BCMB 412: Homologous Recombination
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
50 Terms
What is recombination?
A process by which pieces of DNA are broken (a spontaneous or induced DBS) and recombined to produce new combinations of alleles in the germ line.
What are three of the benefits of recombination between homologous DNA sequences?
Genetic variation: important evolutionarily (a lack of recombination means mutations would accumulate in a species lineage, which is bad for the species).
A mechanism for DNA repair
Can restart stalled replication forks
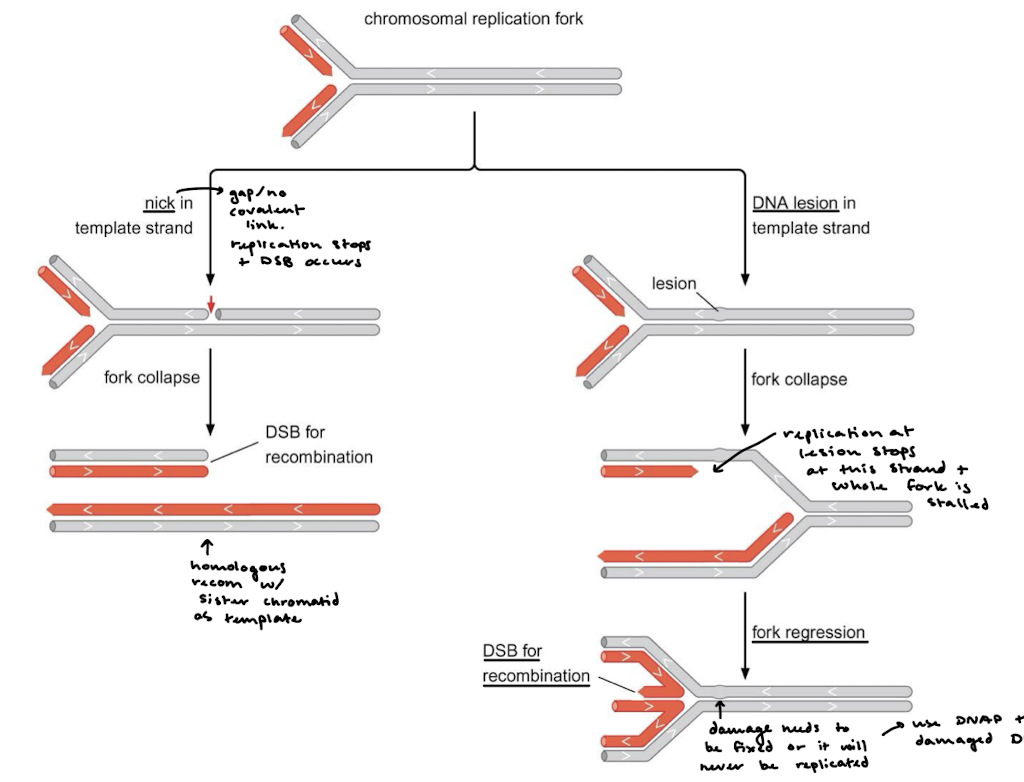
How do DSBs form from a nick in the template strand at a chromosomal replication fork? From a DNA lesion?
A nick in the template strand means there is a gap/no covalent link, so replication will stop, the fork will collapse and the DSB occurs for recombination with the sister chromatid. A DNA lesion will result in fork collapse, but replication stops only on the strand with the lesion, stalling replication. Fork regression occurs instead. The DNA damage still needs to be fixed or it will never be replicated (translesion DNA replication).
What are the five key steps in homologous recombination?
Alignment of two homologous DNA molecules.
Introduction of breaks in the DNA (or it is spontaneous)
Strand invasion
Holliday junction formation
Holliday junction resolution (resolution may lead to genetic recombination or it may not)
Do homologous chromosomes have the same sequences?
No, they both will have proper base pairing, but they will have different nucleotides since they are different alleles.
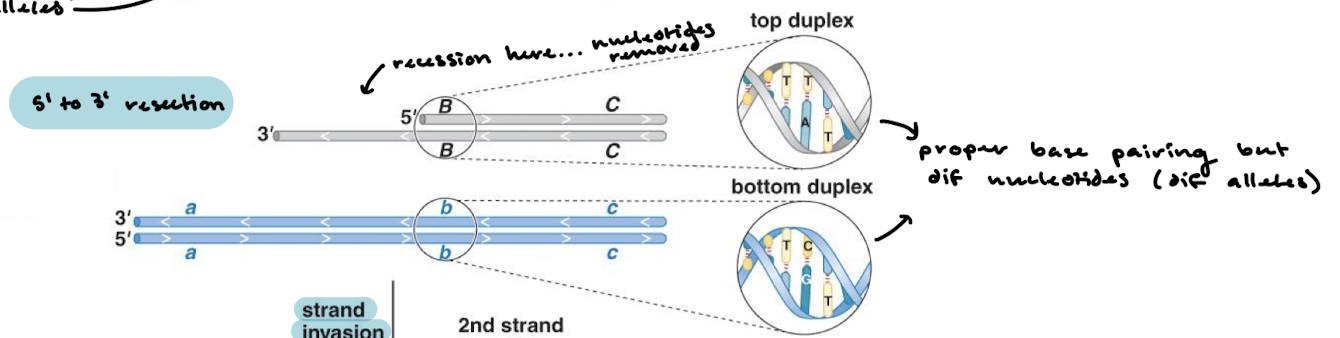
What happens in between the homologous chromosomes aligning and strand invasion?
5’ to 3’ resection of one of the duplexes
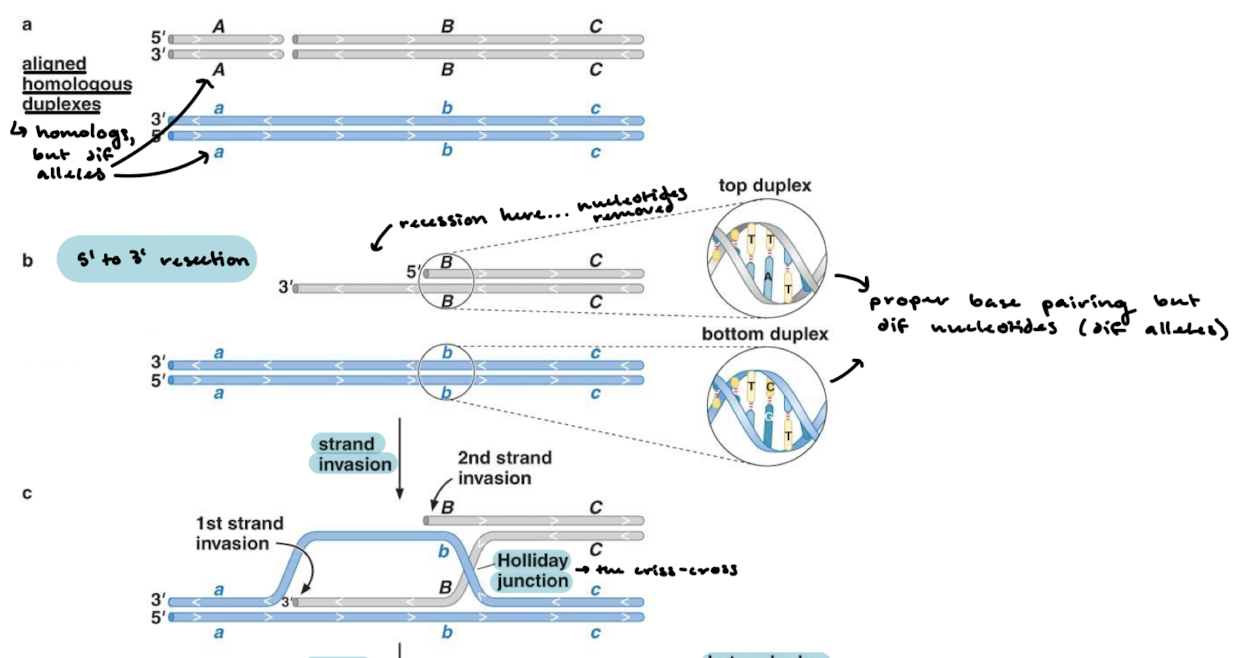
What is strand invasion? Where does the holliday junction form?
There are two strand invasions where the strands cross over. The criss cross is the Holliday junction.
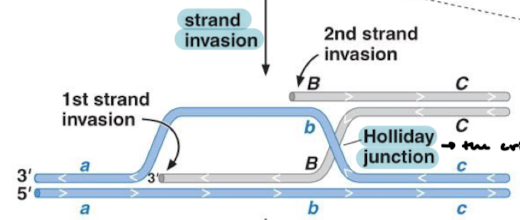
What happens immediately after formation of the holliday junction? What are top and bottom duplexes now called?
Branch migration in which the junction moves away from the original position. The duplexes are now heteroduplexes because they are hybrid duplexes.
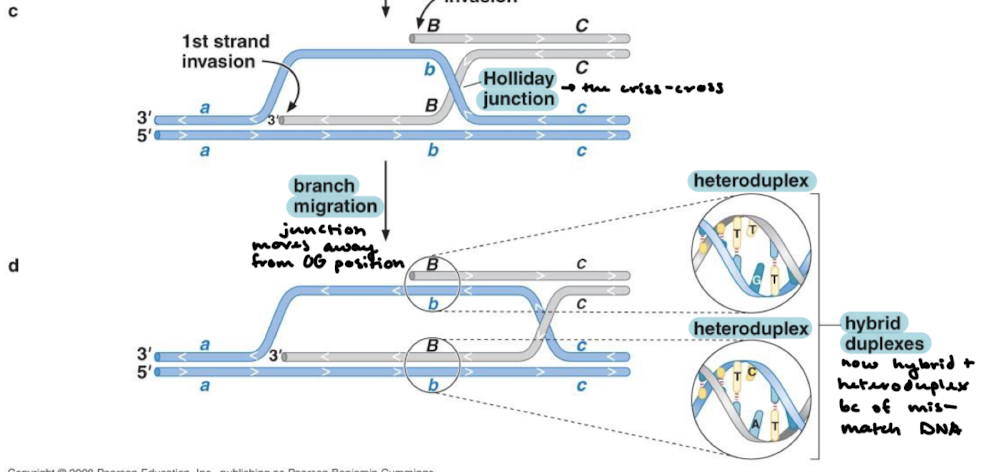
What are the steps for the resolution of Holliday junctions?
The junction is adjusted to a cross form where the strands are still crossed.
The strands are twisted so that they are no longer crossed.
DNA cleavage resulting in either crossover or noncrossover products (crossover resulting in reassortment and noncrossover resulting in no reassortment)
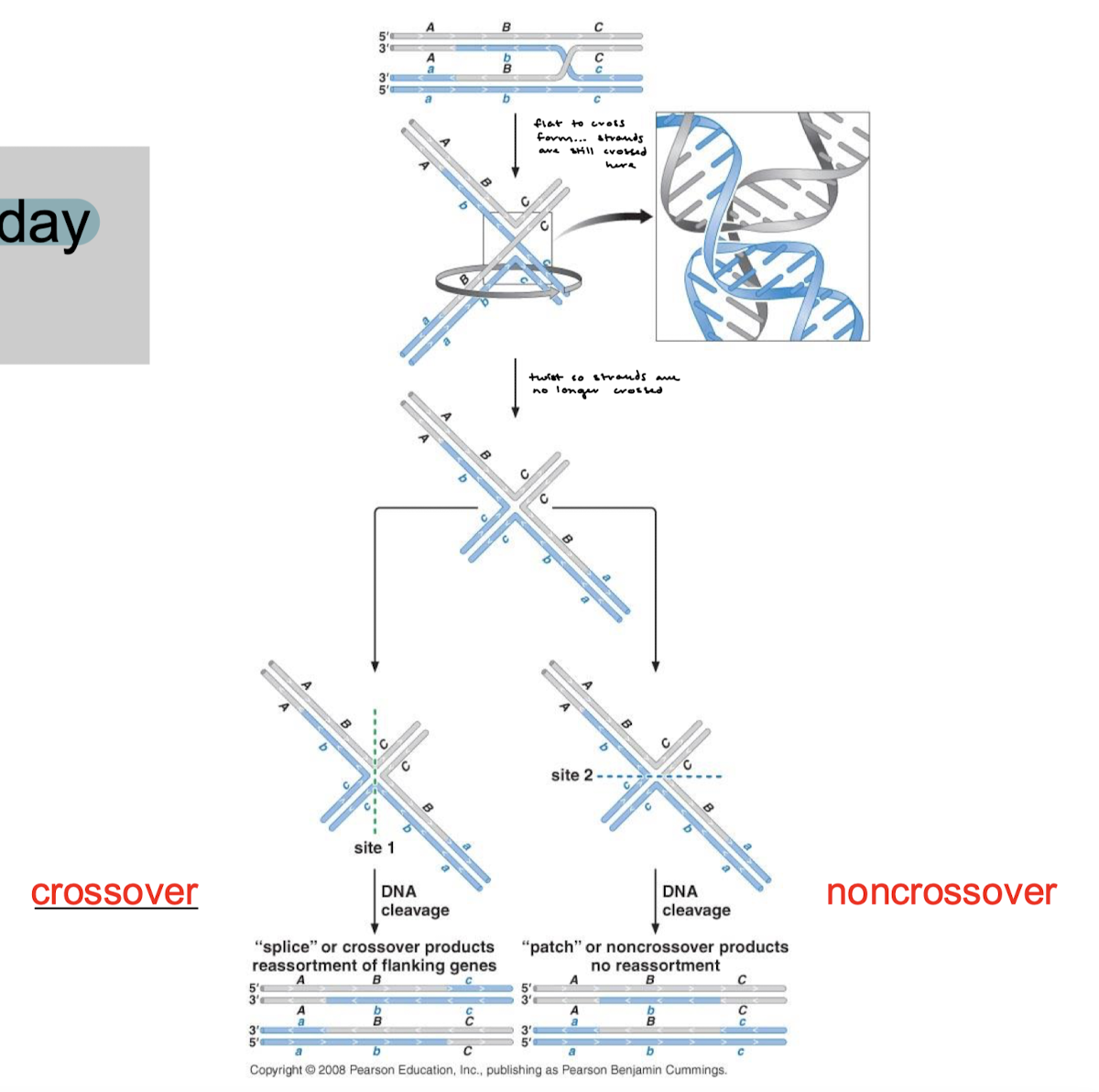
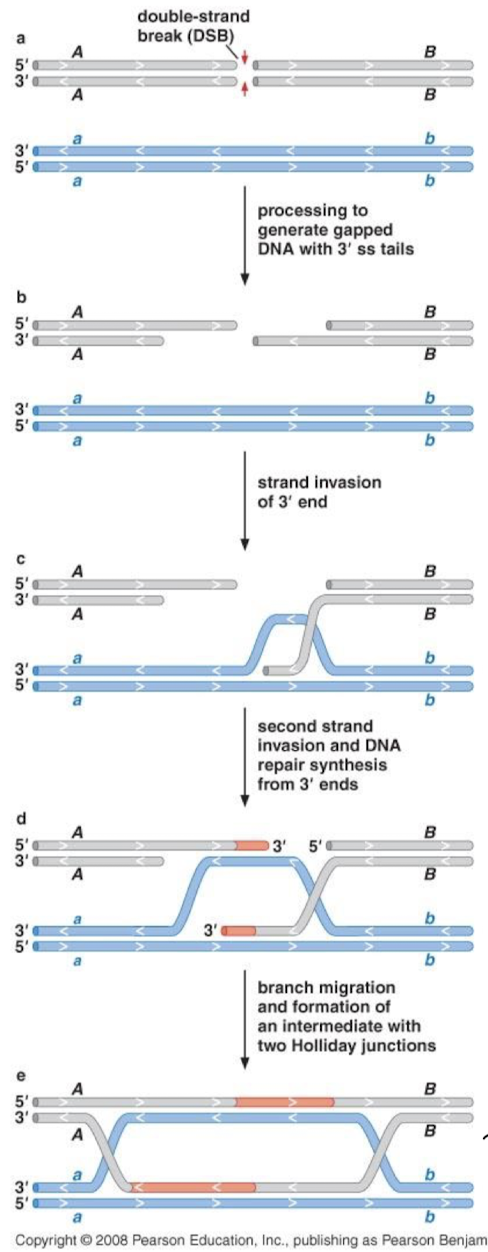
Explain the double-stranded break repair pathway (DSBR pathway).
5’ to 3’ resection, two-strand invasions, and DNA repair synthesis from 3’ ends using the homologous chromosome as a template. Branch migration and formation of an intermediate with TWO Holliday junctions at both ends that need to be resolved.
“gene conversion that is non-reciprocal” refers to what process?
DSBR pathway
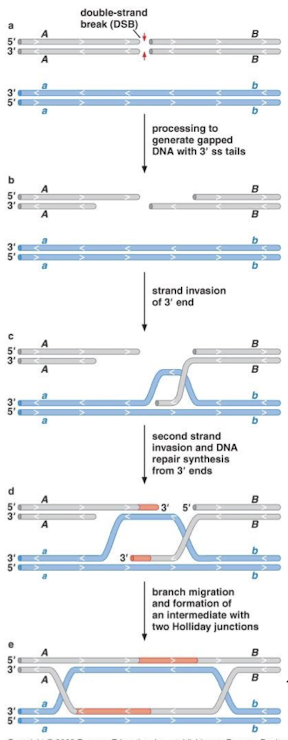
When resolving double Holliday junctions (dHJs), which cleavage mechanism results in the noncrossover (no recombination) products and which results in the crossover products (genetic recombination).
Noncrossover products result from resolution at the crossovers at both Holliday junctions. Crossover products result from cleavage vertically across all four strands at one of the junctions, then a cleavage just at the crossover for the other junction.
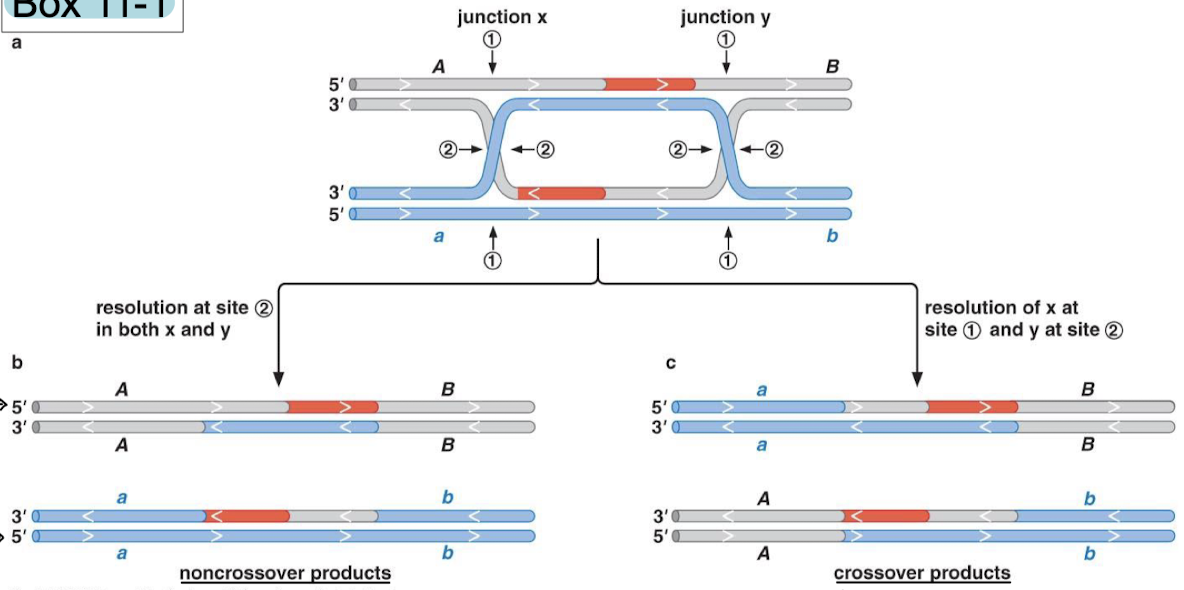
What are the activities of RecBCD? For what recombination step is it used? What organisms have RecBCD?
RecBCD is a helicase and nuclease. It processes DNA breaks to generate single strands for invasion (5’ to 3’ resection) in bacteria. It also processes DNA ends for recombination AND degradation.
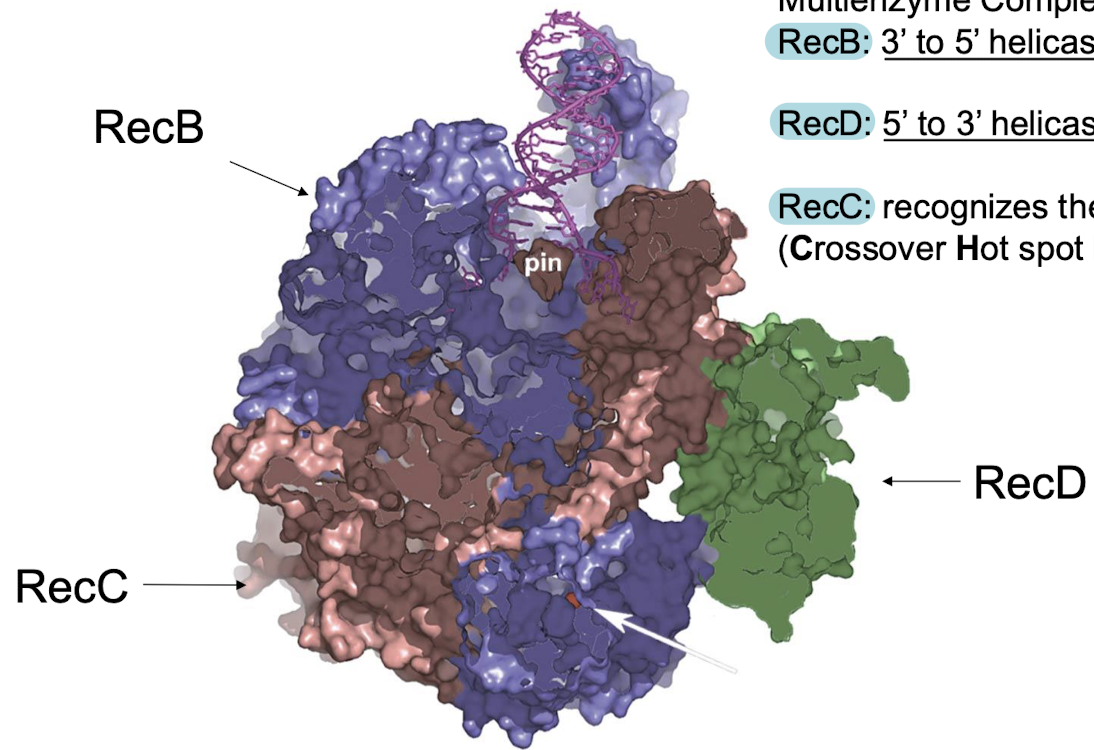
RecBCD-DNA complex is a multienzyme complex. Explain the functions of RecB, RecD, and RecC.
RecB: 3’ to 5’ helicase and nuclease
RecD: 5’ to 3’ helicase
RecC: recognizes the chi site (DNA sequence for facilitation of recombination, crossover hot spot instigator).
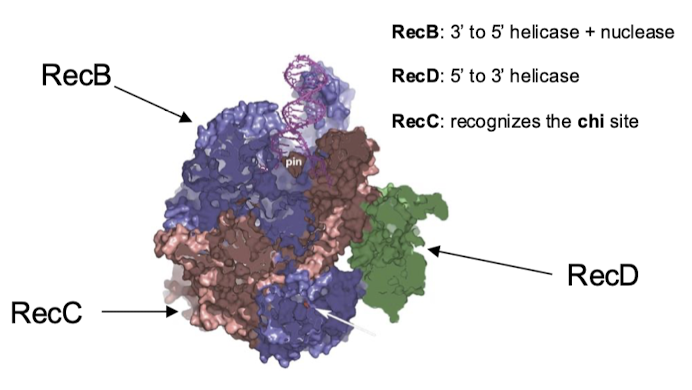
What enzyme in RecBCD recognizes the 5’ end? The 3’ end?
The complex recognizes the DBS…
RecD: 5’ recognition and helicase activity
RecB: 3’ recognition and helicase + nuclease activity
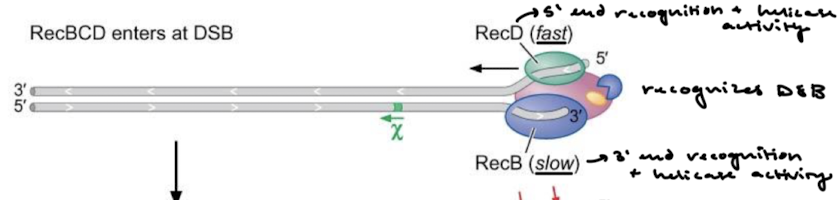
Is RecB or RecD the slower helicase? Why might that be?
RecD is fast while RecB is slow. Rec B lags compared to the other strand. It might have to do with the nuclease activity of RecB working to degrade the 3’ end and some of the 5’ end.
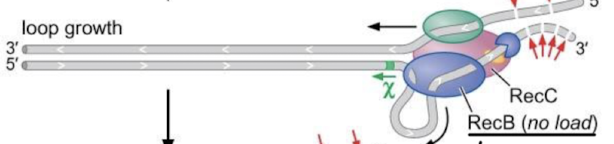
What causes RecBCD to pause?
As the RecC will recognize the Chi site and the complex will pause.
What happens after RecBCD has paused? What is the only issue with the end product?
Loop shortening, where the chi site is bound to RecC, but RecD and RecB keep moving slowly to shorten the loop. This creates a 3’ overhang that allows for strand invasion and recombination. The only issue is that the 5’ end is single-stranded and will be recognized as damaged and targeted for degradation. The 5’ end needs a mechanism for strand invasion.
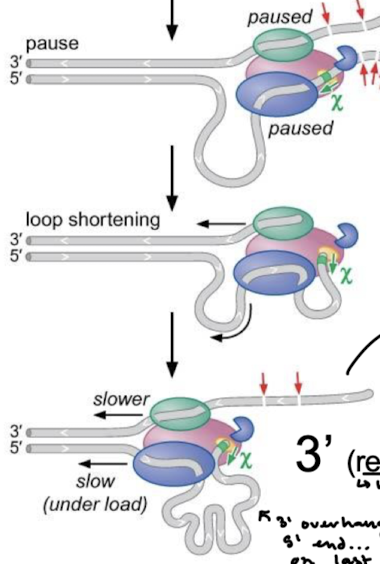
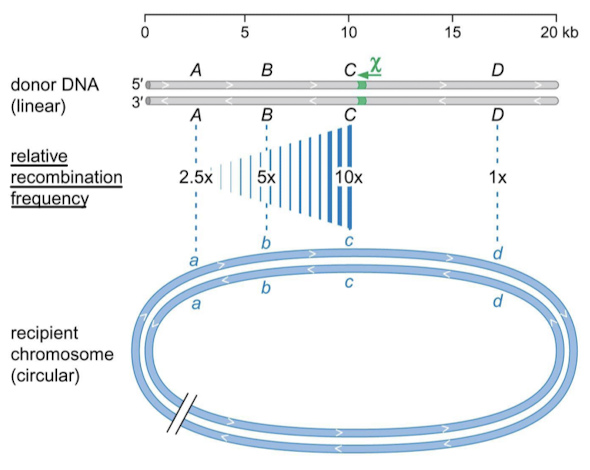
The closer sequences are to the chi site, the higher the probability of ________. In what organism are chi sites overrepresented?
recombination. Overrepresented in E. coli: 1009 (80 expected). If E. coli DNA enters an E. coli cell it will most likely recombine. If non-E. coli DNA enters and E. coli cell, it will most likely be degraded by RecBCD (RecBCD would not stop at the chi site).
What recombination step requires RecA? In what organisms?
Pairing homologous DNAs and strand invasion in E. coli.
What protein family is RecA a member of? What does RecA do?
RecA is a member of the family called strand-exchange proteins. They promote searches for sequence matches and generate base pairing between them (in other words, they catalyze the pairing of homologous DNA molecules).
Is RecA-induced strand exchange spontaneous or non-spontaneous?
spontaneous
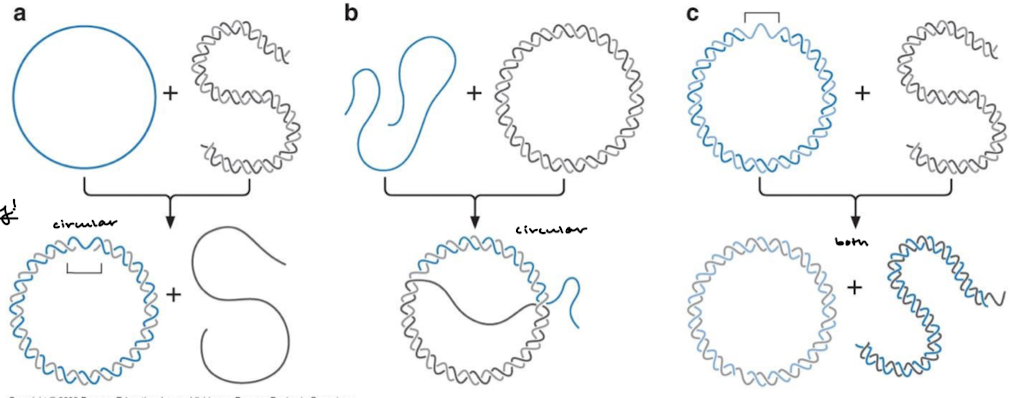
What is the active form of RecA? What is the size of these?
RecA filaments are variable in size (100 subunits to 300 nucleotides).
How many ssDNA strands are accommodated by RecA filaments?
1, 2, or 3, though 1 and 3 are most common in recombination intermediates.
Where does homolog search and DNA strand exchange occur?
Within the Rec A filament.
What happens to the length of the DNA by RecA filaments?
In RecA filaments, the DNA distance between two adjacent nucleotides is extended to 5 A instead of the 3.4 A normally found in B DNA. In a RecA filament, the length of a DNA molecule extends by 1.5 fold.
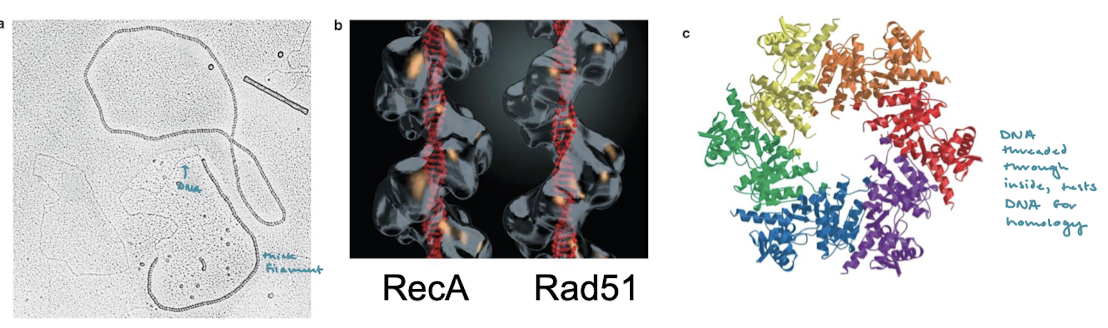
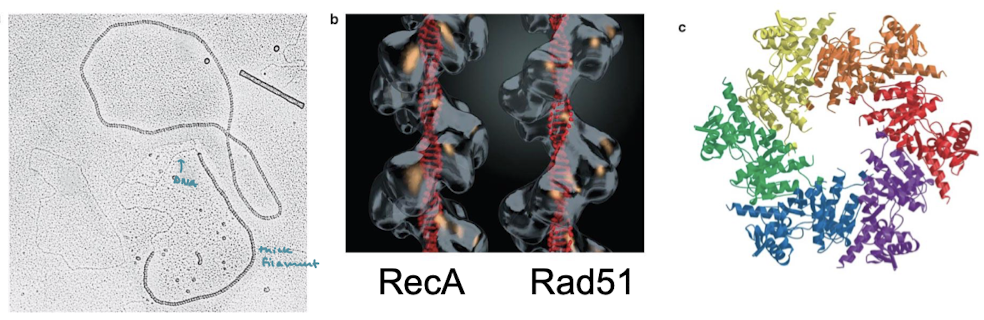
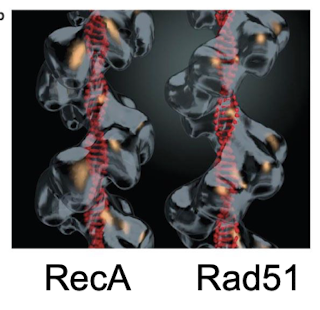
What is the eukaryotic equivalent of RecA? What forms of life have RecA-like filaments?
Rad51. RecA-like filaments are formed across all forms of life including humans, E. coli, and archaea.
What is the polarity of RecA filaments? What concept explains this phenomenon?
No/slow filament formation in the 3’ to 5’ direction. Fast, active filament formation in the 5’ to 3’ direction. RecA binds cooperatively on ssDNA.
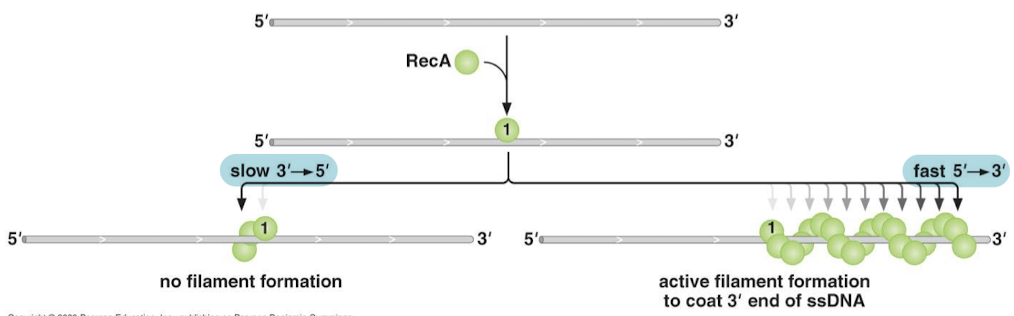
Molecules with what will not form RecA filaments?
5’ ssDNA extensions
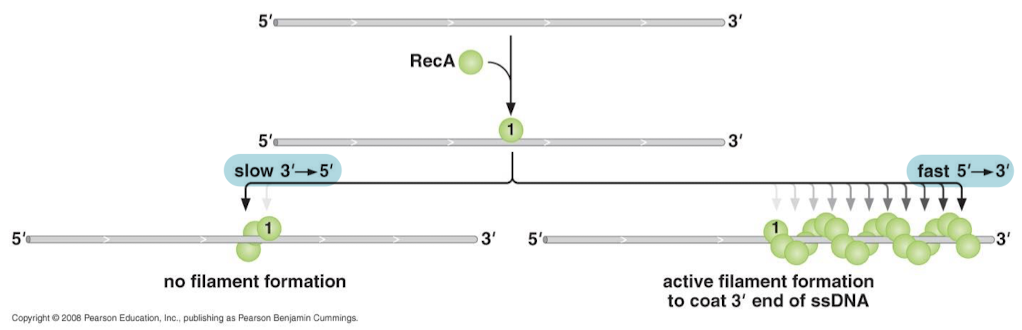
What is it called when RecA protein checks base pairs for homology and recombines ssDNA like a zipper?
Strand-exchange reaction
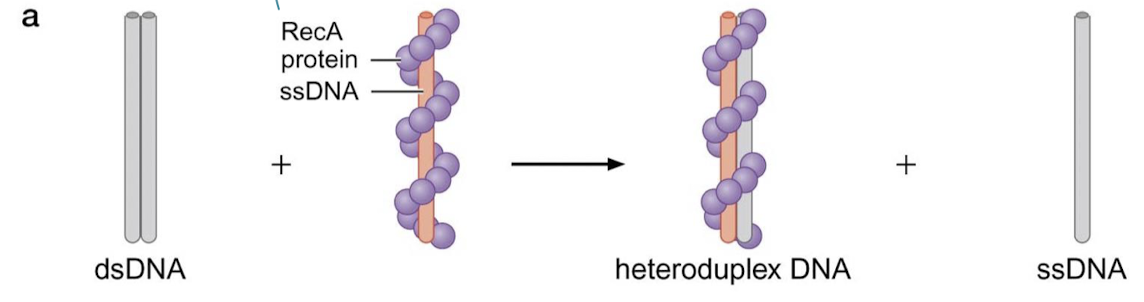
The RuvAB complex is used for what recombination step? In what organisms?
Holliday junction recognition and promotion of branch migration in E. coli (bacteria)
RuvC complex is used for what recombination step? In what organisms?
Resolution of Holliday junctions in E. coli (bacteria)
What is the specific function of RuvA? RuvB?
RuvA binds to Holliday junctions and recruits RuvB. RuvB is an ATPase similar to hexameric helicase involved in replication.
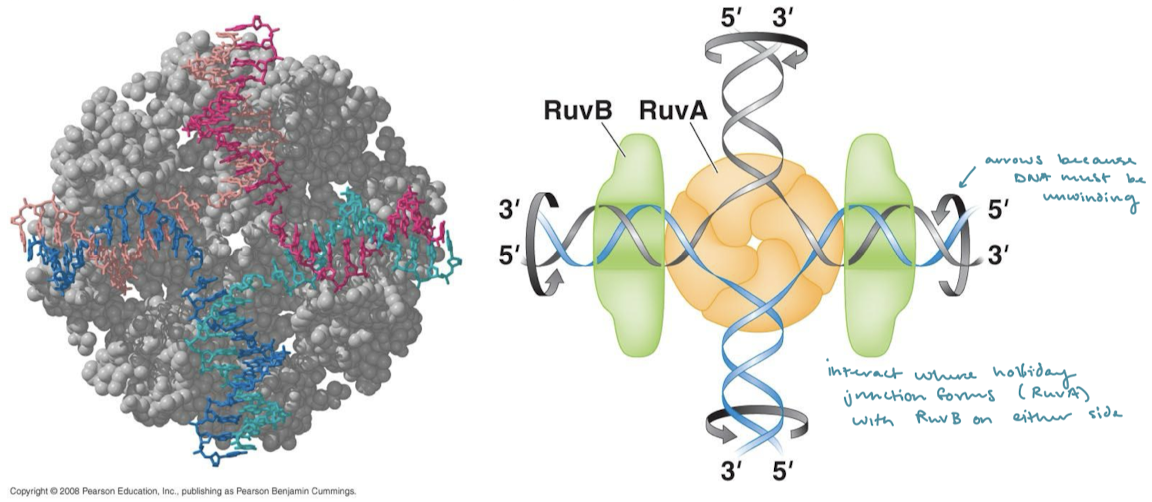
What does Ruv stand for (RuvAB complex and RuvC)?
Resolvase of UV-induced DNA damage
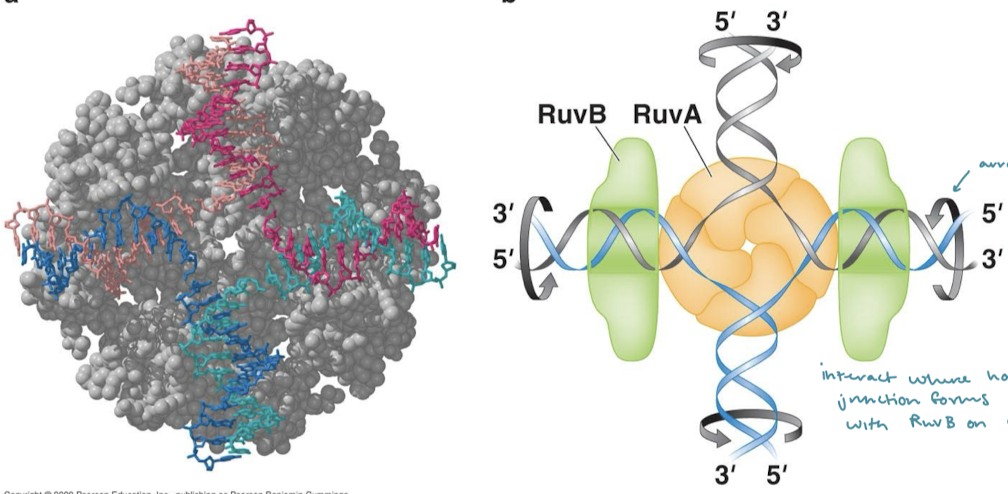
How does RuvC resolvase resolve Holliday junctions?
RuvC in complex with RuvAB nicks two strands with the same polarity (on the same plane/parallel to each other).
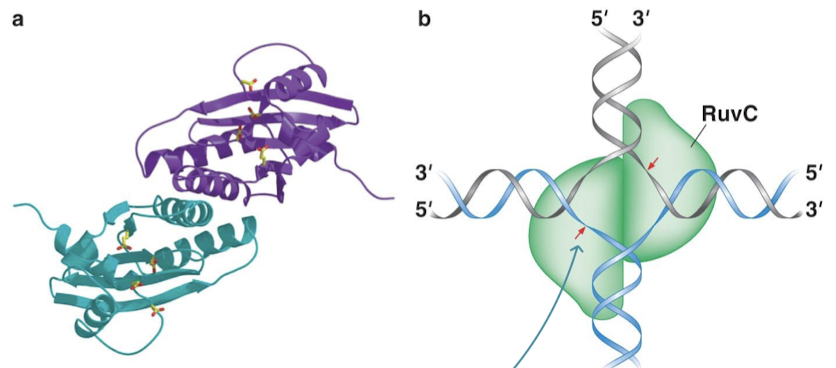
What is the sequence that RuvC cleaves? How often does this sequence appear?
5’-A/T-T-TG/C-3’. This sequence appears every 64 nucleotides on average, which is quite frequent.
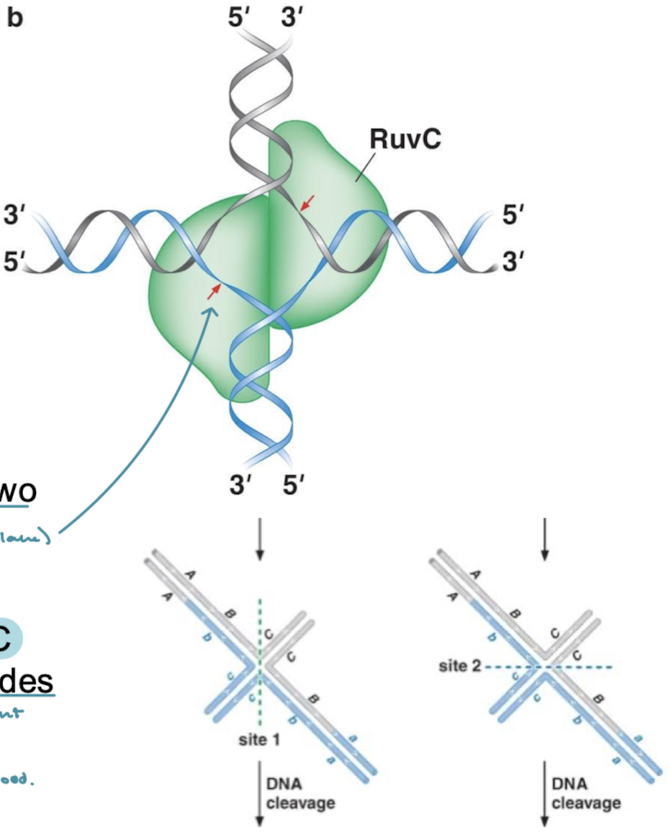
How are Holliday junctions resolved in eukaryotes?
The mechanism in eukaryotes is not well understood.
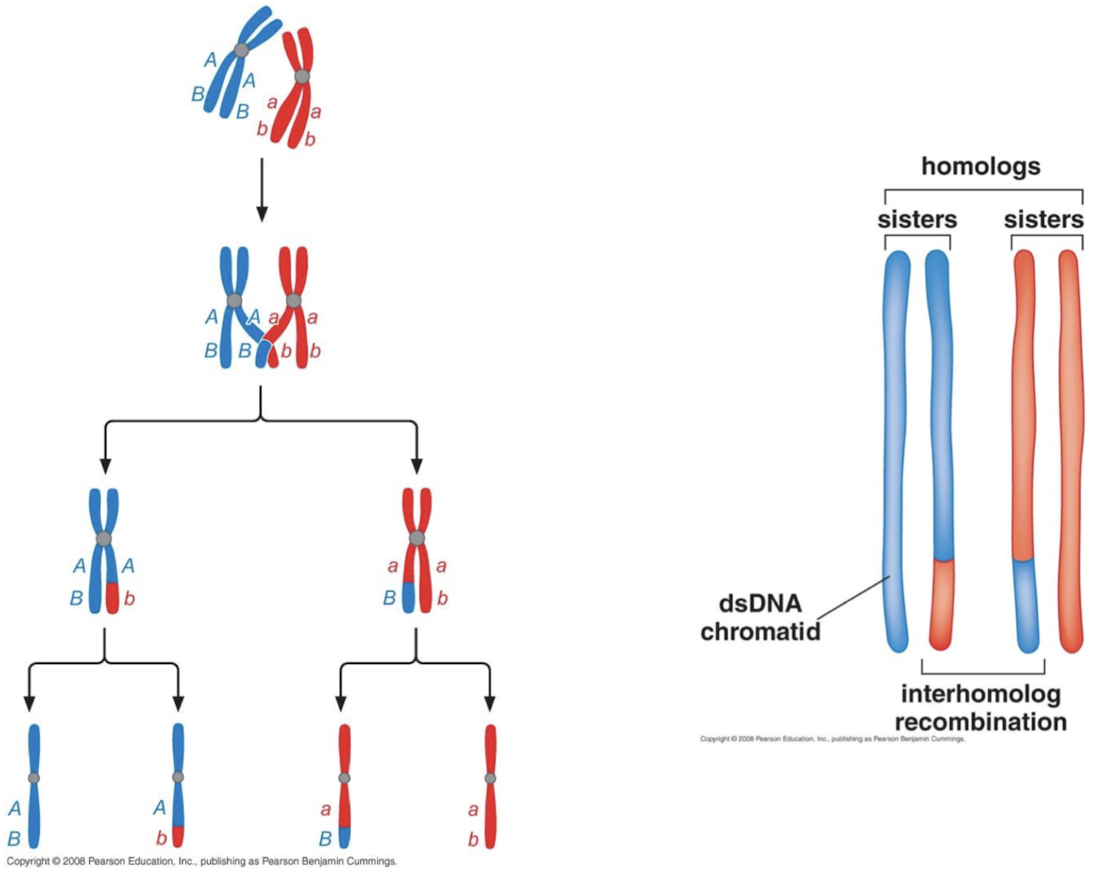
When does genetic recombination occur in cellular division?
In meiosis, meiosis I, prophase I, crossing over and genetic recombination occur when homologous chromosomes align. The site of recombination is the chiasmata, the homologous chromosomes separate in anaphase I, reducing the number of chromosomes.
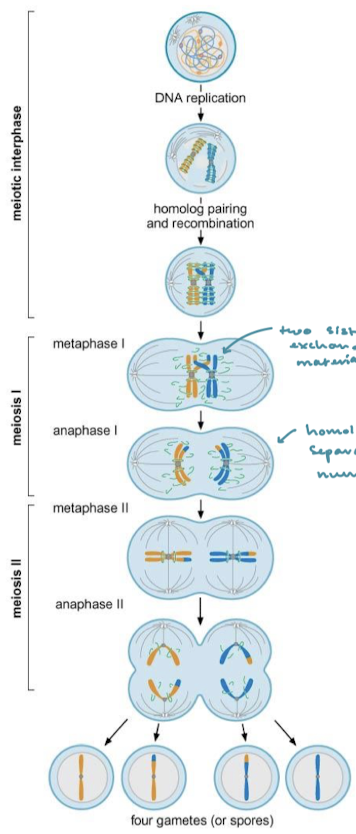
What is required for homologous chromosome segregation during meiosis? How many recombined germ line cells result from this process?
Meiotic recombination. One recombination event leads to two non-recombined germ line cells and two recombined germ line cells because the process occurs between sister chromatids.
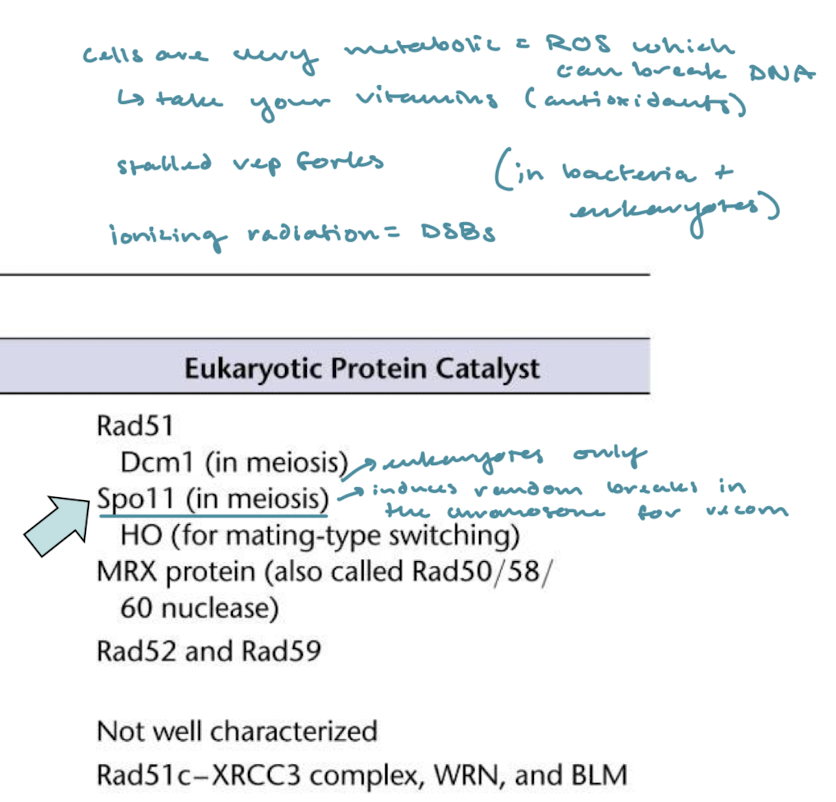
What are the main ways in which a DBS can occur without a catalyst? What is the catalyst for DBSs and in what organisms?
Cells are very metabolic and produce ROS which can induce DBSs (take your vitamins/antioxidants!) as well as stalled replication forks and ionizing radiation (prokaryotes and eukaryotes). In eukaryotes only, Spo11 introduces random DBSs in the chromosome for recombination in meiosis (eukaryotes only).
Meiotic crossovers are initiated by…
DSBs induced by the Spo11 endonuclease.
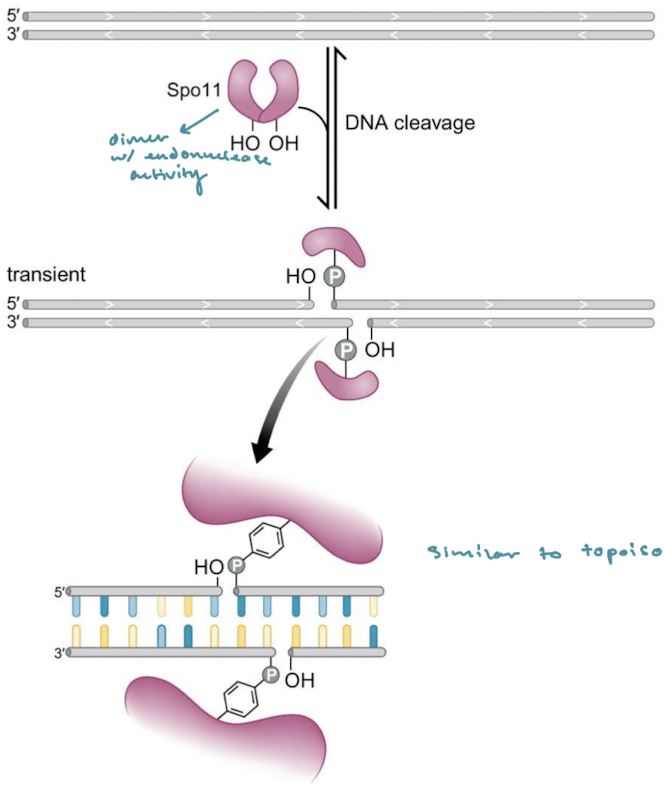
Explain the mechanism behind Spo11 introducing DSBs. What enzyme is this similar to?
Spo11 is a dimer with endonuclease activity. Both dimers contain tyrosine sites that attack the phosphodiester linkage on the backbone two nucleotides apart. This is similar to topoisomerase.
Where does the “spo” come from in Spo11 endonuclease?
Comes from yeast genetics and refers to sporulation
What is the MRX (Mre11/Rad50/Xrs2) complex?
Equivalent (but not homologous) to RecBCD.
Rad51 and DMC1 have what kind of activity?
RecA-like activity
DMC1 is specific to…
DMC1 is meiotic cell-specific and mediates recombination preferentially between non-sister homologous chromatids.
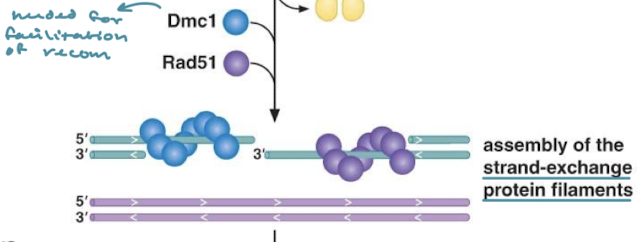
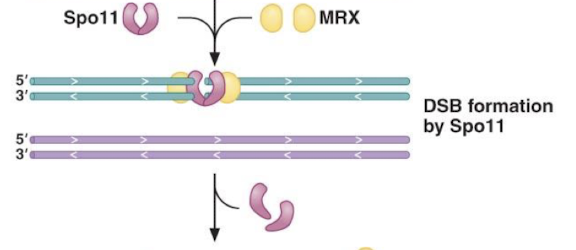
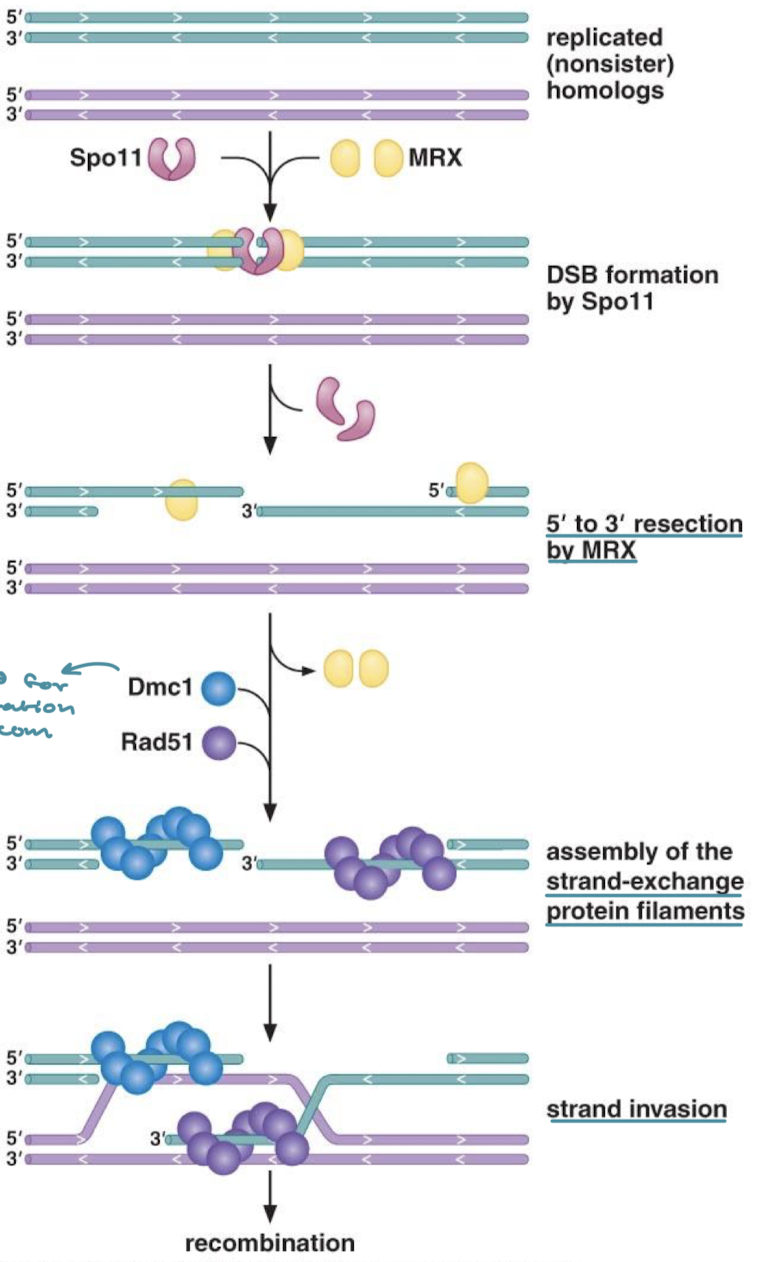
Explain the meiotic recombination pathway (Spo11, MRX, Dmc1, Rad51)
Spo11 induces DSB and MRX induces 5’ to 3’ resection. DMC1 and Rad51 assemble and facilitate strand invasion for recombination in meiosis.
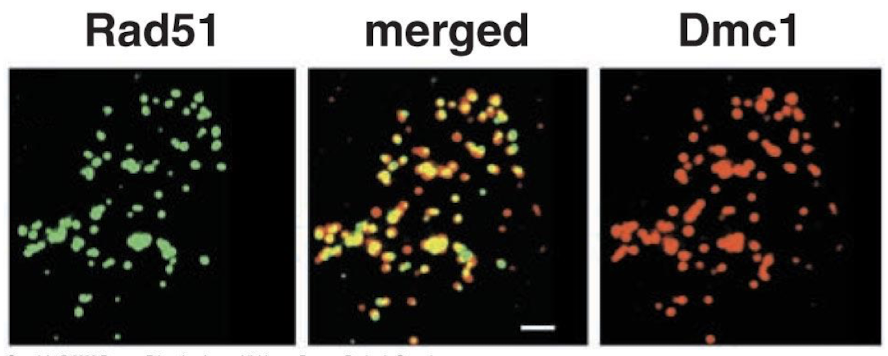
How do Rad51 and DMC1 work together in meiotic recombination?
The colocalize! Large protein-DNA complexes participate in the recombination process forming recombination factories.
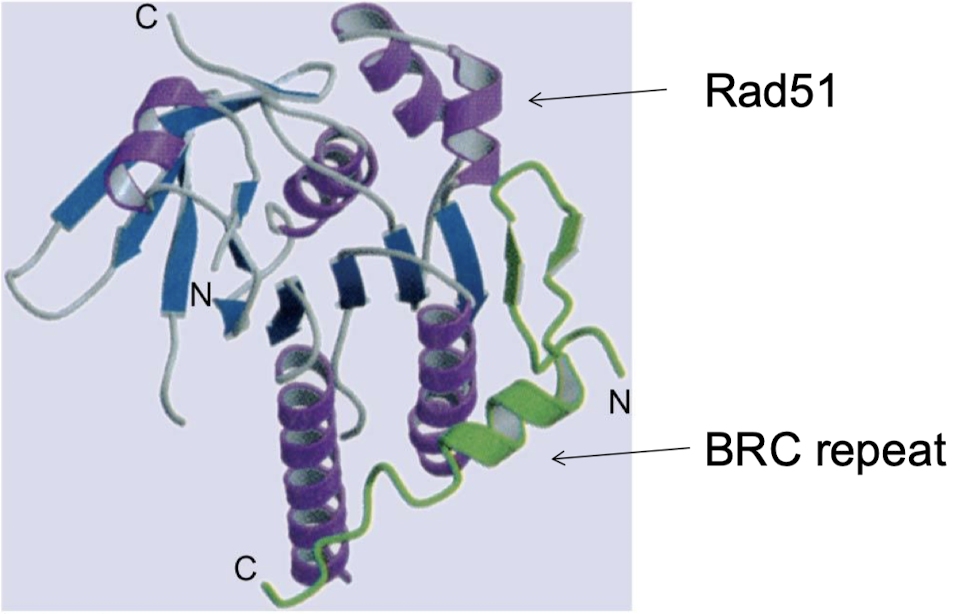
What gene does Rad51 interact with to repair DSBs (think cancer)?
Rad51 interacts with tumor suppressor gene BRCA2 in repairing DSBs. BRCA2 mutations are responsible for half of familial types of breast cancers.
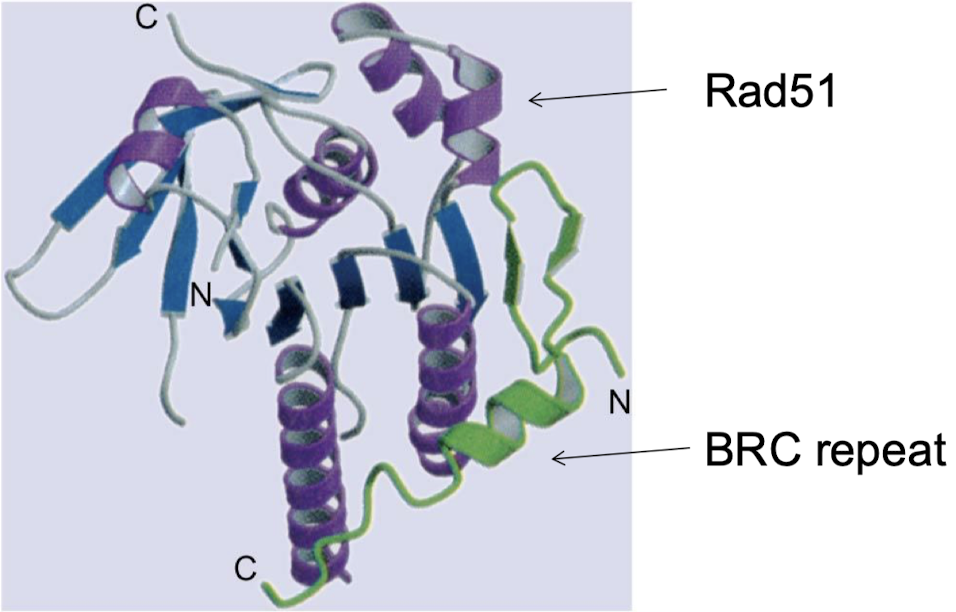
What mutations in BRCA2 would lead to cancer?
Two mutated and nonfunctional copies of BRCA2 would develop when DNA repair does not work. Creates a susceptibility to breast cancer.