Lecture 33: Viral Genomics/ DNA Viruses
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
Viral genome sizes vary. How much?
~1000 fold.
What is the smallest virus?
Circovirus, 1.75-kilobase single strand
What is the largest virus?
Pandoravirus, 2.5-megabase pairs
Infects some marine amoebae
Genome is larger than some bacteria
Which genomes are typically smaller RNA or DNA viruses?
RNA.
What is the Baltimore virus classification scheme based on?
Based on viral genome composition and mechanism of mRNA synthesis.
Includes seven cases.
I. dsDNA
II. ssDNA
III. dsRNA
IV. +RNA
V. -RNA
VI. Retroviruses
VII. Reverse transcribing DNA viruses
When and how did viruses first appear?
•Since viruses require a host cell for replication, viruses may have arisen after cells, ~4 billion years ago
•Therefore, viruses may be remnant cell components that evolved an ability to replicate with the help of a living cell
What is the second theory of viral evolution?
However, also hypothesized that viruses may have arisen from primitive “virocells” that contained RNA genomes; selection for reduced cell size and genomic demands then led to the evolution of viruses
Therefore, viruses might be relics of the “RNA World” (= period in evolution when RNA is hypothesized to have been the sole carrier of genetic information)
Proteomics data support which hypothesis?
virocell
Why are viruses not on the universal tree of life?
Lack of ribosomes prevents placement of viruses on the universal tree of life constructed using rRNA sequences
What does proteomics suggest about viruses?
Proteomics analyses comparing the proteomes of viruses and cells suggests viruses originated from ancient cells called “virocells” that contained segmented RNA genomes
What evidence is in support ofthe virocell hypothesis?
•These cells are hypothesized to have existed before the last universal common ancestor (LUCA) of modern cells during the era of the “RNA World”
•Evolutionary pressure for size reduction perhaps eliminated these cells, leaving a protein shell to protect the genome, and forced dependence on hosts for replication functions
•Proteomic analyses indicate RNA viruses are older than DNA viruses, with dsRNA viruses being the oldest of all
•Many dsRNA viruses contain segmented genomes like the hypothesized virocells
Why was the conversion from RNA to DNA selected?
•DNA is more stable than RNA (not cleaved as readily)
•RNA is more susceptible to spontaneous hydrolysis —>spontaneous mutation rate of RNA is much higher than DNA
•Darwinian selection would have eventually selected for cells with DNA genomes; cells with RNA genomes would have been driven to extinction
Why do RNA viruses still exist?
•The high spontaneous mutation rate of RNA allows these viruses to stay one step ahead of evolving host defenses (need different flu vaccine every year)
•RNA viruses predominate in organisms with highly evolved adaptive immune systems (e.g. animals)
Describe the phylogenetic trees of virus.
•Universal phylogenetic tree of life constructed from a combination of protein sequences and protein structural features
•Viruses end up positioned at the root of the tree, with RNA viruses preceding DNA viruses
•Tree contains a long branch leading to the three domains of cellular life, which branch out in a manner very similar to trees bases on rRNA
Majority of viruses that infect prokaryotic cells (both Bacteria and Archaea) are what kind of viruses?
dsDNA Virus
DNA viruses also infect eukaryotes; important eukaryotic DNA viruses include which one?
the DNA tumor viruses
What are some examples of single stranded DNA?
(+)ssDNA viruses infect plants, animals, and bacteria and share molecular similarities; we’ll look at ɸX174 and M13 (E. coli host)
What are some examples of dsDNA?
dsDNA bacteriophages are some of the best studied viruses (include T4 and lambda); we’ll look at Mu (E. coli host).
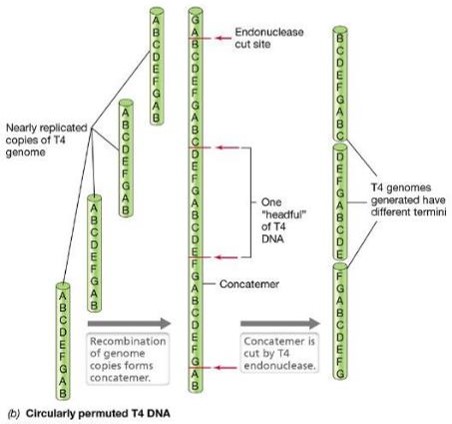
Describe T4: dsDNA linear DNA
T4 genome is first replicated as a unit, then several genomic units recombine to form a concatemer
Then endonucleases cut into individual genomes packaged into progeny viruses
What is a concatemer?
a continuous DNA molecule that contains multiple copies of the same DNA sequence linked in series
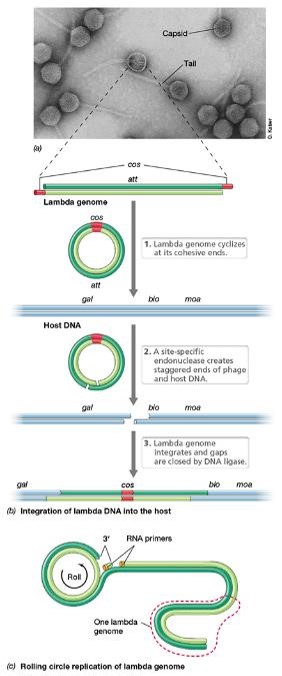
Describe bacteriophage lambda λ:
•dsDNA linear
•The genome has 12-bp complementary single-stranded regions at the 5’ end of each strand = cohesive ends
•Cohesive ends base-pair upon genome entry to form the cos site —> circularizes the linear genome
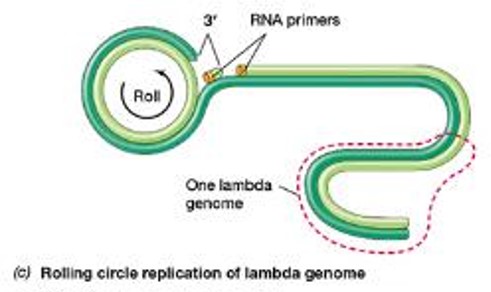
•If λ enters the lytic pathway, the genome is copied using rolling circle replication:
•One strand of the circularized phage genome is nicked
•3’ end of the nick serves as a primer, unnicked strand serves as the template for leading strand synthesis
•The leading strand is rolled out and used as a template for the synthesis of the complementary strand
The resulting concatemer is cut into genome-sized lengths at the cos sites and packaged into phage heads
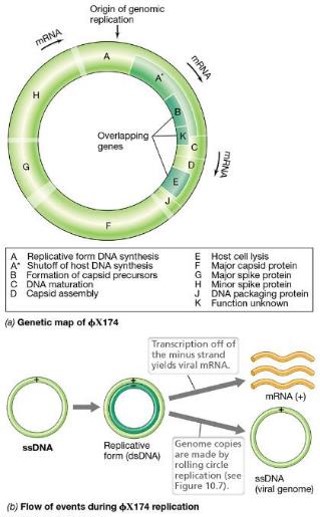
Describe Single-Stranded DNA Bacteriophage: ɸX174
•Virions consist of a circular 5386-nucleotide (+)ssDNA genome inside a tiny (25nm) icosahedral capsid
•Attachment occurs via interactions between spikes on the capsid and LPS components of the host cell’s outer membrane
Genome enters the host cell
Inside the host cell, complementary DNA strand is made —> dsDNA replicative form used as template for transcription and genome replication
Very small genome with overlapping genes; during transcription, parts of the genome are read in different reading frames to conserve genome space
what is the role of rolling circle replication in ɸX174?
Rolling circle replication produces additional genomes for progeny virions in the same manner
How does lysis occur in ss DNA ɸX174?
•Lysis occurs through inhibition of peptidoglycan synthesis —> leads to weakness in newly synthesized cell wall material —> osmolytic cell lysis —> release of progeny virions
Describe Single-Stranded DNA Bacteriophages: M13
•(+) ssDNA filamentous bacteriophage with helical symmetry; long thin virions and circular genome
•Attaches to host pilus for entry
•Filamentous phages like M13 are released from the host cell without cell lysis; infected cells continue to grow; typical plaques not formed of a lawn of infected cells
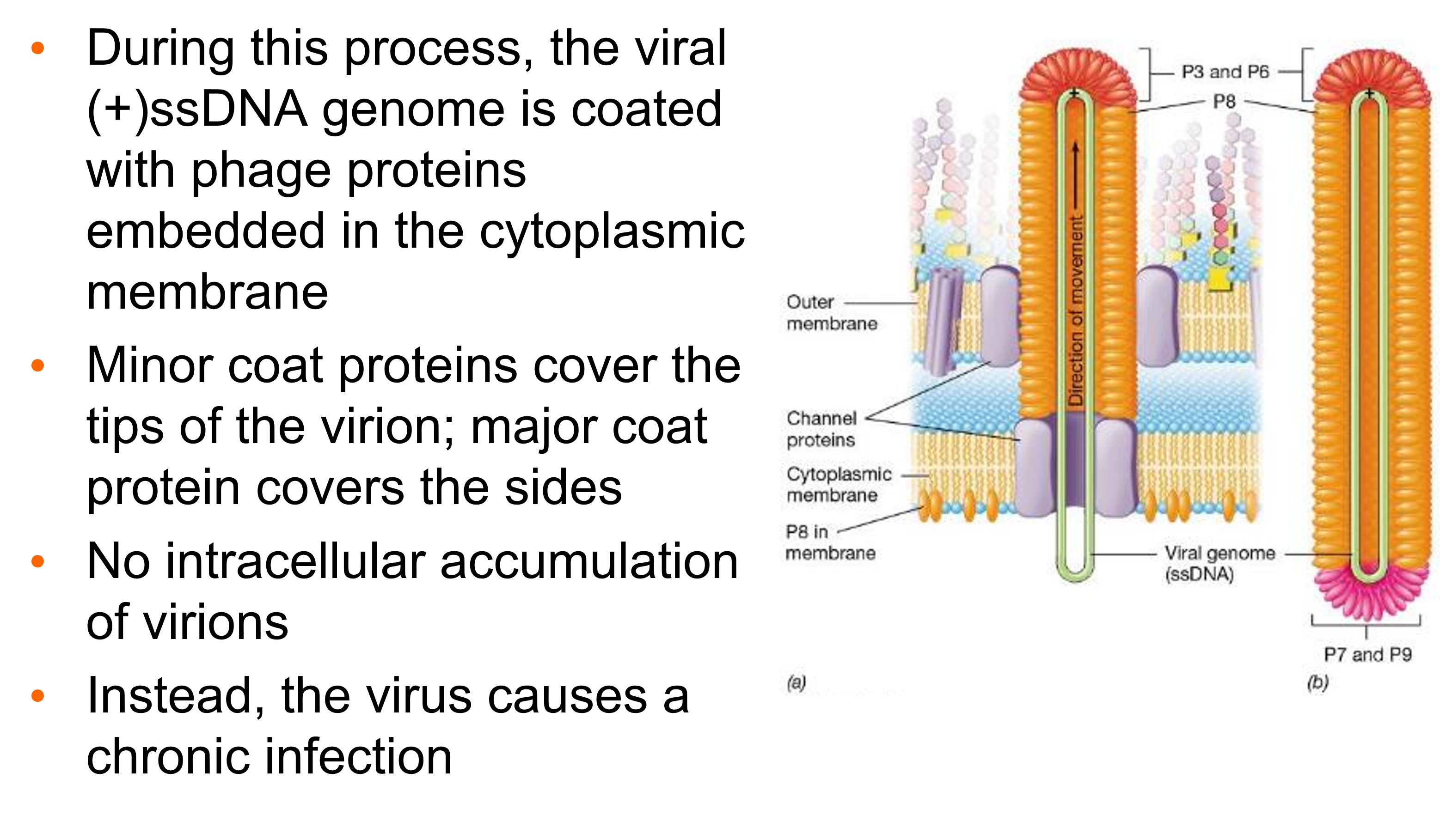
How does M13 exit? What happens during this process?
•To facilitate nonlytic exit, M13 DNA is covered with coat proteins as it exits the cell through a channel constructed from virus-encoded proteins
During this process, the viral (+)ssDNA genome is coated with phage proteins embedded in the cytoplasmic membrane
Minor coat proteins cover the tips of the virion; major coat protein covers the sides
No intracellular accumulation of virions
Instead, the virus causes a chronic infection
Describe the Mu virus.
•Phage Mu is a temperate phage
•Mu replicates by transposition
•Transposons are DNA elements that can move from one location to another in a host genome using a transposase enzyme
•Named Mu because it generates mutations when it integrates into the host genome
•Useful in bacterial genetics experiments as a way to generate mutations in the lab
•Icosahedral head, a helical tail with six tail fibers, and a linear dsDNA genome ~36.7 kbp long
•Host range is controlled by tail fibers: One type allows infection only of E. coli; other allows infection of several other enteric bacteria
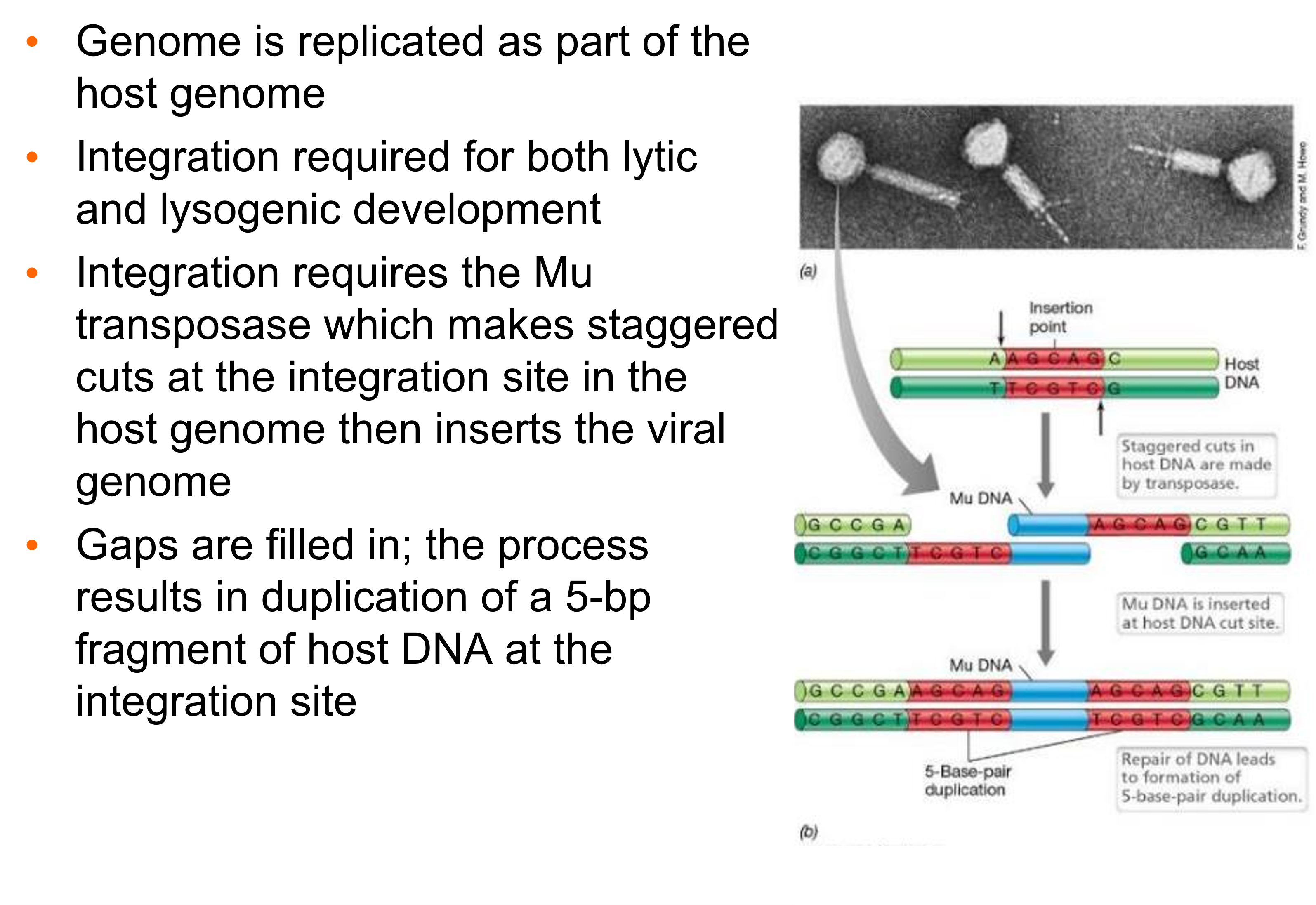
Describe how the mu genome in replicated.
•Genome is replicated as part of the host genome
•Integration required for both lytic and lysogenic development
•Integration requires the Mu transposase which makes staggered cuts at the integration site in the host genome then inserts the viral genome
•Gaps are filled in; the process results in duplication of a 5-bp fragment of host DNA at the integration site
Describe the two pathways of mu.
•Mu can enter the lytic pathway upon initial infection if its repressor is not made
•Alternatively, Mu can form a lysogen if its repressor is made
•In either case, Mu DNA is replicated by repeated replicative transposition of Mu to multiple sites on the host genome
•If the lytic cycle is triggered, only the early genes of Mu are initially transcribed, then head and tail proteins are synthesized; following self-assembly, the cell is lysed and Mu virions are released
•Lysogeny requires sufficient Mu repressor protein to prevent transcription of the integrated Mu genome