Conservation Genetics 1+2 and Genetic Diversity
1/46
Earn XP
Description and Tags
These flashcards cover key concepts and vocabulary related to conservation genetics and genetic diversity, essential for understanding population genetics.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
47 Terms
Transmission genetics covers the ….
(classical genetics)
- Basic principles of heredity and how traits are passed from one generation to the next
- Relationship between chromosomes and heredity, arrangement of genes on chromosomes, and gene mapping
Conservation Genetics
Part of conservation biology is concerned with population genetic variation, viability, and the future evolution of species.
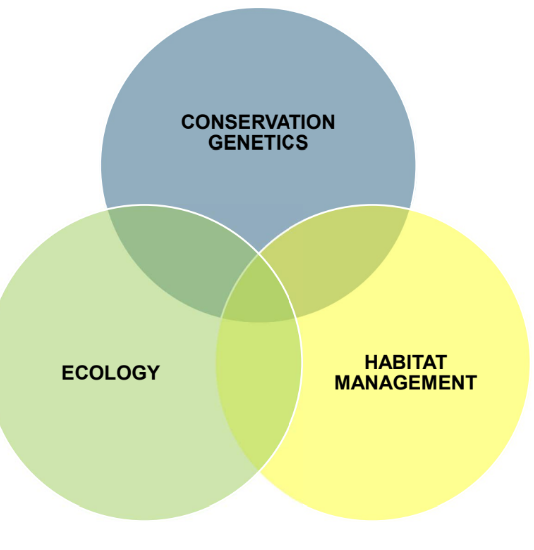
Conservation genetics, ecology, and habitat management together provide the technical underpinnings of conservation biology, a crisis-oriented science of biodiversity management
what are the 3 main concerns of conservation genetics?
• Preserving the genetic info contained in a species
• Assessing the conservation status of species/populations through their genetic characteristics
• Deciding which populations to manage and how and why
Population Genetics
The genetic composition of populations – how that composition changes geographically & overtime
- Fundamental to the study of evolution
what is genetic diversity?
Refers to the variation of genetic characteristics within a species or population.
critical for the adaptability and resilience of species in changing environments.
what groups are likely to show genetic diversity?
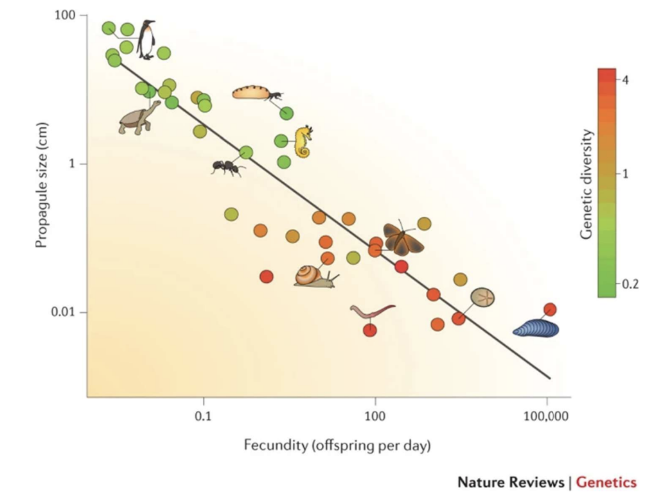
Vertebrates that produces many gametes (as shown in the red circles) experience higher genetic diversity
why is genetic diversity important for conservation genetics?
Although organisms cannot plan for environmental change, the
more variation that exists in a population, the better prepared
that population will be to adapt to change when it does occur.
• Indeed, genetic variation must be present for evolution to occur – so knowledge of it is extremely important
how is genetic diversity measured at the population level?
Relative genotype frequency
Relative allele frequency
Heterozygosity
Nucleotide diversity
Relative genotype frequency
the percentage of individuals in a population that have a specific genotype
Relative allele frequency
the percentage of all copies of a certain gene in a population that carry a specific allele
Heterozygosity
the proportion of individuals in a population that are heterozygous (Aa) at a particular locus
Nucleotide diversity
the mean number of nucleotide substitutions per site between any two randomly selected DNA sequences in a population
what factors increase / decrease genetic diversity?
Increase: mutation, recombination, gene flow, hybridisation
Decrease: inbreeding, genetic drift, habitat fragmentation
How does genetic diversity become reduced?
Changes in migration, selective pressures, geologic isolation, n and founder effects (since the founders of a new population will carry only a
portion of the genetic diversity that was present in the source
population)
New alleles (diversity) can be brought into a population through gene flow, hybridisation or mutation (rare).
Gene flow and genetic drift are opposing forces, so that an equilibrium can be reached if populations are interconnected
Habitat fragmentation makes populations more susceptible to
genetic drift (e.g. desert topminnows
why is info on genetic diversity within/between populations useful?
important for understanding how populations are structured.
This can be extremely important in conservation efforts, as higher genetic diversity often correlates with greater adaptability and resilience to environmental changes.
why is info on genetic diversity between species useful?
important for understanding the relationships between species and how species or particular traits have evolved
What causes Inbreeding Depression?
Low genetic diversity can lead to inbreeding depression
• Results in potential difficulties in dealing with environmental changes
• Genetically similar individuals have a higher risk of producing offspring that have hereditary diseases
what are some examples of how low genetic diversity i.e low heterozygosity causes detrimental effects?
In small, isolated populations there can be rapid losses in
heterozygosity (i.e. reduced genetic variation)
• Associated with fitness declines including:
• Poor competitive ability
• Poor growth rate
• Poor developmental stability
• Poor resistance to parasites
eg in desert topminnows
which chromosome in humans experiences high levels of genetic variation?
Chromosome 7 - is associated with various traits and diseases, (major histocompatibility complex) including those linked to the immune system and certain developmental disorders.
what are some reasons for low genetic diversity in cheetahs?
• Great difficulty in captive breeding
• High degree of juvenile mortality in captivity and in the wild
• High frequency of spermatozoal abnormalities in ejaculates
Gene Flow
The transfer of genetic variation from one population to another, which can increase genetic diversity and adaptability.
how is understanding gene flow important in fisheries management?
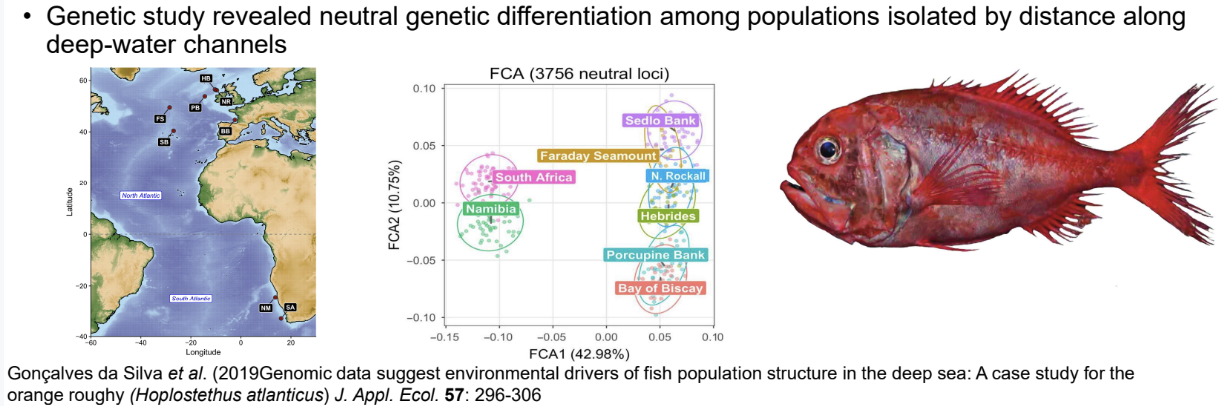
Fisheries often use genetic data for management decisions: e.g. orange roughy (Hoplostethus atlanticus) is “vulnerable to exploitation” due to slow maturity times
• Marine fish populations often have large census sizes and few obvious boundaries to gene flow
• Mistakenly assuming panmixia can lead to ineffective management and population decline
• Genetic study revealed neutral genetic differentiation among populations isolated by distance along
Neutral Variation
Genetic variation that does not affect an organism's/population’s fitness (no adaptive value)
Detrimental Variation
Genetic variation that negatively impacts the fitness of an organism.
Adaptive Variation
Genetic variation that confers an advantage in fitness to an organism.
what do these 3 types of variation depend on?
Depends upon the type of variation considered, the environment, population size, genetic background, etc.
For example, an adaptive allele that provides resistance to an infectious disease in one environment may be detrimental when the pathogen is absent because of fitness costs eg sickle cell anaemia can confer greater protection against malaria however it can also lead to health issues in non-malarial areas.
Genetic Load
reduction in the mean fitness of the population due to a the presence of deleterious variants in its gene pool.
Reflects the burden of low-quality alleles that may hinder reproductive success and survival.
what is the ratio of non-synonymous to synonymous substitutions used for ? (dN/dS)
is a useful measure of the strength and mode of natural selection acting on protein-coding genes
Significantly higher non-synonymous than synonymous rates are evidence for positive selection (dN/dS > 1)
Using such techniques several genes appear to show adaptive variation
what is effective population size Ne?
Effective population size is (approximately) the average number of
individuals in a population that reproduce successfully in each
generation.
• In population genetics what matters is the chance that two copies of a gene will be sampled as the next generation is produced, which is affected by the breeding structure of the population.
• Thus, in population genetic theory Ne is the size of an idealised population that would experience the same rate of genetic drift as the real population
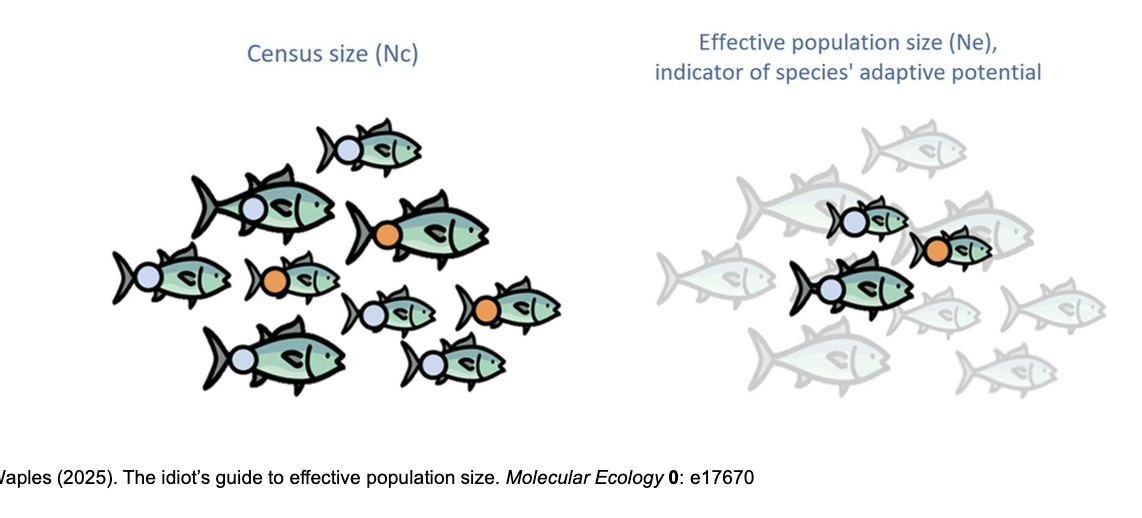
When thinking about Ne and the population genetics of small and
isolated populations, there are 3 major factors to consider:
I. Genetic Drift
II. Inbreeding
III. Gene flow
how does genetic drift effect Ne ?
Occurs when reproductive success within a population is variable – therefore not all alleles will be passed on with the same success
Each generation a random fraction of the population passes on genes to the next generation
Chance alone can result in changes in allele frequency
Effects of drift are most pronounced in small populations.
Without selection, drift drives allele to either fixation or extinction within a relatively short period
In larger populations it takes longer for effects to become pronounce
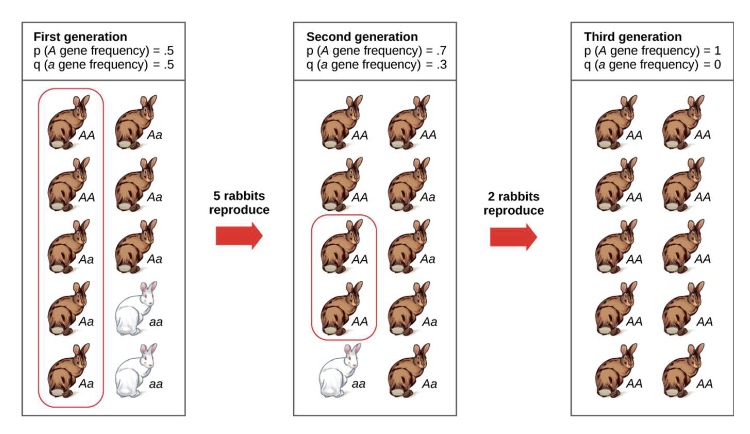
what is an example that illustrates how genetic drift causes more variation in a certain allele in small populations?
• Consider a population at HWE with p=0.5 and q=0.5.
For no drift, frequencies of the alleles in successive generations must remain at 0.5. Suppose we sample 5, 50 or 500 individuals:
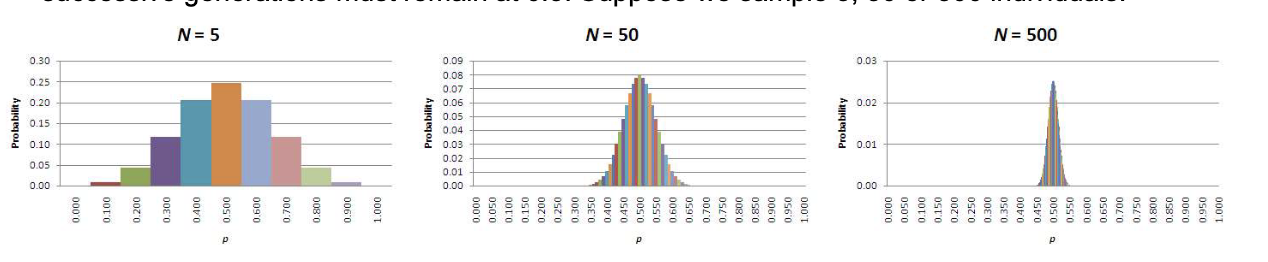
• BUT change due to sampling error (drift) decreases as the population size increases i.e. is inversely proportional to population size, so small populations are prone to random changes
• Direction of the change is unpredictable.
• In a small population, without other factors, p will drift to either 0.0 or 1.0. The time taken depends on the starting allele frequencies and population size
what are some effects of what happens to alleles after many generations of genetic drift and some disadvantages?
Some alleles go to fixation while others are lost by this random process of sampling
• Once the population reaches such a fixation state, it is stuck.
Only new mutations or migrants can reintroduce variation
We want to avoid this! Those alleles may be adaptive now or in the future.
• Also, if detrimental alleles become fixed through chance, populations can be put at risk
• Plans are needed to increase genetic diversity – captive breeding programmes
What are 3 reasons for why Ne often much smaller than the actual number of individuals in a population?
1. Uneven sex ratios
2. Variation in reproductive success
3. Fluctuating population size
How do uneven sex ratios affect the Ne ?
The ratio of breeding adults may be unequal
e.g. in elephant seals there’s competition between males for access to
harems, therefore, only a handful of dominant males in each population will contribute their genes to the next generation
• By comparison, the majority of females reproduce, therefore the result is an effectively female-biased sex ratio
how can uneven sex ratios be calculated?

Thus, we can see uneven sex ratios reduce the effective population size.
what is an example of how variation in reproductive success causes a decreased Ne?
• In steelhead trout (Oncorhynchus mykiss) populations in the Hood River, Oregon, USA, when the population is at high density, females have increased competition for males, spawning sites and other resources
• Successful competitors will produce large numbers of offspring, whereas the less successful may fail to reproduce (i.e. there is variation in reproductive success)
• Ne/Nc was estimated to be 0.17–0.40, with large variance in reproductive success among individuals being the primary cause of the reductio
variation in reproductive success is thought to be high in which types of species?
High in broadcast spawners that release gametes into the ocean.
• Millions of small eggs develop rapidly, without parental care, into planktonic larval stages that suffer high early mortality (Type III survivorship).
• Yet, large marine populations generally have only fractions of the genetic diversity expected from their sheer abundance
how can the low level of molecular genetic diversity be explained in high fecundity animals (e.g bivalves, corals, polychaetes, crabs etc) ?
Release eggs into a temporally and spatially patchy planktonic environment which leads to high mortality
• Ne/Nc is typically <0.01
• Explains the observed levels of molecular genetic diversity that are much lower than expected, given enormous census sizes and “chaotic”
genetic patchiness, which occurs on small spatial scales
• Referred to as “Sweepstakes Reproductive Success” with few “winners” and many “losers.
how is the fluctuating population size an important driver of reduced Ne?
Fluctuating population sizes can lead to variation in reproductive success and genetic drift, where certain alleles may become more common or lost entirely due to chance events.
This results in decreased effective population size (Ne) as fewer individuals contribute to the gene pool.
what is the ‘bottleneck effect’?
The ‘bottleneck effect’ is a significant reduction in genetic diversity that occurs when a population undergoes a rapid decrease in size due to environmental events or human activities, leading to a loss of alleles and increased inbreeding.
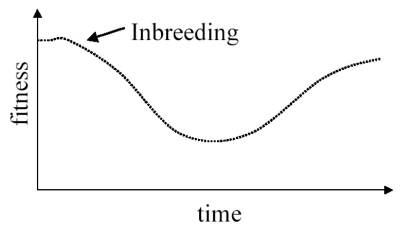
how does inbreeding effect population genetics in small or isolated populations?
Inbreeding can lead to increased homozygosity within the population
Inbreeding and heterozygosity are negatively correlated
Inbreeding coefficient F = 1 - (Hobs/Hexp)
Favourable alleles at a locus are usually dominant and the deleterious alleles have been maintained within the population because they are recessive
Maintaining of deleterious alleles, resulting in reduced fitness, reproductive success, population decline
What are some examples of how inbreeding depression can effect species?
The population of 40 Swedish adders (Vipera berus) isolated from other populations through farming activities produced higher proportions of stillborn and deformed offspring
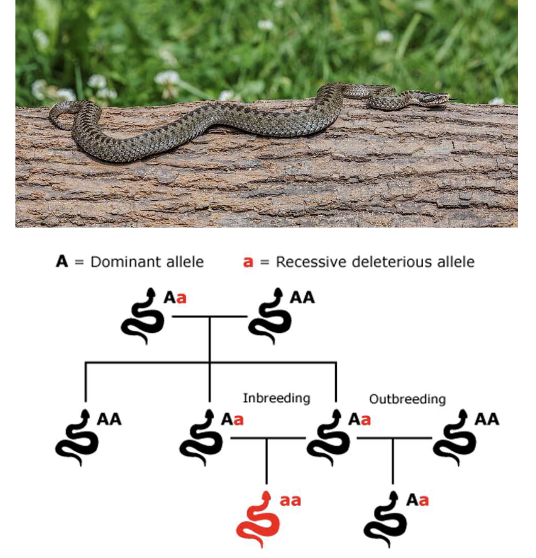
Introduction of adders from other populations — an example of outbreeding — the isolated population recovered and produced a higher proportion of viable offspring
What is the effect of purging on deleterious alleles ?
This process is particularly effective against alleles that are lethal in the homozygous state
• Therefore, in habitually inbreeding species/populations deleterious recessives are expected to have been purged by selection
• Purging is unlikely to have much effect on mildly deleterious alleles - these alleles may become fixed in a small population following drift
•It cannot get rid of the effects of inbreeding that are associated with heterozygote advantage
• Effectiveness of purging remains debatable and can lead to the removal of harmful alleles from the gene pool over generations.
what is an evolutionary significant unit?
An evolutionary significant unit (ESU) is a population or group of populations that is considered important for conservation because it has a distinct genetic identity and evolutionary history.
ESUs are often used to prioritise management strategies for species conservation.
what are some considerations for selecting an ESU?
• Substantial reproductive isolation that represents an important component in the evolutionary history of the species
• Discontinuous, significant genetic divergence, and geographic isolation
• Have a wholly unique heritable trait
• Populations that have been in isolation for a long time
• ecological and genetic exchangeability together with isolation for a sufficient period of time
• A full ESU requires differentiation for both neutral and adaptive variation
How is conservation genetics done in practice?
• Define populations that are threatened or endangered
• Form hypotheses about relationships between populations and/or species and test these hypotheses by examining the genetics characteristics of the organisms
• Determine the rate at which the exchange of genetic info (gene flow) is occurring
• Develop a management strategy i.e conservation. strategies.