glycosylated
what is oligosaccharyltransferase
integral membrane protein associated with the translocon complex
a glycoprotein to look at :)
Source of Oligosaccharides
the oligosacc is built initially ___
partway through construction it is moved ___
construction is finished ___
the entire oligosacc is then transferred ___
in the cytoplasm on a lipid carrier (dolichol)
across the membrane by transport-like proteins to the ER lumen
on the ER lumen side
to the protein during translation (co-translocationally) by oligosaccharyltransferase
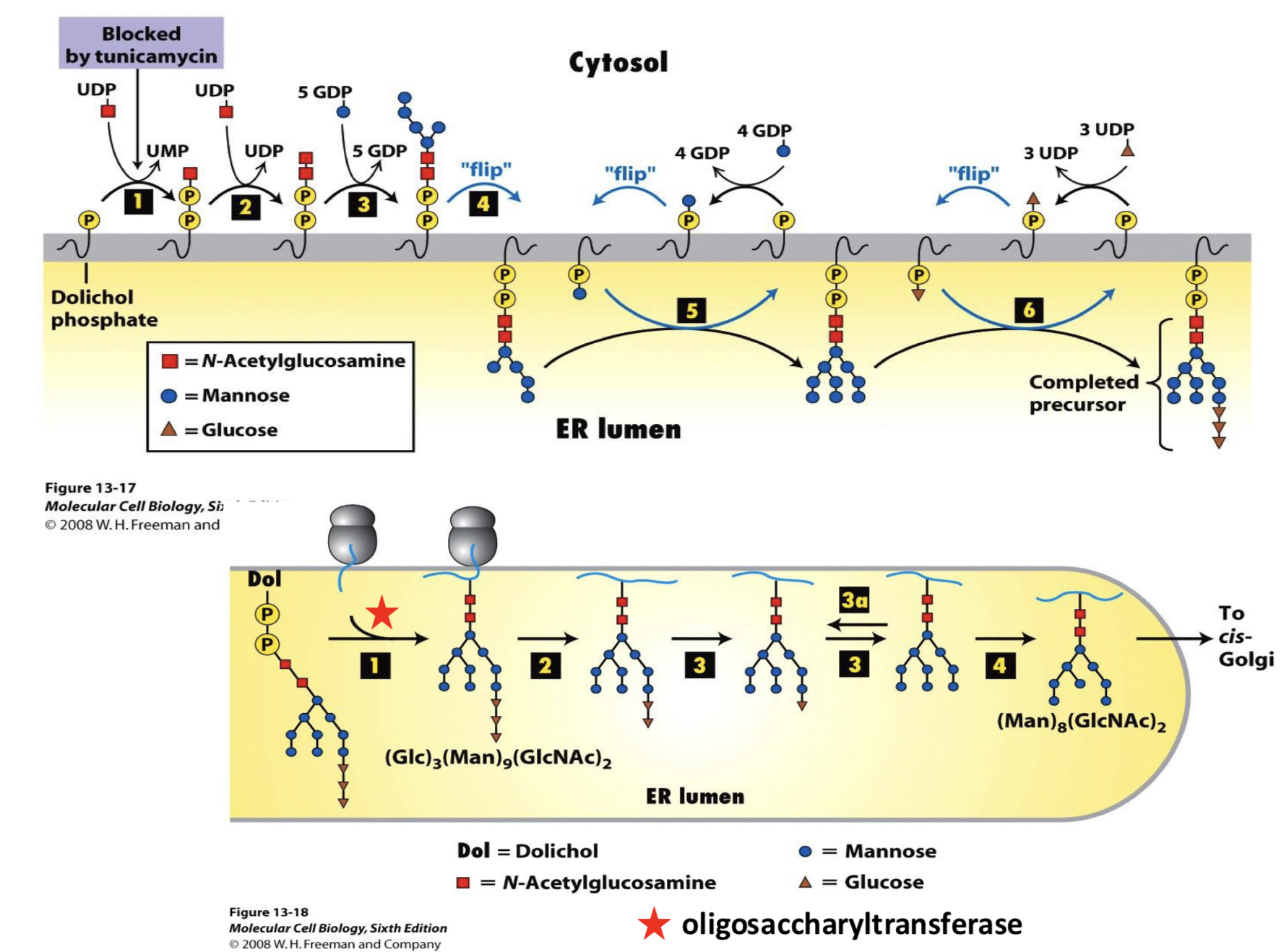
cotranslational glycosylation happens via ___
oligosaccharyltransferase
how does ADP “pull” a polypeptide chain through the translocon in post-translational translocation
thermal ratchet mechanism
GPI
glycosyl-phosphatidyl-inositol
some proteins are held in the membrane by ___
lipid anchors
Lipid anchored proteins
GPI anchoring is done in the ___
begins as TM protein; an enzyme cleaves the ___ and attaches it to the ___ at the same time
GPI-anchored membrane proteins face the ___ or ultimately the ___ if in plasma membrane
these proteins can be rapidly released from membranes by ___
ER
the protein; GPI anchor
lumen; extracellular space
cleaving the anchor
Protein folding in the ER
__ in the ER lumen aid in folding
__ - the family of proteins that either aid in protein folding or protect the cell from unfolding in response to heat stress (heat, chemical)
__ are constitutively expressed and bind unfolded or misfolded proteins in this compartment
Chaperone proteins
Heat shock proteins (Hsc, Hsp)
Similar ER localized chaperones such as BiP (Hsp70 family member)
what in the ER lumen helps aid in protein folding
chaperone proteins
Protein folding in the ER
Protein disulfide isomerase (PDI)
the lumens of ___ are harsher, more oxidizing environments that the cytoplasm
__ in oxidizing conditions can help to stabilize protein structure
proteins have evolved differently depending on where ___
PDI catalyzes ___
the “shuffling” of disulfide bonds by PDI gives proteins ___
endomembrane compartments, and the extracellular world
Disulfide bonds S-S that form between cysteines
they are going to fold!
a chance to be properly folded
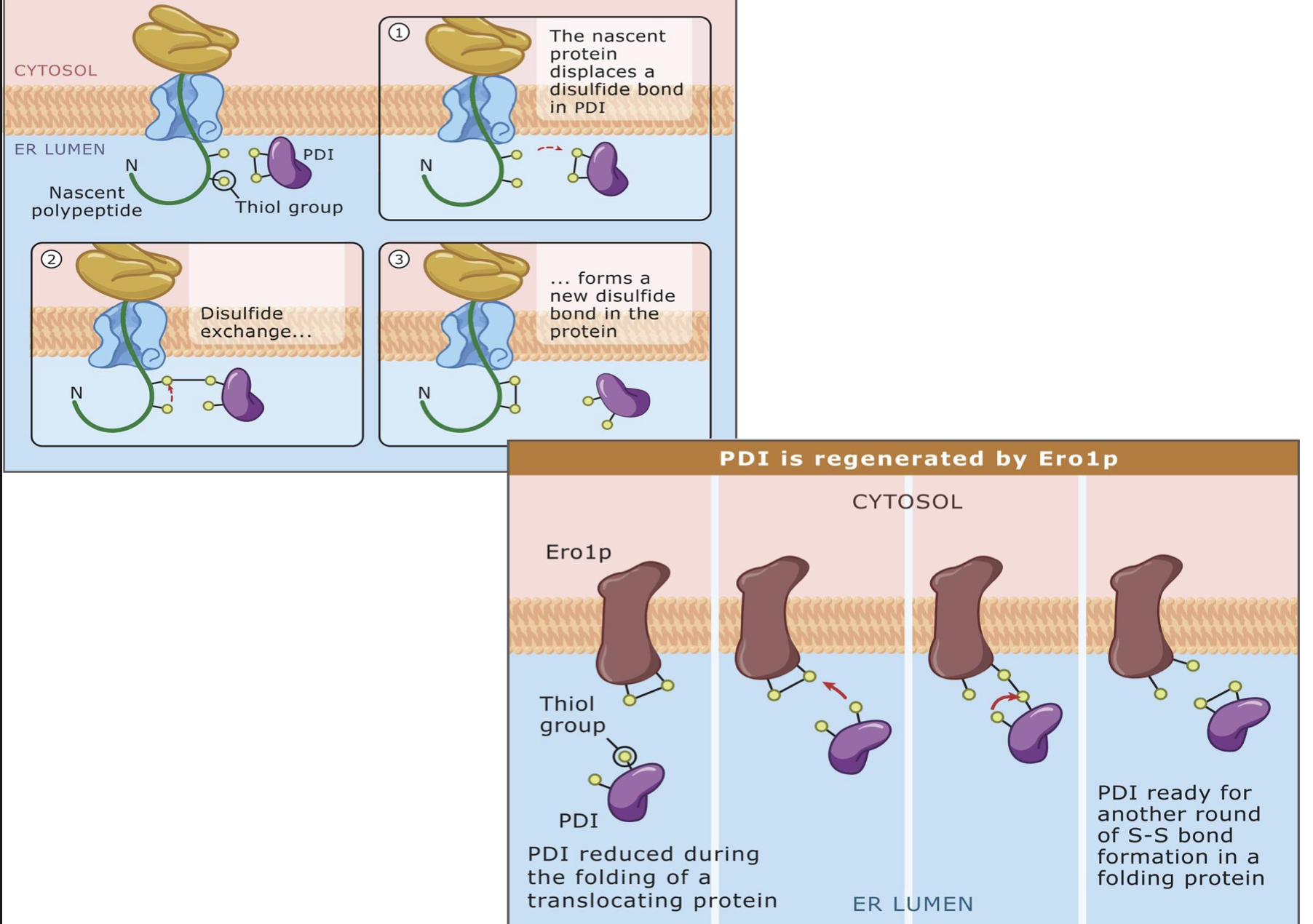
PDI
protein disulfide isomerase
proteins that fold incorrectly are recognized as ___
“WRONG”
ERAD
ER-associated protein degradation
(think of “eradicate”)
what does it mean for something to be polyubiquitinated
monomers of the small protein molecule ubiquitin are added
ER-associated protein degradation (ERAD)
misfolded proteins are ___, ___, and ___ back across the ER membrane to the cytoplasm
during ___ the proteins become polyubiquitinated; ___
the cytoplasmic proteasome recognizes ___ and ___
recognized, modified, “dislocated”
dislocation; monomers of the small protein molecule ubiquitous are added in a chain by an E3-ubiquitin ligase
polyubiquitin; degrades the protein
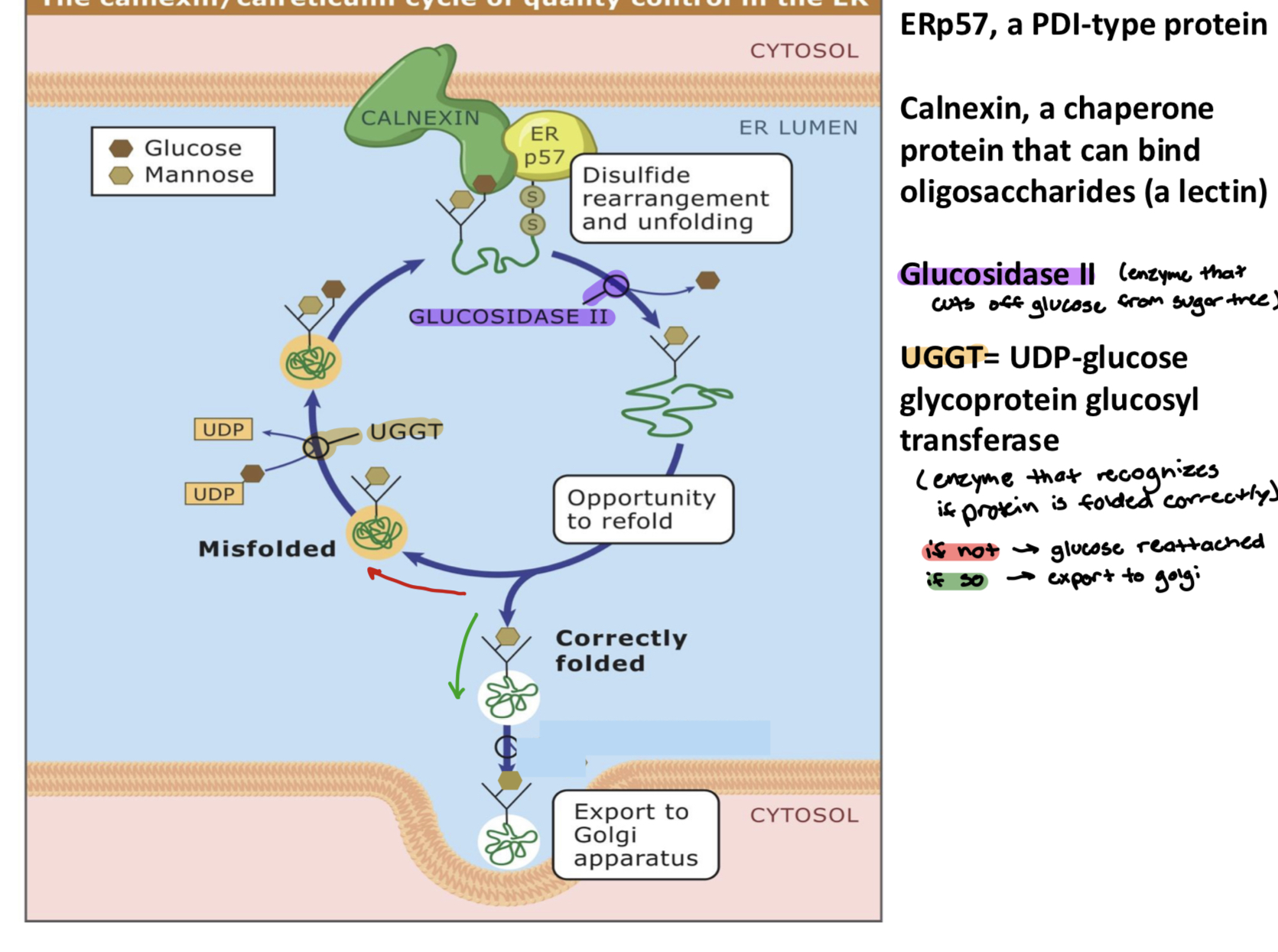
the calnexin/calcreticulin cycle of quality control in the ER
ERp57, a ___
Calnexin, a ___
Glucosidase II, an ___
UGGT, a ____
a PDI-type protein
a chaperon protein that can bind oligosaccharides (a lectin)
enzyme that recognizes if protein is folded correctly
if not: glucose reattached
if so: export to golgi
the accumulation of misfolded proteins in the ER triggers ____, including increased transcription of genes encoding ___
an unfolded protein response; chaperones and dislocation machinery