Macromolecules: Nucleic acids & proteins
1/90
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
91 Terms
What do membranes do?
form semipermeable barriers
Membranes
essential structures that define the boundaries of cells and organelles, playing crucial roles in transport, signalling, and adhesion.
Polysaccharides function
cell energy, structure, and signaling
polysaccharide
polymer consisting of sugars and sugar derivatives linked together by glycosidic bonds.
Nucleic acids function
store, transmit, and express genetic information (instructions for life)
Proteins function
do almost everything else that cells need
-Virtually everything a cell is or does depends on the genes it express and the proteins it contains
e.g proteins can be enzymes, structural proteins, motility proteins, Regulatory proteins, Transport proteins, Signalling proteins, defensive proteins, and antibody proteins
What is DNA the molecule of?
Heredity
Heredity
The process by which characteristics are passed down from a parent to child through genes
Genes are “written” in what?
DNA
What is the genome?
The genome (all genes in an organism) provide all of the instructions for making that organism
Gene
The basic unit of heredity passed down from parent to child. Genes are made up of sequences of DNA and are arranged, one after another, at specific locations on chromosomes in the nucleus of the cell. They contain information for making specific proteins that lead to the expression of a particular physical characteristic or trait, such as hair colour or eye colour, or to a particular function in a cell.
What does DNA do?
Stores and transmits biological information
What does DNA transmitting information mean?
it carries the instructions (genes) for building and operating a living thing, encoded in its base sequence, and passes these instructions accurately from parent to offspring during reproduction, ensuring continuity and traits from one generation to the next
What does RNA do?
RNA is chemically similar to DNA, but is used to “express” genetic information
Gene expression:
A genes DNA sequence is used to create a functional product, usually a protein or functional RNA, through two main steps: Transcription and Translation
Transcription
process by which RNA polymerase utilizes one DNA strand as a template for guiding the synthesis of a complementary RNA molecule.
Translation
Process by which the base sequence of an mRNA molecule guides the sequence of amino acids incorporated into a polypeptide chain; occurs on ribosomes.
Nucleic acids:
Linear polymers of nucleotides
DNA:
deoxyribonucleic acid
RNA:
ribonucleic acid
Nucleotides:
Basic building blocks of nucleic acids
Nucleotides consist of:
Phosphate group Five-carbon sugar (ribose or deoxyribose) Nitrogen-containing aromatic base ▪ Purine or pyramidine
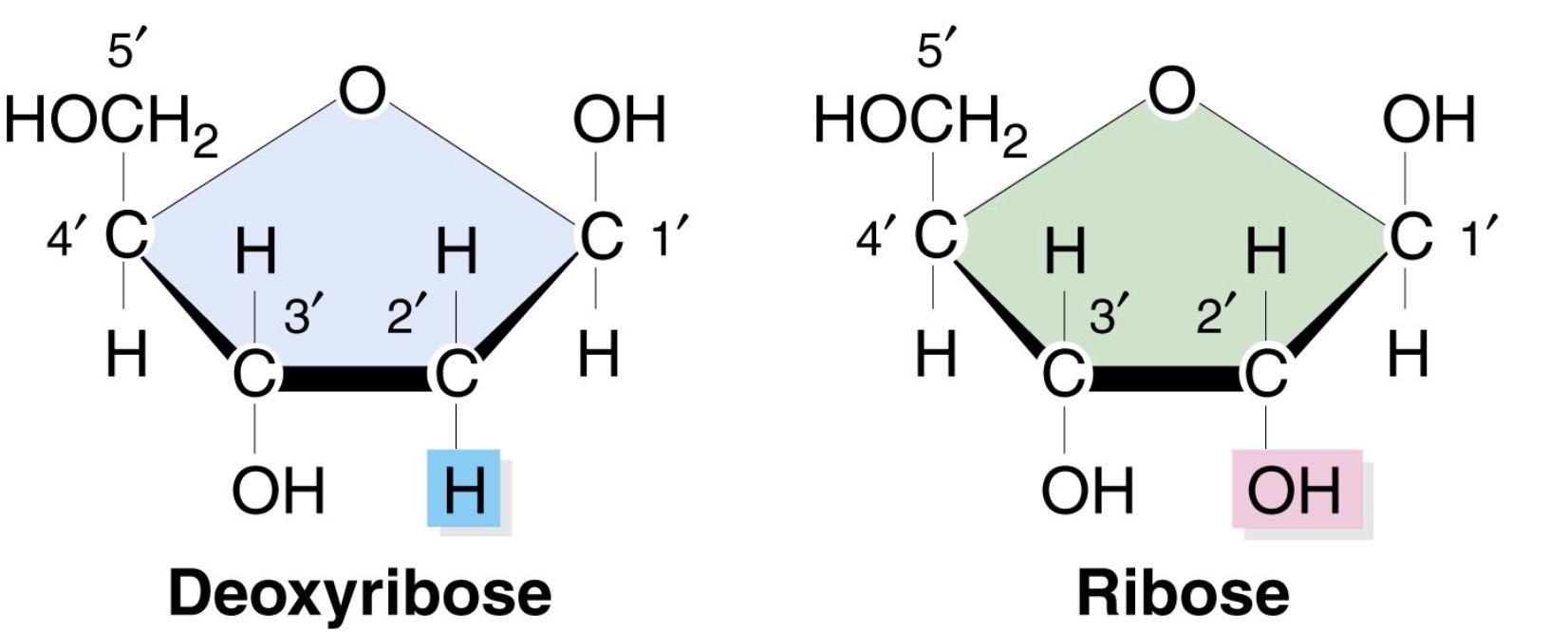
What is the difference in the sugars of DNA and RNA?
RNA contains the five-carbon sugar ribose in each of its nucleotides, whereas DNA contains the closely related sugar deoxyribose
-Ribose has OH group at C 2’
-Deoxyribose has H group at C 2’
Purine
two-ringed nitrogen-containing molecule; parent compound of the bases adenine and guanine.
Pyrimidine
single-ringed nitrogen-containing molecule; parent compound of the bases cytosine, thymine, and uracil.
DNA purines:
Adenine, Guanine
Dna Pyrimidines
Cytosine, Thymine
RNA purines
adenine, guanine
Rna pyrimidines
Cytosine, Uracil
Nucleoside
molecule consisting of a nitrogen-containing base (purine or pyrimidine) linked to a five-carbon sugar (ribose or deoxyribose); a nucleotide with the phosphate removed.
What can each purine and pyrimidine exist as?
The free base, a nucleoside, or the nucleotide
Adenine; Nucleoside, Nucleotide, Deoxynucleoside, Deoxynucleotide,
Nucleoside: Adenosine
Nucleotide: Adenosine monophosphate (AMP)
Deoxynucleoside: Deoxyadenosine
Deoxynucleotide: Dexoyadenosine monophosphate (dAMP)
Guanine: Nucleoside, Nucleotide, Deoxynucleoside, Deoxynucleotide,
Nucleoside: Guanosine
Nucleotide: Guanosine monophosphate (GMP)
Deoxynucleoside: Deoxyguanosine
Deoxynucleotide: Deoxyguanosine monophosphate (dGMP)
Cytosine: Nucleoside, Nucleotide, Deoxynucleoside, Deoxynucleotide,
Nucleoside: Cytidine
Nucleotide: Cytidine monophosphate (CMP)
Deoxynucleoside: Deoxycytidine
Deoxynucleotide: Deoxycytidine monophosphate (dCMP)
Uracil: Nucleoside, Nucleotide, Deoxynucleoside, Deoxynucleotide,
Nucleoside: Uridine
Nucleotide: Uridine monophosphate (UMP)
Deoxynucleoside: N/A
Deoxynucleotide: N/A
Thyamine: Nucleoside, Nucleotide, Deoxynucleoside, Deoxynucleotide,
Nucleoside: N/A
Nucleotide: N/A
Deoxynucleoside: deoxythymidine
Deoxynucleotide: deoxythymidine monophosphate (dTMP)
a nucleoside monophosphate
has one phosphate group attached to it
Can be diphopshate (2 phosphate groups) Triphosphate (3 phosphate groups)
example: Adenosine Triphisphate (ATP) : 3 phosphate groups, Adenine base, Ribose sugar.
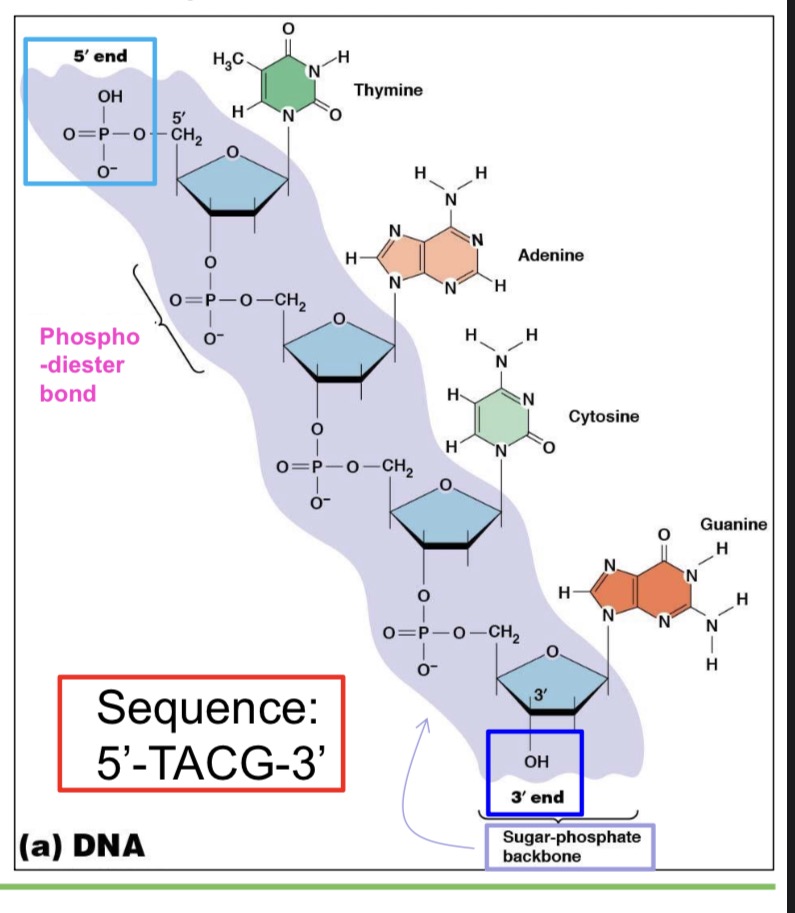
Nucleic acids
Nucleic acids are linear polymers of nucleotides
-formed by linking each nucleotide to the next through a phosphate group
3ʹ,5ʹ phosphodiester bonds:
Nucleotides linked by 3ʹ,5ʹ phosphodiester bonds:
▪ 1 phosphate group linked to 2 adjacent nucleotides via two phosphodiester bonds
What are Nucleotide in polymers are linked by?
phosphodiester bonds
Directionality of nucleotide polymers:
5ʹ phosphate at one end
▪ 3ʹ hydroxyl at the other end
▪ Nucleotide sequences written in 5ʹ to 3ʹ direction
Nucleic acid synthesis requires what?
Information and energy
Information:
A pre-existing template
template:
a nucleic acid whose base sequence serves as a pattern for the synthesis of another (complementary) nucleic acid.
phosphodiester bridge
covalent linkage in which two parts of a molecule are joined through oxygen atoms to the same phosphate group.
To provide the energy needed to form each new phosphodiester bridge:
each successive nucleotide enters as a high-energy nucleoside triphosphate
Energy:
Nucleotides enter as high energy triphosphates
RNA: nucleotide triphosphates:
(NTPs; e.g., ATP)
DNA: deoxynucleotide triphosphates:
(dNTPs)
In nucleic acid synthesis, what ensures correct order?
Correct base pairing between the template and the incoming nucleotide ensures correct order
base pairing
complementary relationship between purines and pyrimidines based on hydrogen bonding that provides a mechanism for nucleic acids to recognize and bind to each other; involves the pairing of A with T or U, and the pairing of G with C.
Base pairs form between:
complementary bases:
▪ A forms two hydrogen bonds with T
▪ G forms three hydrogen bonds with C
How many hydrogen bonds between Cytosine and Guanine?
3
How many Hydrogen bonds between Thymine and Adenine?
2
Base pairing is
Complimentary
between purine (little name big base) and Pyrimadine (big name, little base)
Describe the structure of DNA
DNA forms a double-stranded helix
One strand base-pairs with the other= Complementary
The two strands are in opposite orientations = Antiparallel
DNA replication
Each strand of Dna acts as a template for DNA replication
Explain Base Pairing and RNA:
▪ RNA is normally single stranded
▪ RNA structure also depends on base pairing ▪ RNA base pairing is usually between bases in different areas of the same molecule and is less extensive than that of DN
Explain Proteins
Polymers of Amino Acids
20 amino acids are used in proteins
Amino Acid Structure
α (central) carbon which the following is attached to:
amino (NH3) group
Carboxyl group
A hydrogen atom
R group (different for each amino acid)
Most have two enantiomers (L and D) ▪ Only the L enantiomer is used in proteins
What do specific properties of amino acids depend on?
the nature of their R groups
Polypeptide:
product of amino acid polymerization
Protein synthesis
The process of elongating a chain of amino acids
A polypeptide does not become a protein until it has assumed a stable, three-dimensional shape and is biologically active
What are the classes of amino acid R groups?
9 amino acids have hydrophobic (nonpolar) R groups
11 amino acids have hydrophilic R groups
In regards to the 11 amino acids have hydrophilic R groups:
6 are polar and uncharged
5 are charged at neutral pH
Acidic amino acids
have negative charges
basic amino acids
have positive charges
What kind of side groups do hydrophobic (nonpolar) R groups have?
hydrophobic (nonpolar) R groups
-e.g phenyl groups, just H, CH, ect
What kind of side groups do hydrophilic R groups have?
the R group is polar e.g has OH, NH2,
R group is acidic or basic and thus is charged at cellular pH e.g NH+, O-
Where are Hydrophilic and hydrophobic amino acids found?
Hydrophilic amino acids tend to be found on the water-facing surfaces of proteins; hydrophobic tend to be on the interior
Amino acids in a polypeptide and are linked by what?
peptide bonds
How do peptides have directionality?
the chain of amino acids formed has an intrinsic directionality because it always has an amino group at one end and a carboxyl group at the other end
The end with the amino group is called the N- (or amino) terminus ▪ Beginning of the protein
▪ The end with the carboxyl group is called the C- (or carboxyl) terminus ▪ End of the protein
peptide bond
a covalent bond between the amino group of one amino acid and the carboxyl group of a second amino acid
he initial folding of a polypeptide into its proper shape, or conformation, depends on what?
several different kinds of bonds and interactions including the covalent disulfide bond and several noncovalent bonds and interactions
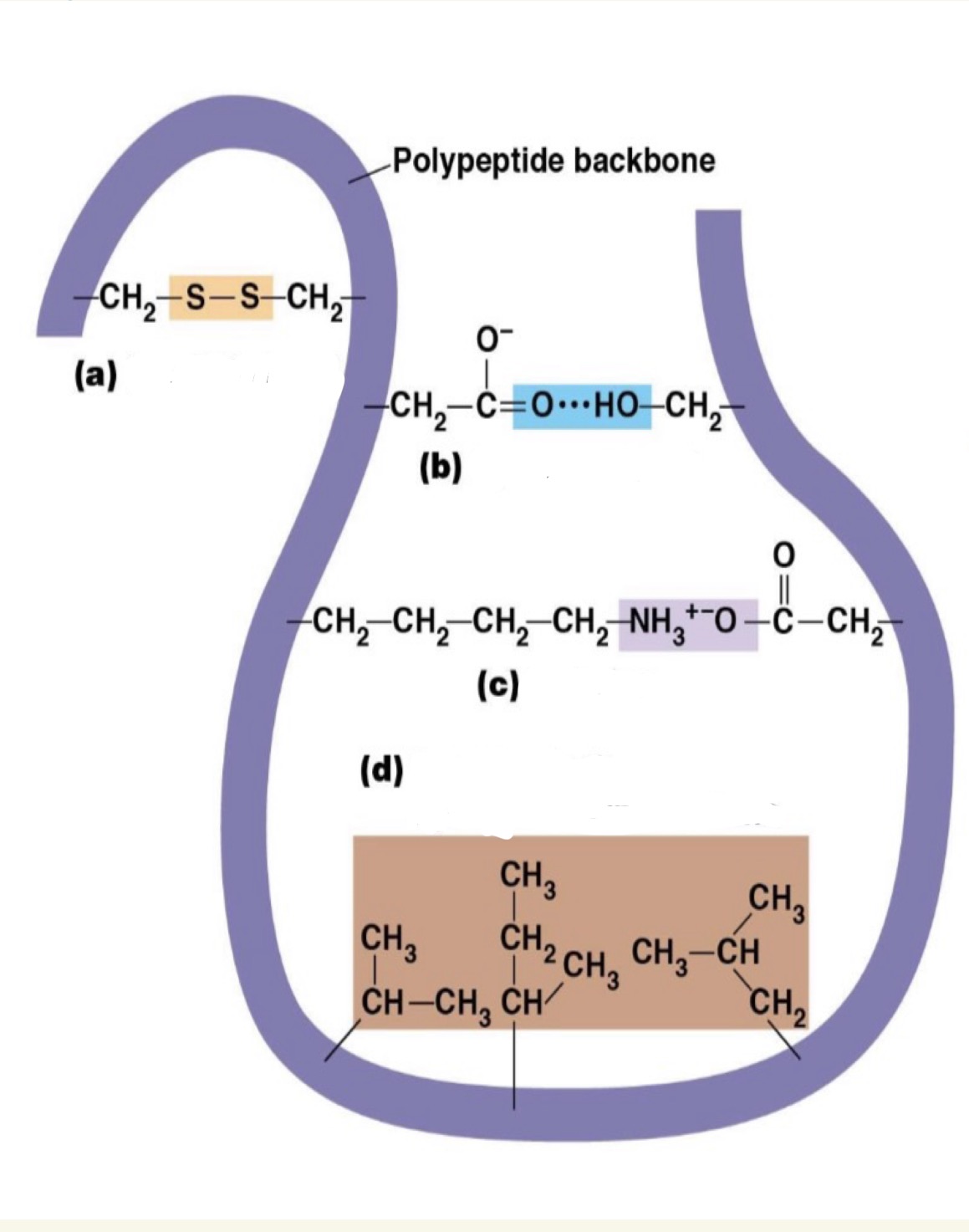
Label a) b) c) d) and explain this figure
a)Disulfide bonds: Covalent bonds. Very stable bond ▪ Formed between sulfur atoms in cystein residues ▪ Intra- or Inter-molecular
b)Hydrogen bonds
c) Ionic bonds
d)van der Waals interactions ▪ Hydrophobic interactions
b, c, and d are all Noncovalent bonds/interactions
Individually weaker than bonds, but collectively strong
What determines the determine the 3D shape of the protein?
Bonds/interactions between R groups
Primary structure basis:
Amino acid sequence
Primary structure bonds/interactions:
Covalent peptide bonds
Secondary structure basis:
Folding into alpha helix or beta sheet, or random coiling *Local folding
Secondary structure bonds/interactions:
Hydrogen bonds between NH and CO groups of peptide bonds in the backbone.
Tertiary structure basis:
Three dimensional folding of a single polypeptide chain
Tertiary structure bonds/interactions:
Disulfide bonds, hydrogen bonds, ionic bonds, van der Waals interactions, hydrophobic interacrions
Quaternary structure basis:
Association of multiple polypeptide to form a multimeric protein
Quaternary structure bonds/interactions:
Disulfide bonds, hydrogen bonds, ionic bonds, van der Waals interactions, hydrophobic interacrions
Secondary Structure
Local regions of structure
Readily predictable, and determined by the primary structure
Results from hydrogen bonding between NH and CO groups along the polypeptide backbone
Two major patterns ▪ α helix and β sheet
Result from localized folding of a segment of the polypeptide ▪ Alpha helices are usually ~10-20 amino acids long ▪ Beta sheets have multiple strands with variable length
Result from localized folding of a segment of the polypeptide ▪ Alpha helices are usually ~10-20 amino acids long ▪ Beta sheets have multiple strands with variable length
The α Helix
Helix is spiral in shape ▪ R groups jut-out from the spiral ▪ Some amino acids have strong propensities toward forming helices; other amino acids disrupt helices
The β Sheet
Sheet-like conformation formed by multiple polypeptide strands ▪ Extensive but variable hydrogen bonding between backbones ▪ R groups jut-out on alternating sides of the (pleated) sheet ▪ Classified based on relative directionality of strands (N-to-C) ▪ Parallel: same directions ▪ Antiparallel: opposite directions
Tertiary Structure:
3D folding of a polypeptide ▪ Depends upon interactions between R groups; prediction enabled by new AI platforms (AlphaFold)
Tertiary Structure is determined by
1. Hydrophobic residues avoiding water 2. Hydrophilic residues interacting with water 3. Repulsion of similarly charged residues 4. Attraction between oppositely charged residues
quaternary structure
level of organization concerned with subunit interactions and assembly
The term only applies to multimeric proteins (composed of two ore more polypeptides)
Some multimeric proteins consist of multiple identical subunits; others, such as hemoglobin, contain two or more types of polypeptides