BICD 110 midterm 1
1/98
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
99 Terms
What are the three pillars of cell theory?
all living organisms are composed of one or more cells
cells are the most basic unit of life
all cells arise only from pre-existing cells
How long ago was LECA?
1.5-2 billion years ago
What did Robert Hooke do?
discovered (plant) cells
named honeycomb pattern in “cork” cells
What did Anthonie Van Leeuwenhoek do?
discovered microorganisms (animalcules)
grandfather of microbiology
discovered RBC, sperm cell → nucleus
observed fertilization
What did Rudolphi/Link do?
discovered cells have independent cell walls
What did Dutrochet do?
“cell is the fundamental element of organization”
What did Schlelden do?
discovered every part of a plant is made of cells
incorrectly spread the idea that cells are made from “crystallization”
What did Schwann do?
discovered every part of a plant + animal is made of cells and/or its products
What did Brown and Flemming do?
used basic stains (hematoxylin) to discover nucleus, chromosomes, and cell division cycle
What is the “black reaction”?
A technique developed by C. Golgi that uses a silver chromate solution to stain nerve cells, identified Golgi apparatus.
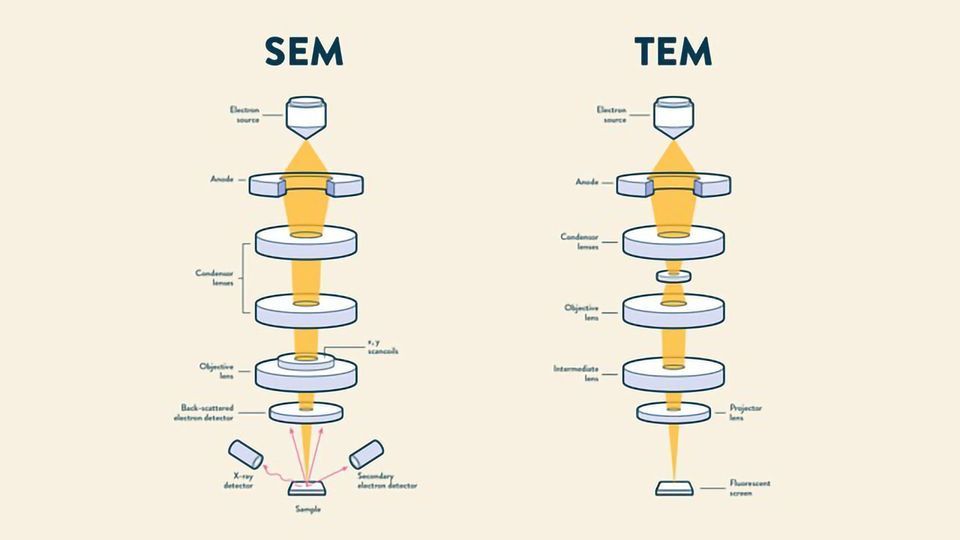
What is TEM and SEM?
TEM (transmission electron microscopy): thin → stained/shadowed with heavy metals. thick → fixed dehydrated, embedded in resin, sectioned, and stained with heavy metals. uses transmitted electrons (electrons that are passing through the sample) to create an image
SEM (scanning electron microscopy): surface of sample is metal shadowed. uses electrons that that are reflected or knocked-off to create an image
What did Virchow do?
discovered cellular pathology “all cells come from other cells”
What is cryogenic electron microscopy (cryo-EM)?
hydrated, unfixed, unstained samples are plunge-frozen leading to formation of vitreous (non crystalline ice) that preserves sample.
What is a tomogram?
A 3D image obtained from combining multiple 2D slices of a sample, providing detailed structural information.
What did Porter, Claude, and Fullham do?
obtained the first electron micrograph of a cell (chicken fibroblasts)
What did George Palade do?
as Claude’s postdoc, got thinnest and most detailed image of cell via electron microscopy
grandfather of molecular cell biology
What is GFP and how is it useful?
GFP (green fluorescent protein), is a bioluminescent protein allows scientists to visualize and track proteins in live cells. FP genes can be fused to a gene of interest to produce recombinant fluorescent protein and track protein of interest
What are some ways to deliver recombinant DNA to cells?
polymers, viral transduction/transfection, microinjection, electroporation, liposomes
What is immunolabeling and how is it used?
Immunolabeling is a technique used to visualize proteins or antigens in cells by using specific antibodies tagged with a marker. first, antibodies must be generated to POI. (antigen). Then, the antibodies are applied to the sample, allowing for detection under a microscope, often using fluorescent markers.
To visualize subcellular structures or location of specific biomolecules, cells/tissue need to be:
fixed (w/ formaldehyde), embedded in paraffin, sectioned, permeabilized to make membrane permeable to detergents, and stained with a marker (fluorescence or gold) that are attached to antibodies so they gain contrast and can be seen
What are the 4 concepts of life?
molecular complementarity, polymerization, chemical equilibrium, energy
What is the order of energy of covalent/noncovalent bonds?
thermal energy < (noncov) van der Waals < (noncov) H-bond < (cov) hydrolysis of ATP < (cov) C-C < (cov) C=C
What is Kd and how does it relate to affinity
dissociation constant, lower Kd = higher affinity
What is the hydrophobic effect?
The hydrophobic effect is the tendency of nonpolar substances to aggregate in aqueous solutions to minimize their exposure to water, leading to an increase in entropy because water is liberated from the cages
What does it mean for a molecule to be amphipathic?
has both hydrophilic (water-attracting) and hydrophobic (water-repelling) regions, allowing it to interact with both polar and nonpolar substances.
How does temperature control membrane bilayer fluidity?
Temperature affects membrane fluidity by influencing the kinetic energy of the lipid molecules; higher temperatures increase fluidity while lower temperatures decrease it.
What is Tm?
Tm is the temperature at which a lipid bilayer transitions from a gel-like state to a more fluid state; 50% of lipids are fluid
How does fatty acid composition control membrane bilayer fluidity?
Fatty acid composition affects membrane fluidity through the degree of saturation; saturated fatty acids pack tightly, making the bilayer less fluid, while unsaturated fatty acids introduce kinks, increasing fluidity.
Which out of PC, PS, PE, PI, PG, SM, CL is a glycerophospholipid?
PC, PS, PE, PI, PG
How does cholesterol control membrane bilayer fluidity?
Cholesterol regulates membrane fluidity by inserting itself between phospholipids, increasing fluidity by disturbing lipid packing, and decreasing fluidity w/ its rigid structure. low temp: inc fluidity, high temp: dec fluidity
What is the permeability like for hydrophobic molecules, small uncharged polar molecules, large uncharged polar molecules, and ions in the phospholipid bilayer?
Hydrophobic molecules easily pass through the bilayer, while small uncharged polar molecules can also cross but more slowly. Large uncharged polar molecules and ions generally cannot pass through without assistance.
What are the special amino acids (C, G, P)?
cysteine (can form disulfide bonds, be phosphorylated)
glycine (only AA with proton as side chain, allows for tight packing within polypeptide)
profile (cyclic side chain leads to kinks and can undergo isomerization)
Which amino acids are the most major phosphorylation sites?
Ser, Thr, Tyr
What are kinases and phosphatases?
kinases add phosphate groups, phosphatases remove phosphate groups
What are the 5 protein structures?
primary structure: linear sequence of amino acids (peptide bond)
secondary structure: a helices, b sheets (h-bond)
tertiary strucure: peptide 3D shape
quaternary structure: association between multipeptide complex
supramolecular complex: consisting of tens to hundreds of subunits
What is coiled coil motif?
A coiled coil motif is a structural feature in proteins formed by the winding of two or more alpha helices around each other, with a hydrophobic residue at position 1 and 4. building block for spike proteins
What is the EF hand/helix-loop-helix motif
A common protein structural motif characterized by a helix-loop-helix configuration, often involved in calcium binding and regulation in various proteins.
What is a zinc-finger motif?
A zinc-finger motif is a structural domain in proteins that binds zinc ions to stabilize its fold, typically consisting of short beta strands and alpha helices, commonly involved in DNA binding and transcription regulation.
Difference between cytosol and cytoplasm?
The cytosol is the liquid component of the cytoplasm, excluding organelles and other insoluble materials, while the cytoplasm includes both the cytosol and the organelles suspended within it
Which organelles have a single bilayer of lipids?
ER, ERGIC, Golgi, TGN, PM, Endo-lysosomal system, peroxisomes
Which organelles have 2 bilayers?
Nucleus and mitochondria
Which organelle has a monolayer?
Lipid Droplets
What is an evolutionary idea as to how organelles formed?
infolding of plasma membrane → ER, nucleus
endocytosis of heterotrophic prokaryote → mitochondria
from a membrane to membrane: organelles exchange content → branches into other organelles
How does the inheritance of chromosomes differ from organelles?
chromosome inheritance is highly organized, and organelle inheritance is more variable and not guided
Contribution of organelles to cell membrane
rough + smooth ER > mitochondria > Golgi > plasma > everything else
Contribution of organelles to total volume
cytosol (50-60%) > mitochondria (20%) > rough ER (10%) > smooth ER (6%) > nucleus (6%) > peroxisome, lysosome, endosome (3%)
What is the function of the rough ER?
main manufacturer for protein synthesis, starting point of secretory pathway, place where protein undergoes folding + post translational mods, protein quality control
What is the function of the smooth ER?
lipid synthesis, cellular detoxification (p450), calcium ion storage
What is the function of the ER-Golgi Intermediate Compartment (ERGIC)?
involved in the transport and processing of proteins and lipids between the endoplasmic reticulum and the Golgi apparatus
What is the function of the Golgi apparatus?
sorting hub, lipid synthesis and transport, remodeling of n-glycans, addition of o glycans, lipidation of proteins,
What is the function of the trans-Golgi network?
shipping + sorting station
What is the function of the plasma membrane?
physical barrier with selective permeability, fluid mosaic model, transport of solutes and macromolecules, endo/phago/exocytosis
What is the function of endo-lysosomal system?
endocytosis/phagocytosis/autopagy (engulfs damaged organelles). low pH activates digestive enzymes
What is the function of the nucleus?
defines eukaryotic cell, storage of genetic material, separates transcription from translation
What is the function of mitochondria?
produce ATP, regulate metabolic activity, involved in apoptosis
What is the function of lipid droplets?
store lipids, connected to secretory pathway
What is the function of peroxisomes?
generation/scavenging of reactive oxygen species from oxygen, breakdown of long chain lipids, D-amino acids, biosynthesis of special membrane lipids, pentose phosphate pathway
What are differences between the structures of plant and animal cells?
Plant cells have cell walls, chloroplasts, large central vacuoles, and plasmodesmata. Animal cells have microvilli
What is George Palade’s experiment about?
tracked the cellular pathway of proteins using radioactive labeling, and SDS-PAGE, revealing the role of the rough endoplasmic reticulum in protein synthesis and secretion.
Who discovered the signal sequence hypothesis and what is it?
Günter Blobel: explained how proteins are directed to their correct cellular destinations by specific sequences within their polypeptide chains
What is the pulse-chase experiment
incubate purified ER membrane briefly with radiolabeled amino acids. isolate microsome with bound ribosome. treat purified microsome with protease: -detergent → protein in ER are protected from protease. +detergent → dissolved ER membrane, protein was digested. conclusion: newly made proteins are inside lumen of rough ER after synthesis
Mechanism of cotranslational translocation (proteins made in the ER lumen)
N-terminal ERSS emerges from ribosome
SRP binds to SS - pause synthesis
ribosome-SRP-SS complex binds to SRPr on membrane
GTP binds to SRP-SRPr
conformational change of translocon → opens, protein enters translocon
GTP hydrolyzed by SRP-SRPr and it dissociates
protein elongation. SS cleaved by signal peptidase
conclude translation, translocon close
3 ways translocons preserve ER membrane integrity
protein plug
hydrophobic pore ring
lateral gate
Type I ER membrane protein
C in cytosol, N in lumen. has terminal SS. can be insulin, LDL, GH receptor
Type II ER membrane protein
C in lumen, N in cytosol. Lacks an N-terminal signal sequence. can be transferrin receptor, golgi ftransferase/ferase
For ER membrane proteins, which is the + sequence in the cytosol or the lumen?
cytosol (II, III)
Tail anchored proteins
are anchored to the membrane by a hydrophobic C terminus, no terminal N SS.
three proteins bind to hydrophobic C tail and transfer to Get3-ATP
protein + Get3-ATP binds to Get1/Get2 receptor on ER membrane
Get3-ATP hydrolysis releases tail anchored protein into membrane
ADP leaves Get3 and ATP is replenished
GPI anchored proteins
protein with C already in membrane is broken off by GPI transamidase in the luman side and transferred to preformed GPI anchor
Type III ER membrane protein
C in cytosol, N in lumen. no terminal SS. cytochrome P450
What amino acid is N-glycan added to
Asparagine
What is the sequence of N-glycans
Asn - Xaa - Ser/Thr
Mechanism for the synthesis of N-glycan precurser
dilichol phosphate + 2 GlcNAc + 5 mannose on cytosol side
flipped to lumen side
+ 4 mannose residues + 3 glucose
What is the processing of the N-glycans when adding to target proteins?
minus 3 glucose one each until 1 being added/removed is cycled, last structure is -1 mannose
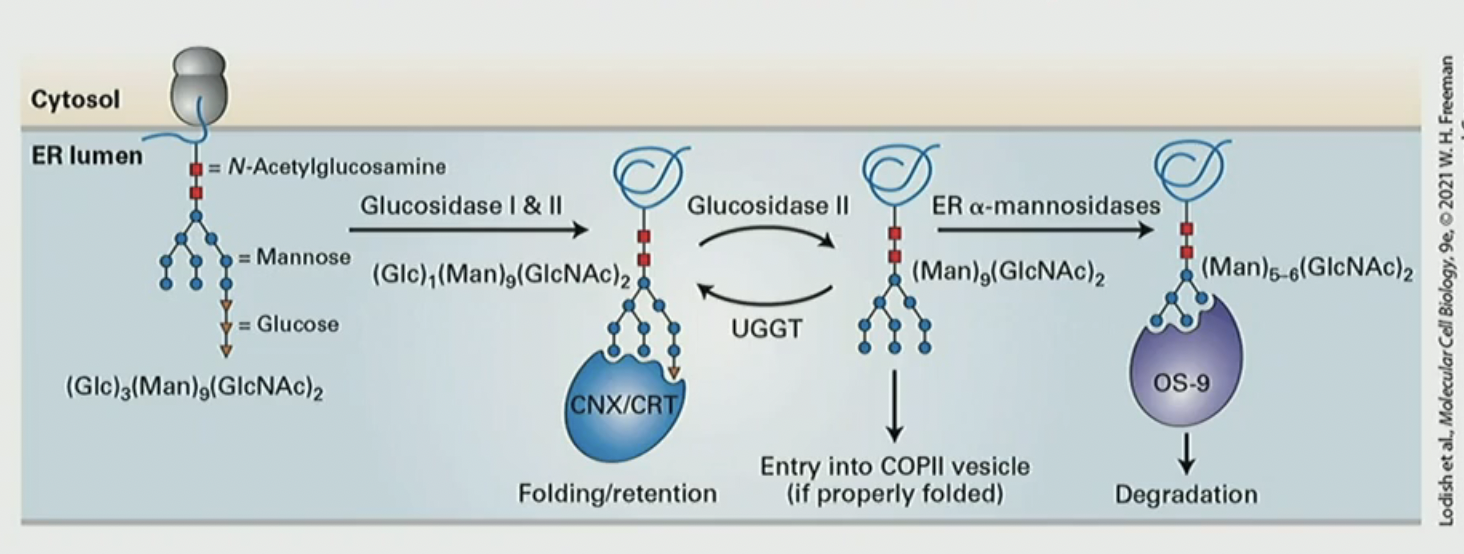
What is the calnexin (CNX)/calreticulin (CRT) cycle?
a quality control mechanism in the ER that assists in the proper folding of N-glycan and buys time for the polypeptide to fold. CNX/CRT repeatedly binds and releases the terminal glucose on N-glycan. after glucosidase II removes glucose. then UGGT adds CNX/CRT again. for degradation, mannosidases remove mannose until there’s 5-6 left OS-9 degrades
Mechanism for PDI (protein disulfide isomerase)
reduced substrate protein comes out of translocon
oxidized PDI with thiols locates thiols on protein
reaction makes reduced PDI and oxidized substrate protein with intramolecular S-S bond
Oxidized Ero1 oxidizes PDI

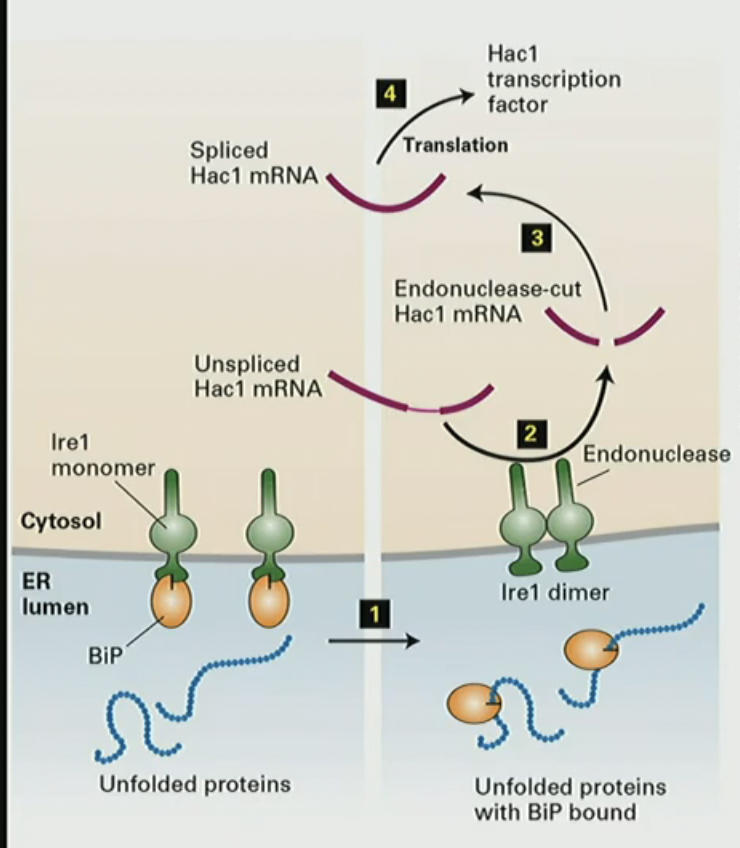
Mechanism for UPR (unfolded protein response)
unfolded protein in lumen bind to BiPs
BiP leaves Ire1 monomer its attached to to help protein fold
Ire1 becomes a dimer
Ire1 dimer cuts unspliced Hac1 mRNA
translation of Hac1 mRNA
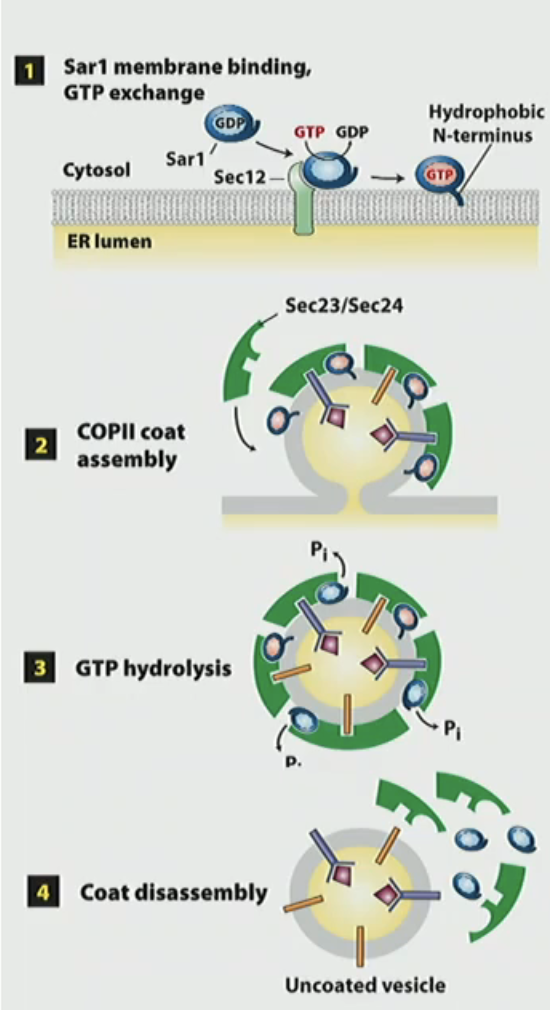
What the requirements for vesicle trafficking
machinery to attract vesicle coat
coat proteins
uncoating of carrier
machinery for fusion
What turns GTPase off?
GAP (GTPase Activating Protein), hydrolyzes GTP into GDP
What activates GTPase?
GEF (Guanine Nucleotide Exchange Factor), exchanges of GDP for GTP
COPII assembly/disassembly mechanism
Sar1-GDP binds to Sec12-GEF on membrane to convert to Sar1-GTP. Sar1 has a conformational change and anchors into membrane
Sec23/Sec24 + Sec13/Sec31 are recruited to make coat protein complex and becomes vesicle
Sec23 GAP hydrolyzes Sar1 GTP
Sar1-GDP is released from vesicle and causes disassembly of coat
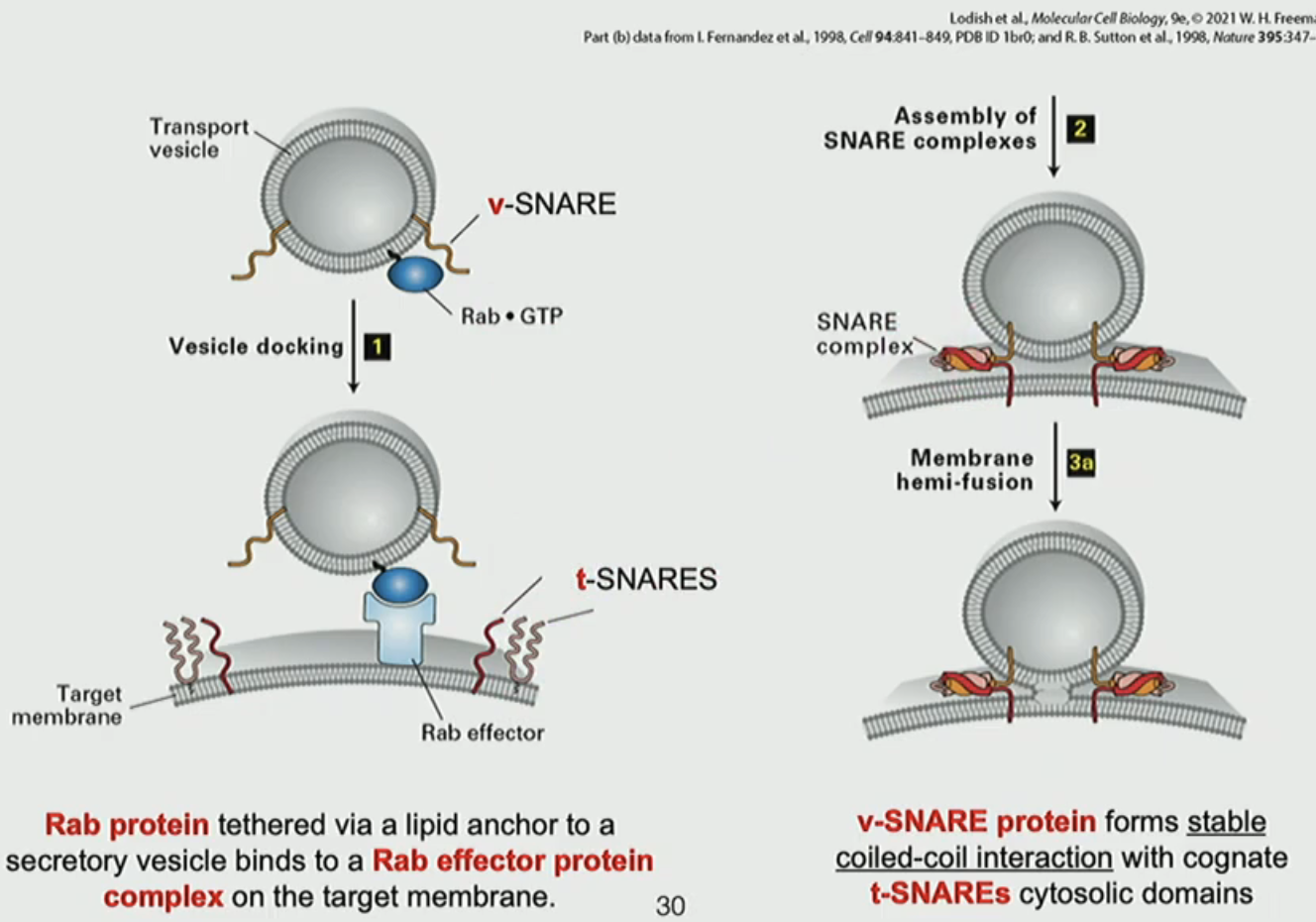
COPII fusion mechanism
Rab1 on COPII vesicles bind to Rab effects on target membrane to melt through “cocoon”
v-SNARE protein on COPII interacts with t-SNARE on membrane to form stable coiled-coil
fusion of vesicle to membrane
NSF ATPase separates 4 helix bundle
COPI assembly/disassembly mechanism
Arf1-GDP binds to p23/p24 ER membrane protein
GBF1 GEF converts to Arf1-GTP to activate and Arf1 has conformational change
Arf1-GTP recruits coatomer (heptameric protein) and begins curving ERGIC/Golgi membrane
Coatomer disassembles when Arf1 is hydrolyzed to Arf1-GDP, releasing the vesicle
What is KDEL?
KDEL is a C-terminal signal sequence found on proteins that ensures they remain in the ER, preventing their transport to the Golgi apparatus. It consists of the amino acids K, D, E, L
Mechanism for remodeling of N-glycan in Golgi apparatus
cis: - 3 mannose
medial: + 1 GlcNAc, - 2 mannose, + 2 GlcNAc, + 1 fucose
trans: + galactose, + 3 NANA
What kinds of lipids are at the cis vs trans Golgi?
cis: PC
trans: sphingolipid + cholesterol
Are retrograde signals acidic or basic?
basic
Are anterograde signals acidic or basic?
acidic and hydrophobic
Which direction does COPI and COPII go
COPII: ER → Golgi (anterograde)
COPI: Golgi → ER (retrograde)
Which mechanism do the cargo-binding proteins ERGIC-53 and the mannose-6 phosphate (M6Pr) have in common?
pH dependent cargo release. reduced pH in target membrane triggers vesicle release
TGN: What are the 5 main types of cargo and their destinations?
retrograde transport → trans Golgi
lysosomal enzyme → lysosome
lysosomal enzyme → late endosome
constitutive secretory protein (PM/ECM protein)
regulated secretory protein (hormone, enzyme, NT)
Which protein is present in regulated exocytosis but not in constitutive exocytosis?
synaptotagmin-1
What does M6P do and what is its mechanism?
M6P (mannose 6 phosphate) is a tag that signals to the TGN that it is going to the lysosome.
1. in TGN, M6P binds to M6Pr
2. CCV will bud off with M6Pr and carry to the late endosome
3. M6Pr will dissociate with the acidic pH of endosome
4. endosome delivers enzymes to lysosome
What does BiP do?
it binds to exposed hydrophobic regions of the nascent chain and stabilizes them until the protein is folded correctly
Why does a receptor not directly activate an effector protein?
signal transduction proteins must amplify signal by several orders of magnitude
Name two membraneless compartments in the cytosol of humans
stress granules, synaptic densities
How are individual chromosomes spatially organized within the nucleoplasm?
they are separated in distinct, nonoverlapping compartments in the nucleus
True/false: in mitochondria, the lumen of cristae is not continuous with the intermembrane space
F, the lumen of the cristae is continuous
Are proteins targeted to peroxisomes translocated in a folded or unfolded state?
folded
Are proteins targeted to mitochondria translocated in a folded or unfolded state?
unfolded