L1: Protein Sorting in the Endomembrane system: The secretory pathway
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
43 Terms
Why do we need protein exchange and sorting
variety of organelles
each with specific
ion transport, signal transduction
biosynthetic processes
need to maintain their protein compositions
even though they are constantly exchaning membrane lipids and proteins
So need this sorting to generate and to maintain compartment identity
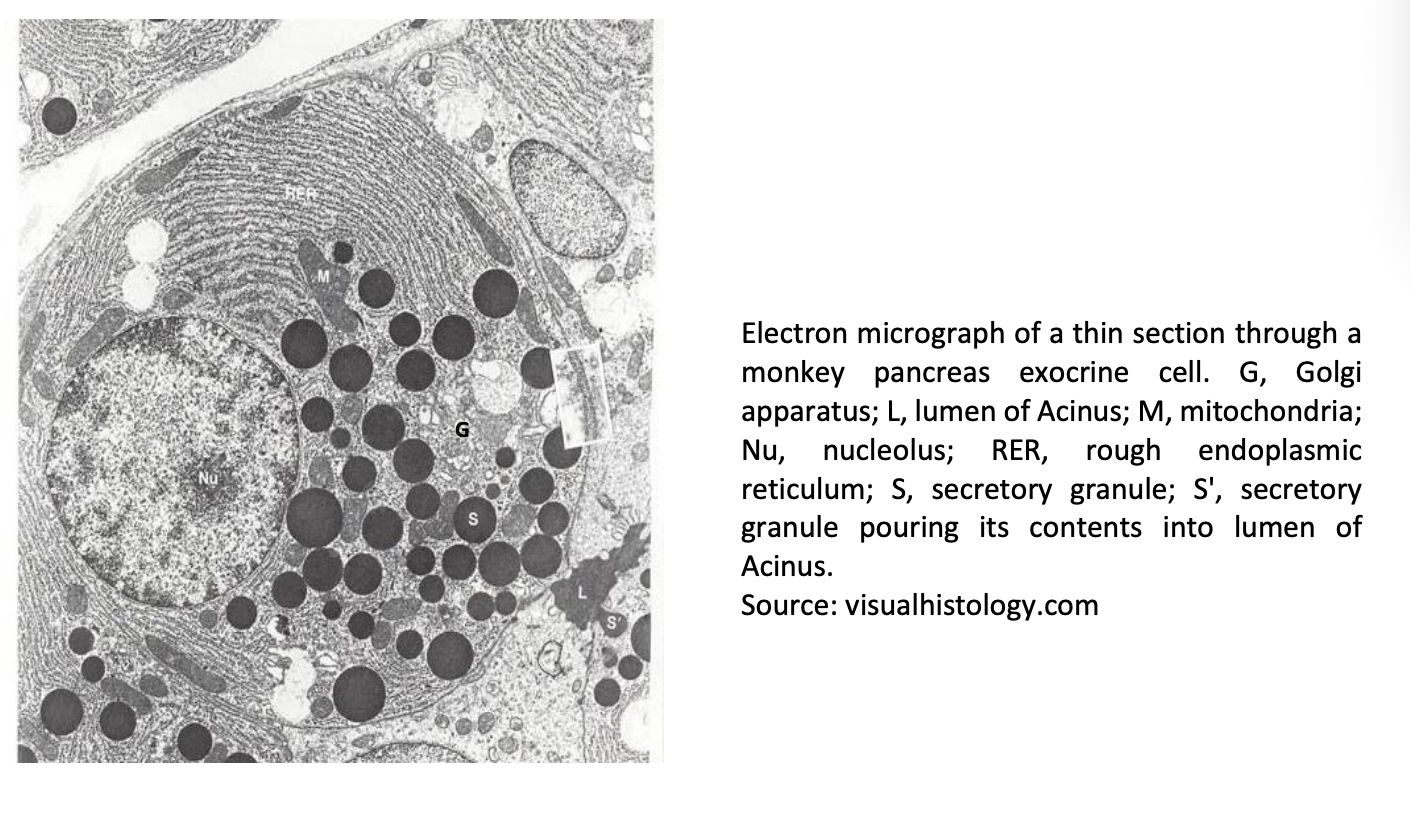
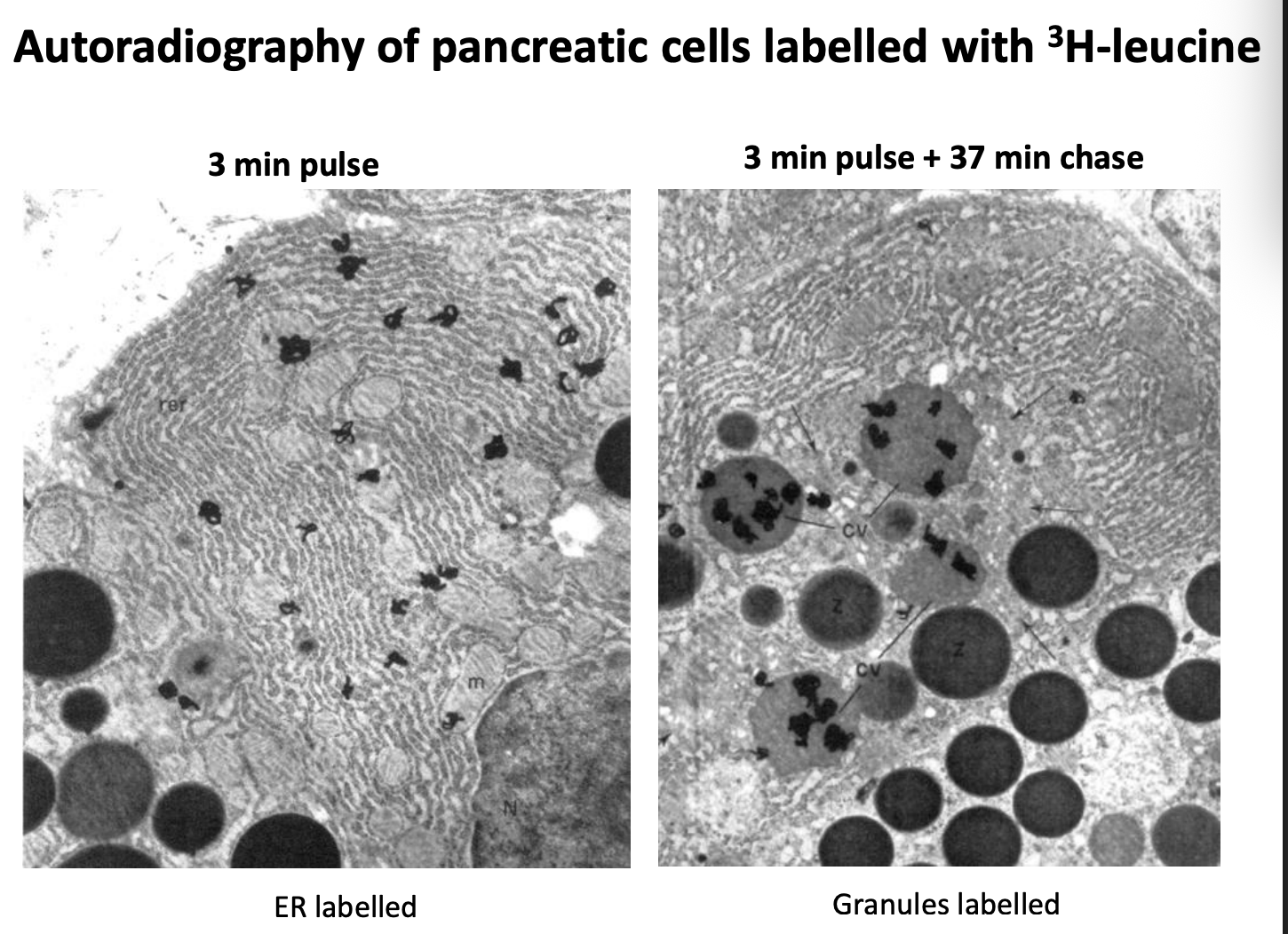
Em thin section through a monkey pancreas exocrine cell
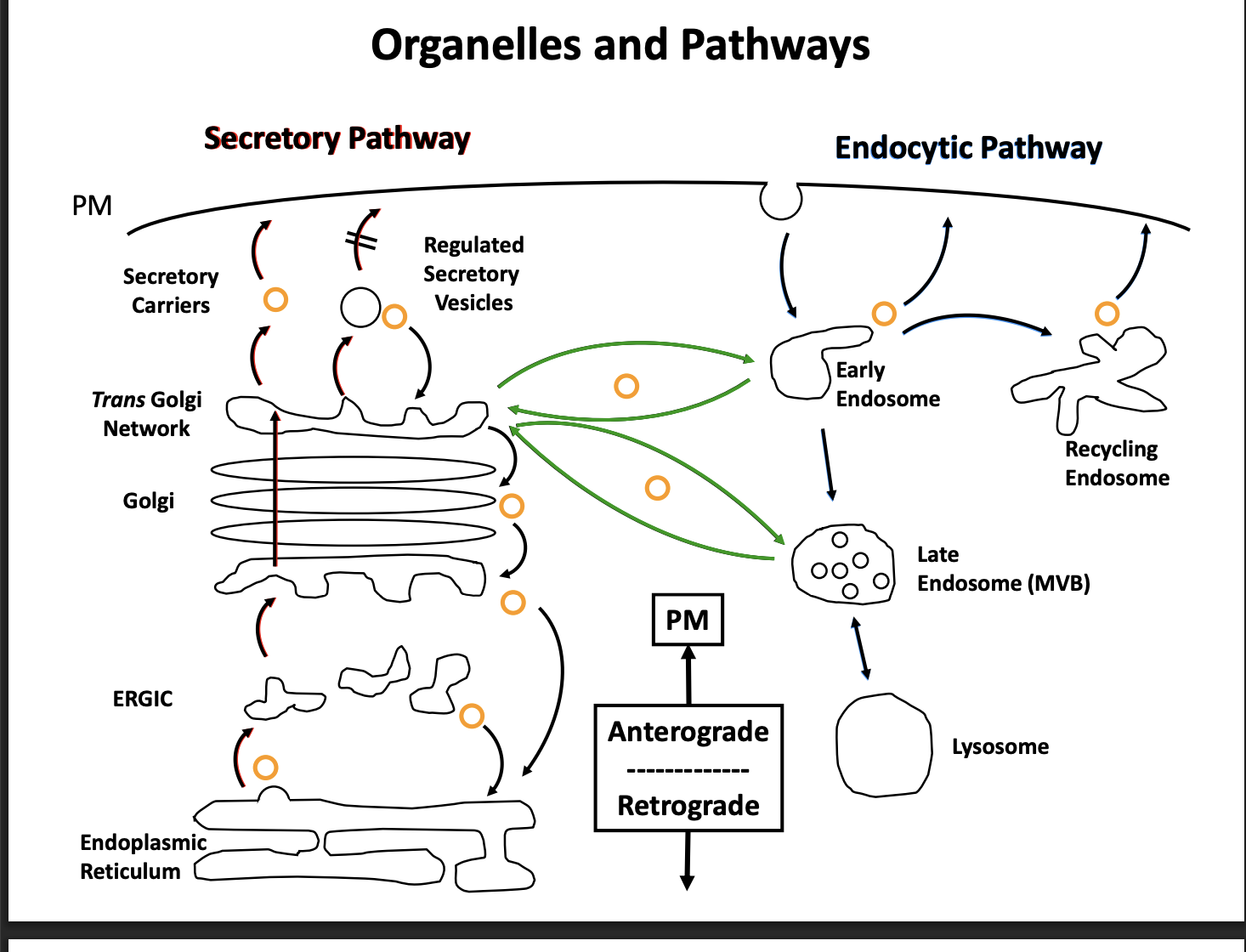
What are the defined routes
Secretory (talked about in this lecture) (aka biosynthetic pathway)
takes newly synthesied protein
ER→ Golgi→ Plasma membrane
Endocytotic
first: internalisation of extracellular material (endocytosis)
then: series of endosomal compartment to the lysosome (or vacuole in fungi and plants)
Are they completely separate pathways?
No
connected via tran-sgolgi network and endosomes
many tracfficking routes are bi-diretional
anterograde and retrograde transport
Why are many trafficking route bi-directional
1. Maintain membrane homeostasis
must balance retro and anterograde traffic
other wise ER would get smaller and smaller
Help with the recycling of the protein
How can you map the secretory pathway: 1
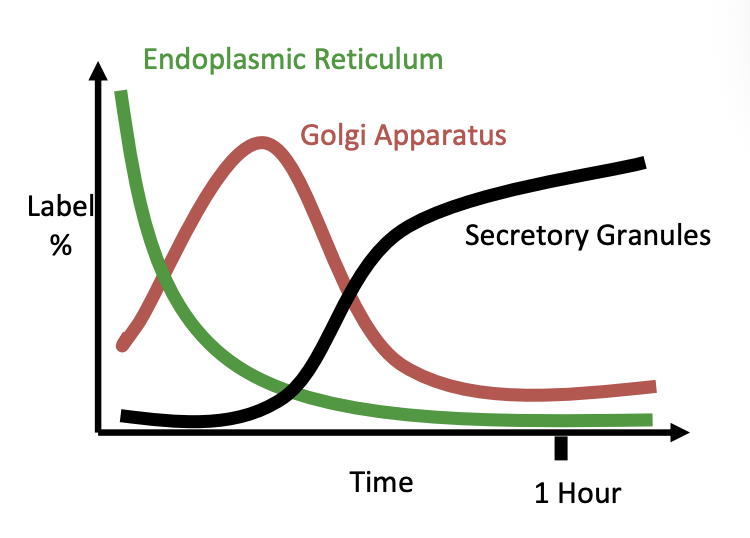
Pulse-Chase Approach
Palade

How does the pulse-chase approach work and what did it show
pancreatic cells incubated with radioactive amino acids for a few mins→ pulse
Cells then incubated in unlabelled medium for variable lengths of time
Analyse by
autoradiography
EM
Results:
Labelled proteins detectable in the ER
then the Golgi
then the secretory granules
Therefore: shows the pathway/ mapping the secretory pathway
The graph shows the autoradiography graph
shows how as decreases from Er→ goes to Golgi and then eventually to the seretory granules
EM of H³-Leucine
New way of mapping the secretory pathway by pulse chase
Live cell imaging of trafficking→ visualsing the movement as more of a wave than just at fixed time points
Use fluorescent proteins→ GFP
or
Use fluorescent lipids
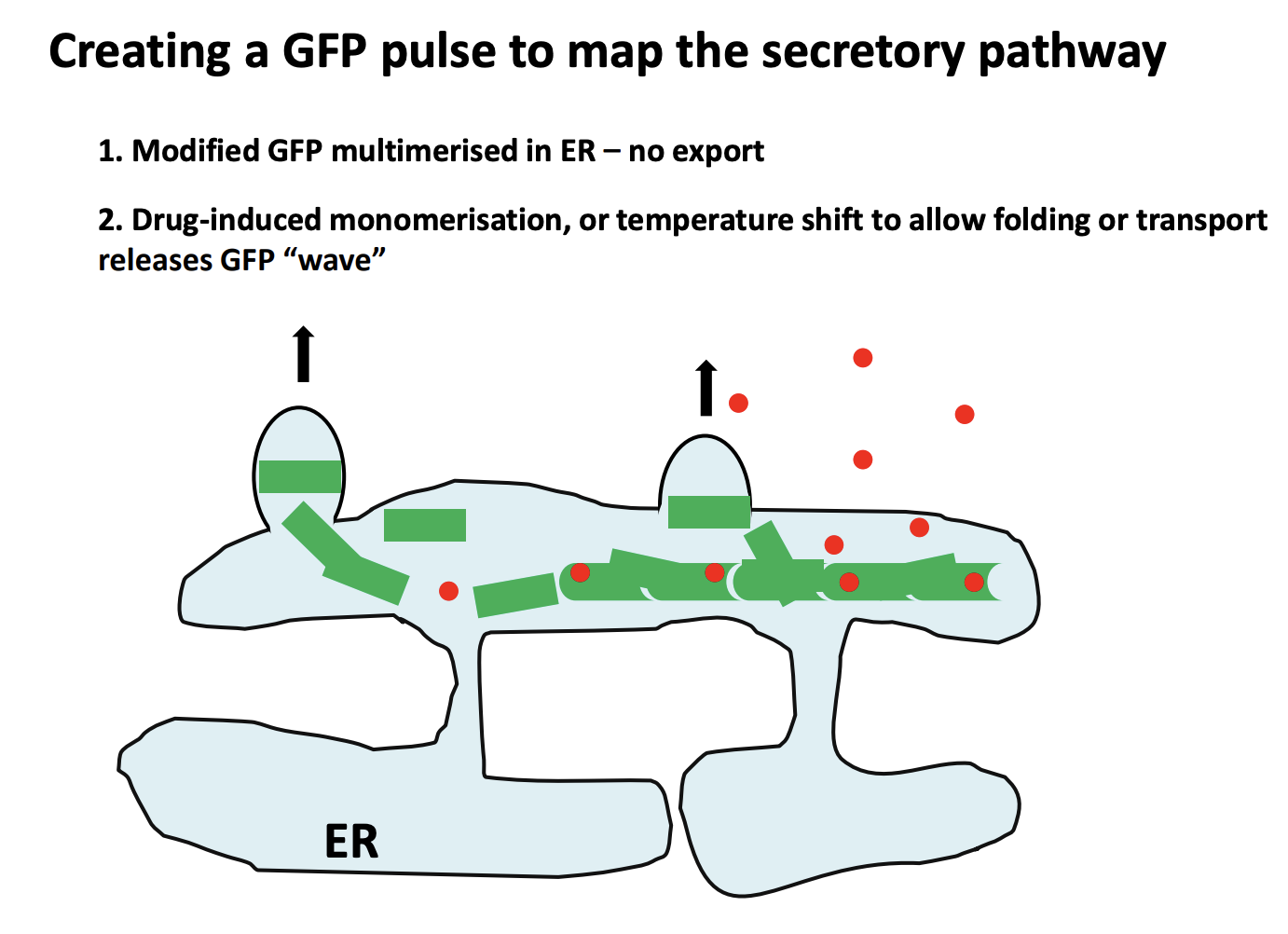
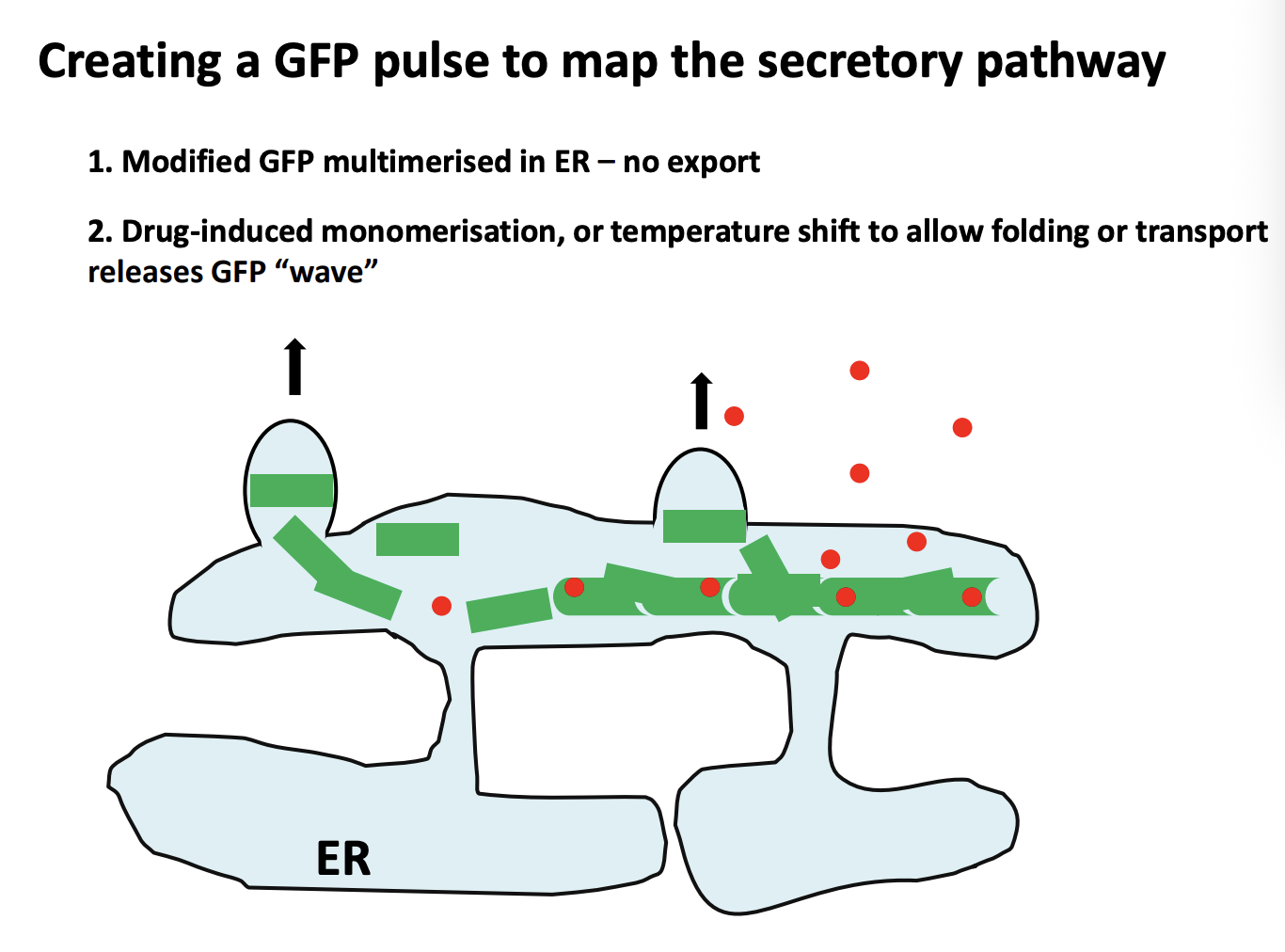
How to create a GFP pulse to map the secretory pathway
Modified GFP multimerised with multimerisation factor in ER→ stops it from leaving the ER→no export
Next, borken up by Drug-induced monomerisation or temp shift to allow folding or transport releases GFP ‘wave’
allows the proteins to move further in the cell→ can then be visualised as a wave
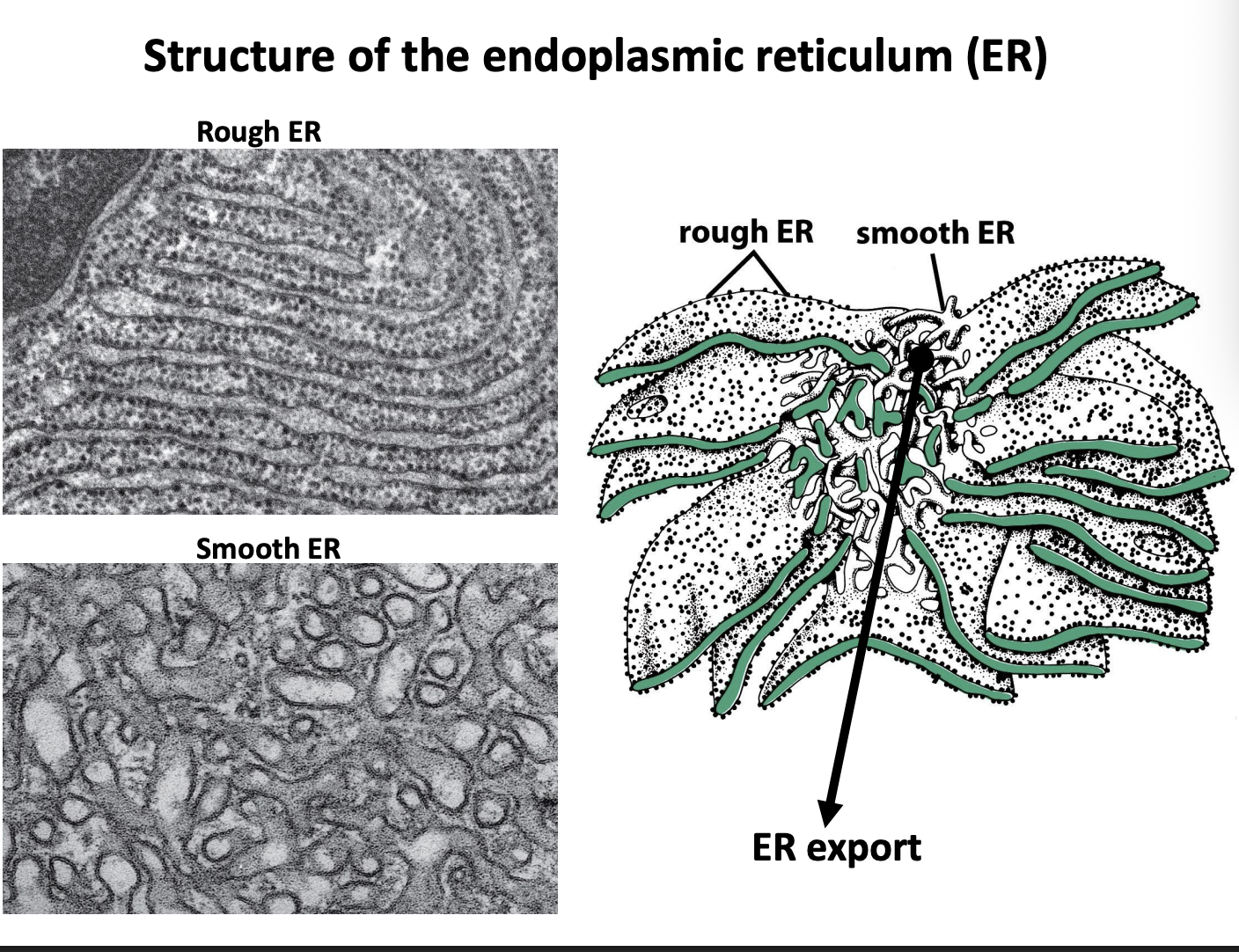
Strucuture of the endoplasmic reticulum
Two functionally and morpholgically distinct domains
Rough
sheet-like
studded with protein-synthesisng ribosomes
Smooth
tubular
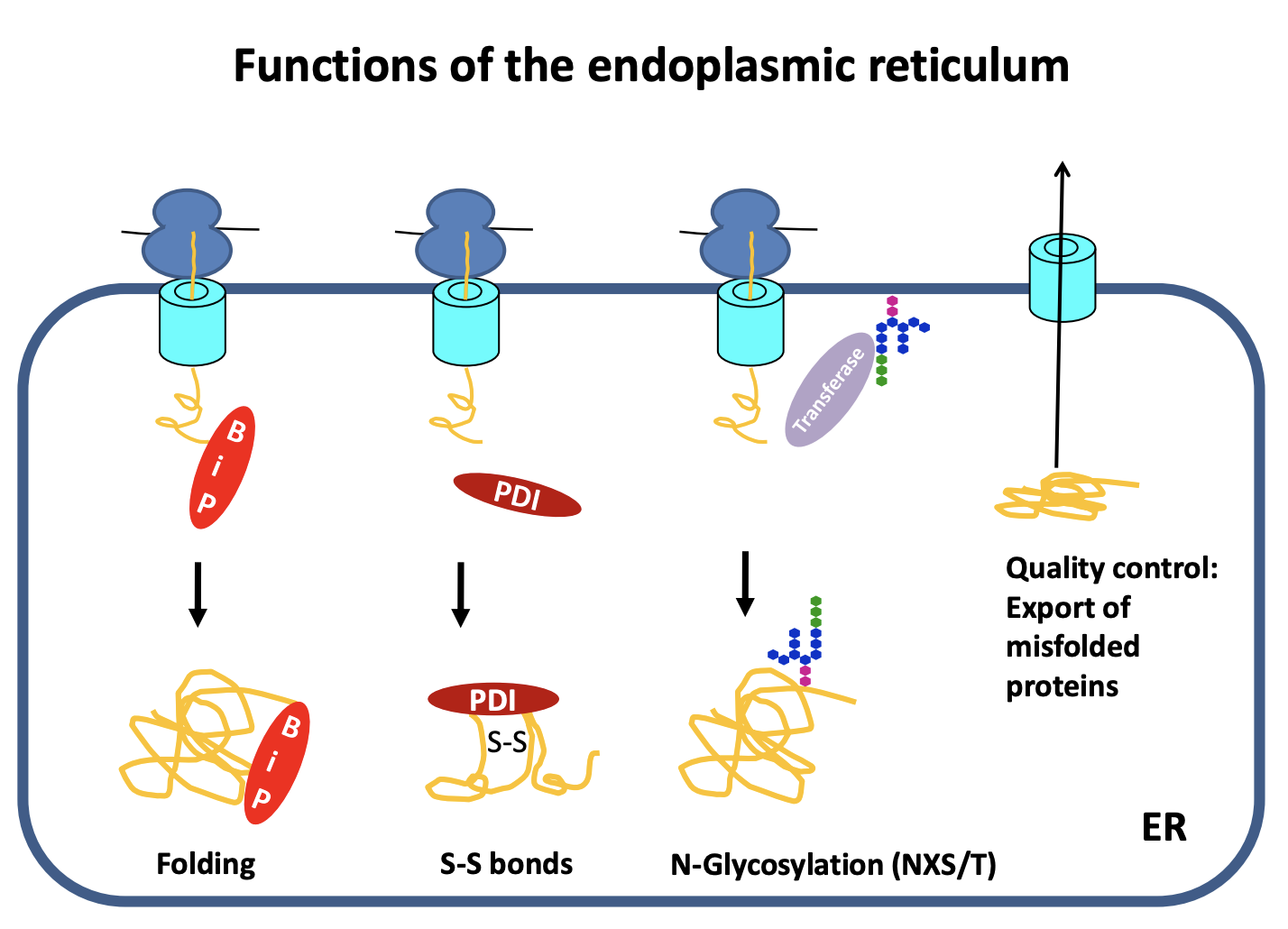
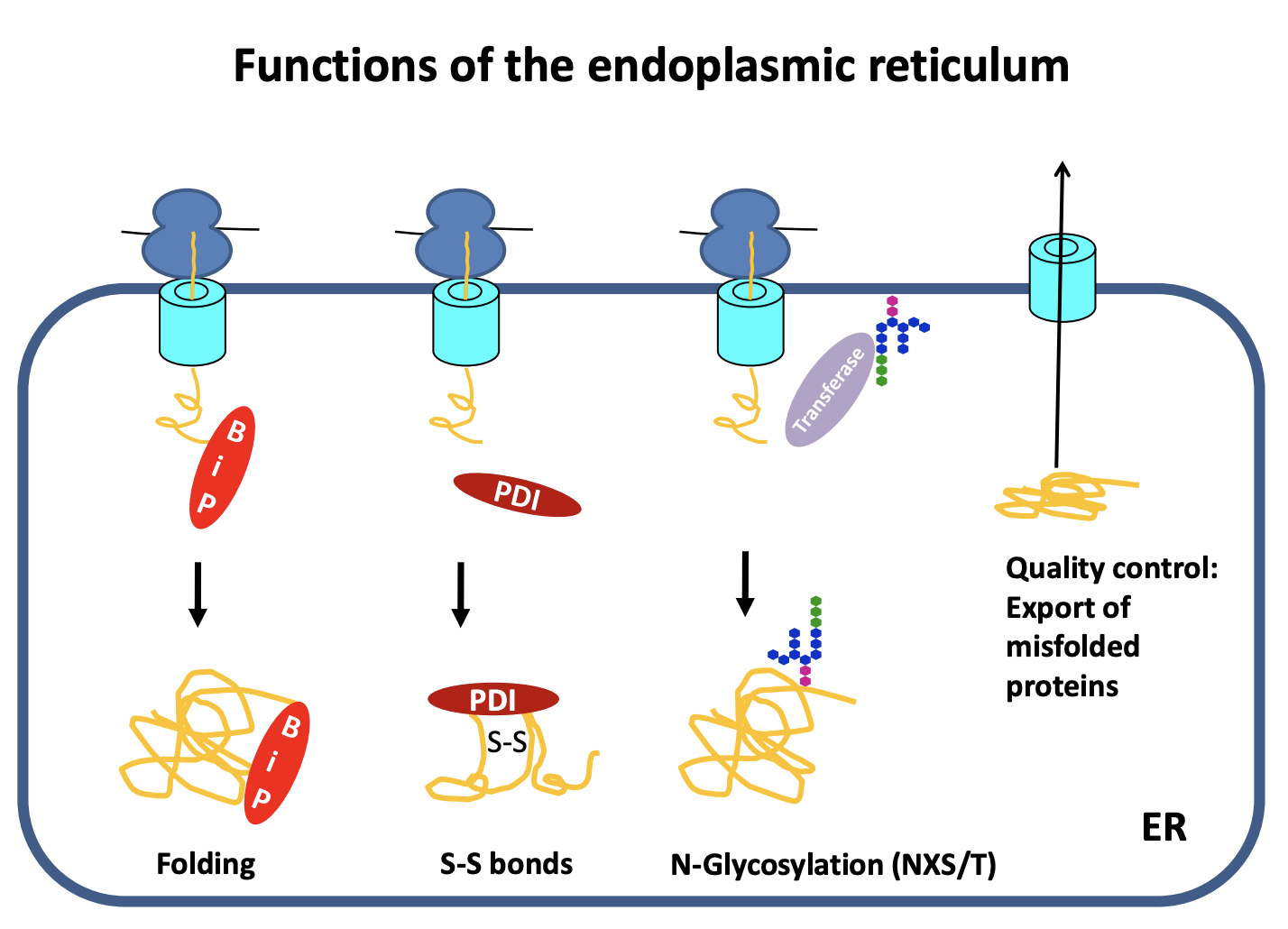
Functions of endoplasmic reticulum
Main site of protein biosynthesis
Protein folding
general
with S-S bonds
Post-translational modifications→ Protein Glycolysation
Quality control
Protein folding
General folding
need chaperone to fold→ Binding Protein ‘BiP’ (e.g Hsp70)
folds and prevents aggregation
Proteins with S-S
In the cytosol→ reducing environemnt→ the S-S forms and folds the protein find
In the cell→ oxidising comparment
sulfydryl groups on cystine reduces
and forms covaneltn bonds
assisted by→ Protein disulfide isomerases (PDI)
helps to cut and rejoin the S-S once it is in a new environement
Glycosylated
invariant glycan is transferred to asparagines (N) of the consensus NXS/T
note: X is any amino acid except proline S and T threonine
transferred in a single step
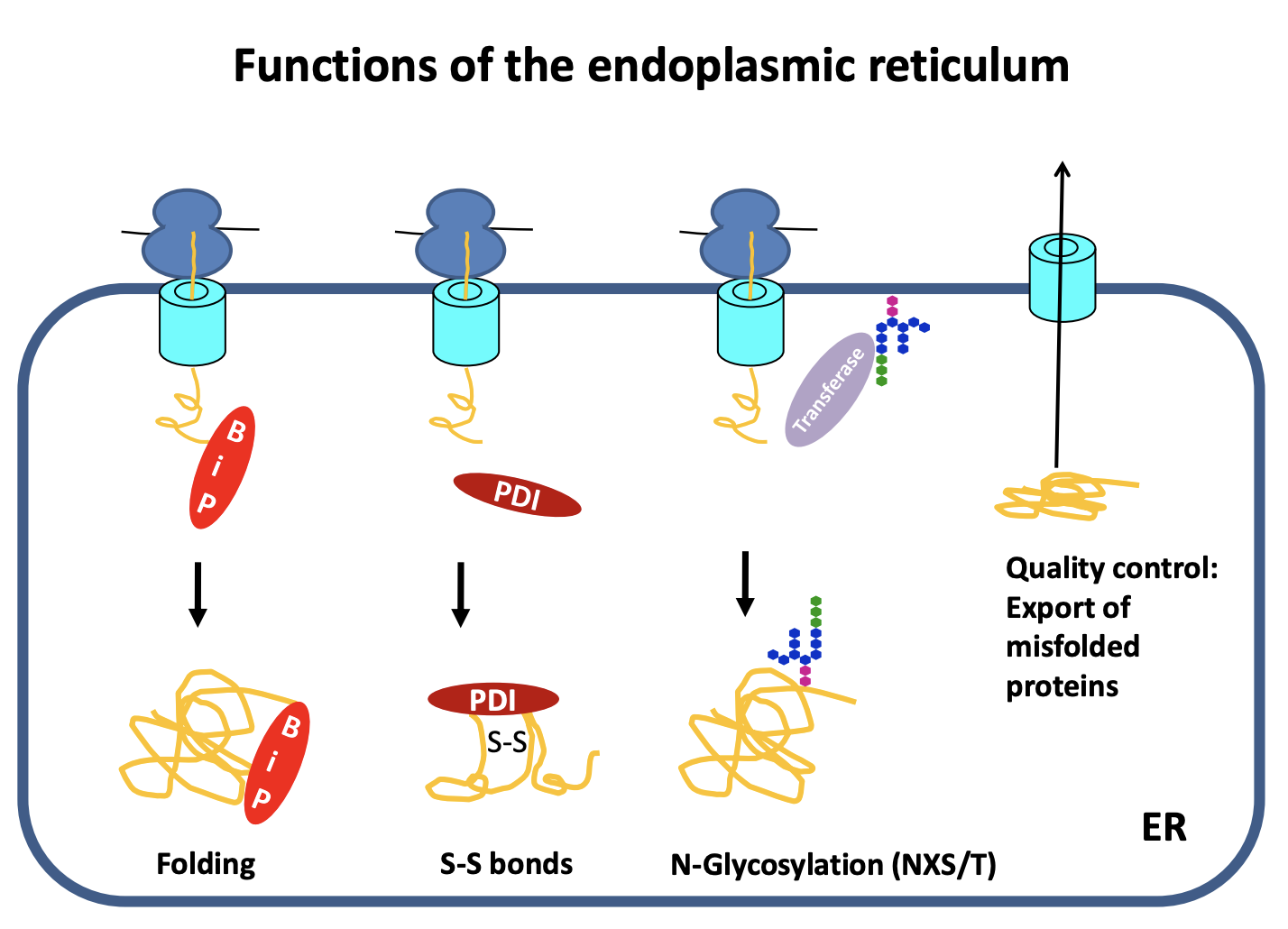
What does glycosylation help do
folding
glycan is hydrophilic and so ensures that the hydrophobic residues are folded into the inside of the protein
further assists chaperone binding
e.g calnexin
Signals the next step→ quality control
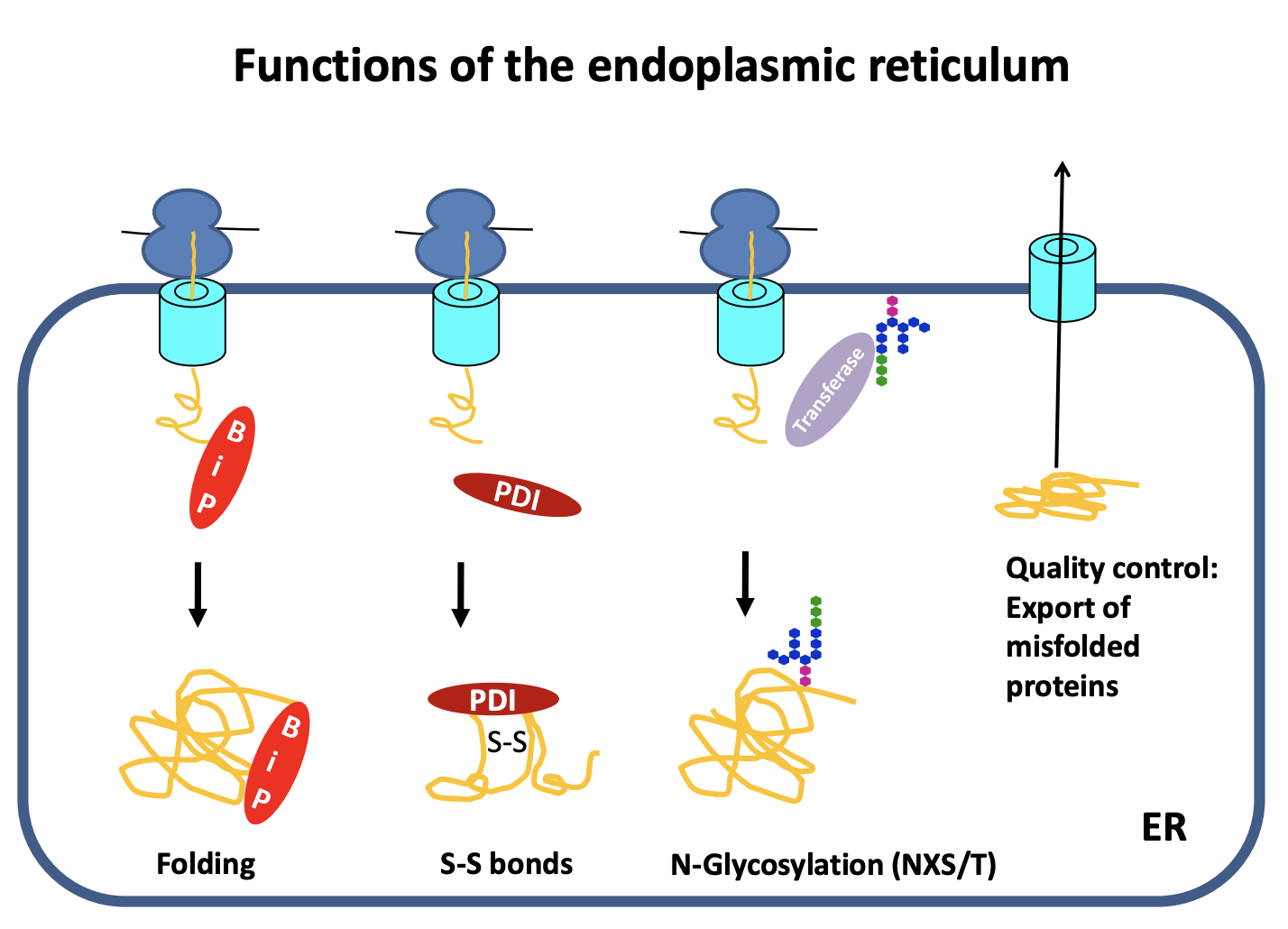
Quality control
ensures only correctly folded proteins are shipped to the golgi
what happens to the others:
exported to the cytosol→ degraded
ER export
Protein made in the rough ER
Moves to the smooth ER
Move to the specialised ER exit sites
Cargo is selectively packed into COPII (coat protein II) vesicles
COPII homotypic fusion (fuse with each other)
form vesicular tubular clusters (VTCs)
These can subsequently undergo further homotypic fusion events
RETROGRADE: This is the first compartment for recyling proteins
proteins that escaped from the ER
trafficking machinery itself
This is with COPI vesicles
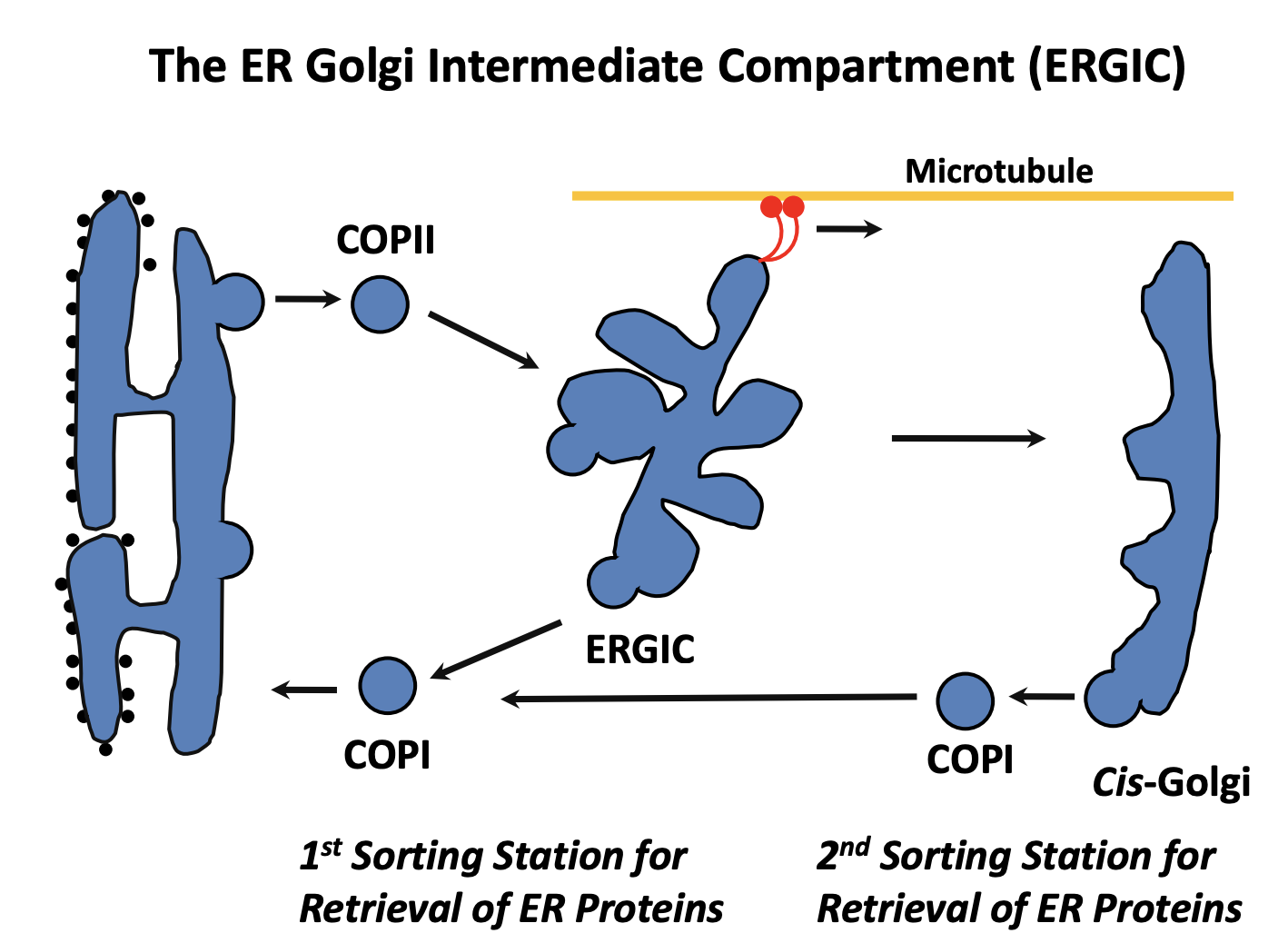
What are the vesicular tubular clusters (VTCs) also known as
ER-Golgi intermediate compartment (ERGIC)
What happens next for the ERGIC itself
becomes attached to MT via dynein motors
Pulled towards the golgi
ERGIC may either
fuse with an existing cis-cisterna
undergo homotypic fusion to form new cis-cisterna
note: this depends on the two models see after
SECOND RETROGRADE: COP1 from cis-golgi back to the ER
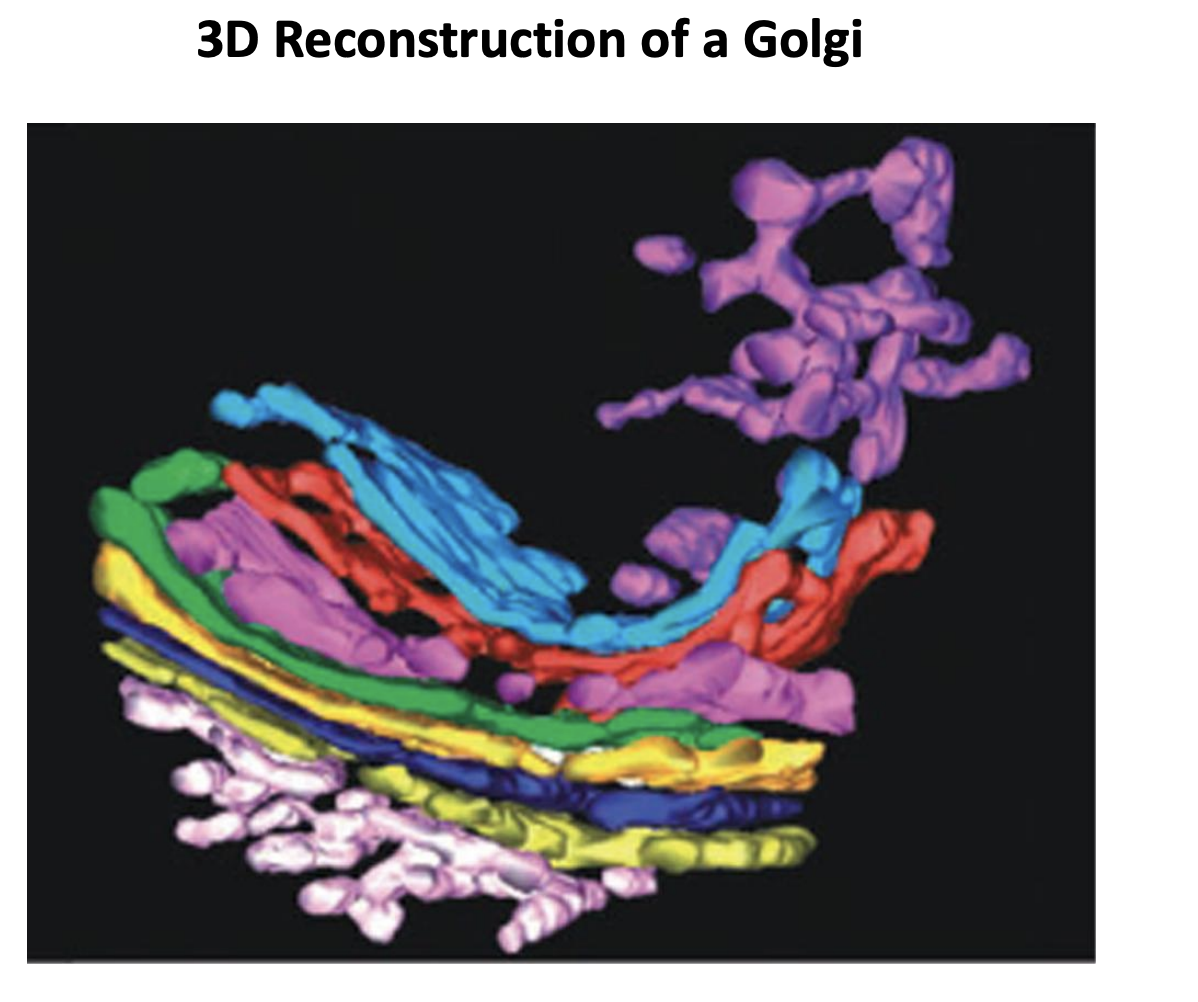
Golgi Apparatus what is it
Found next to the nuclear envelope
stack of flattened fenestrated (with holes) membrane sacks (cisternae)
each with its own lumen
number of cisternae→ variable
depends on how much protein secretion the cell does
6 or 3 or 20
Have polarity:
Cis→ medial→ trans
cis (closest to the nuclear envelope)
with different functions and partly distinct protein and lipid complements
In which direction do the proteins traverse the golgi
cis to tans direction
What is the cis-golgi-network
area near the cis-most cisterna
where ERGICs fuse
Cis and trans are network
cis is more tubular
with fenestrations
transport and fasiculations
no network in the medial cisternae
What is the point of going through the golgi
Post-translational protein modification
maturation and sorting
What modifications take place
trimming
addition or extension of attached glycans
sugar phosphorylation
proteolytic cleavage
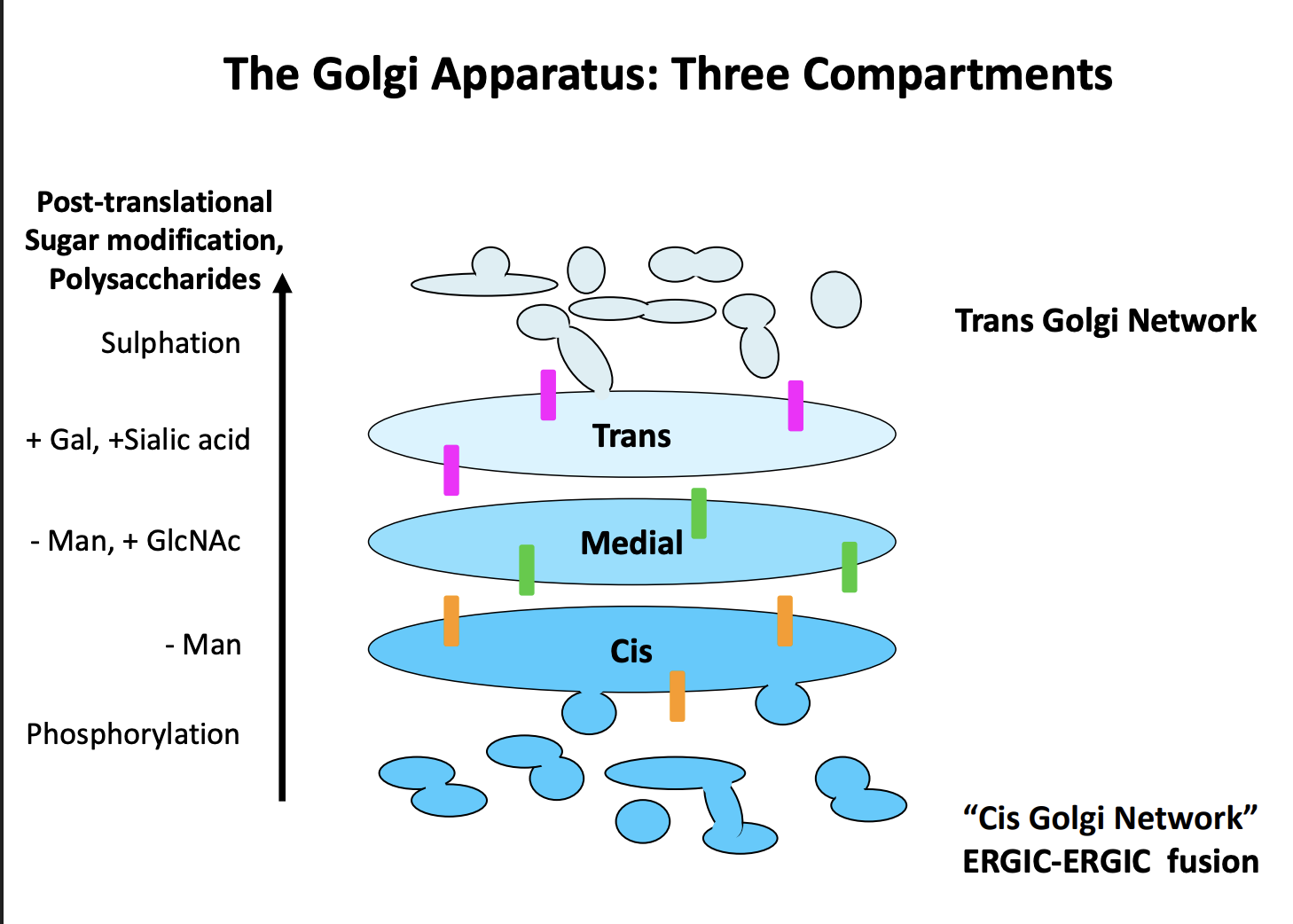
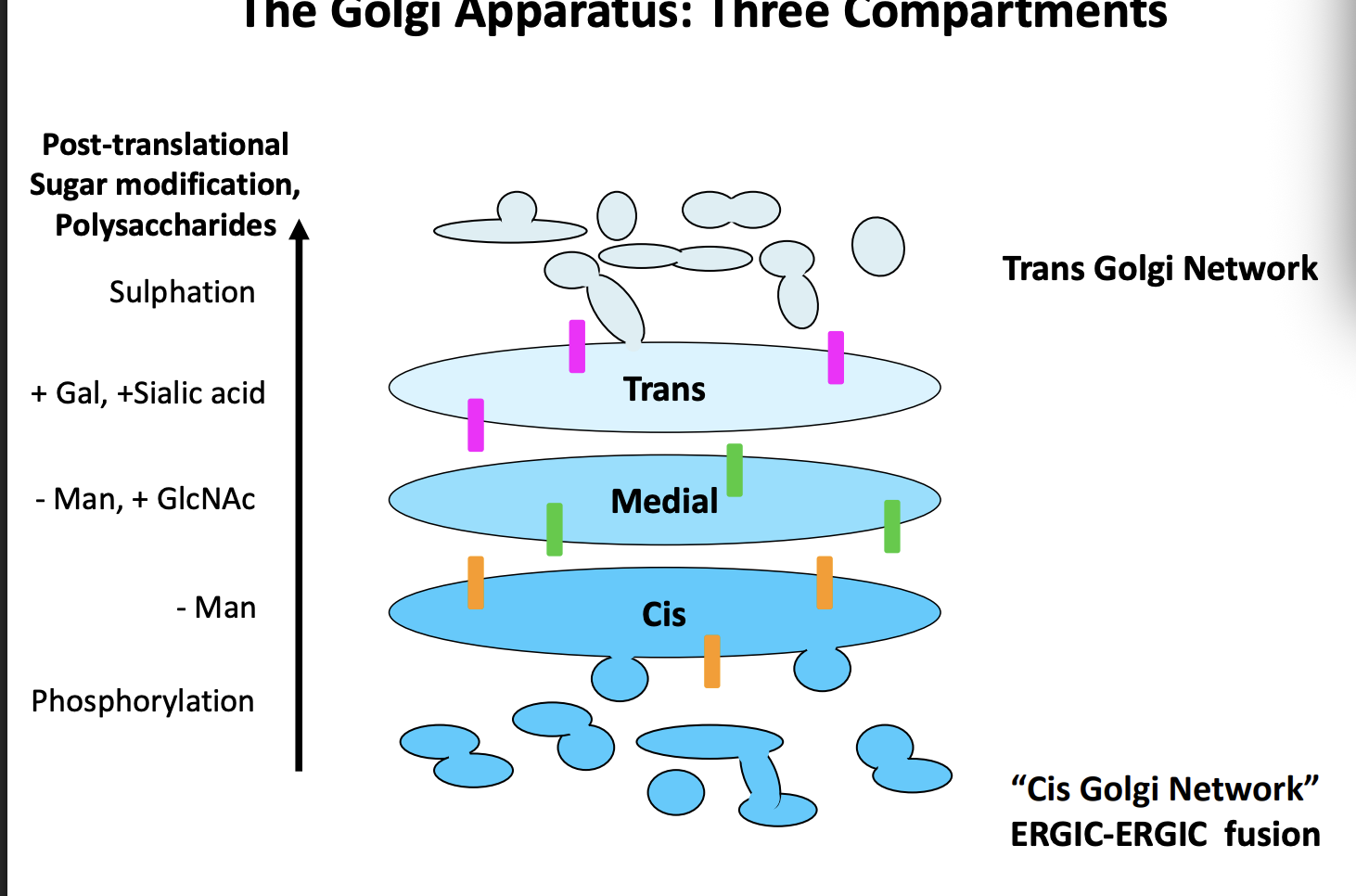
In order to do this→ golgi have three functional domains
Cis
remove Manose
Medial
- Man, + GlcNAc
Trans
+Gal, + Sialic acid, Sulphation
enzymes found in these compartments are different and used to monitor the progress of secretory proteins
Other functions of golgi
Synthesis of
extracellular polysaccharides
glycosaminoglycans
plant and fungal cell wall polysaccharides
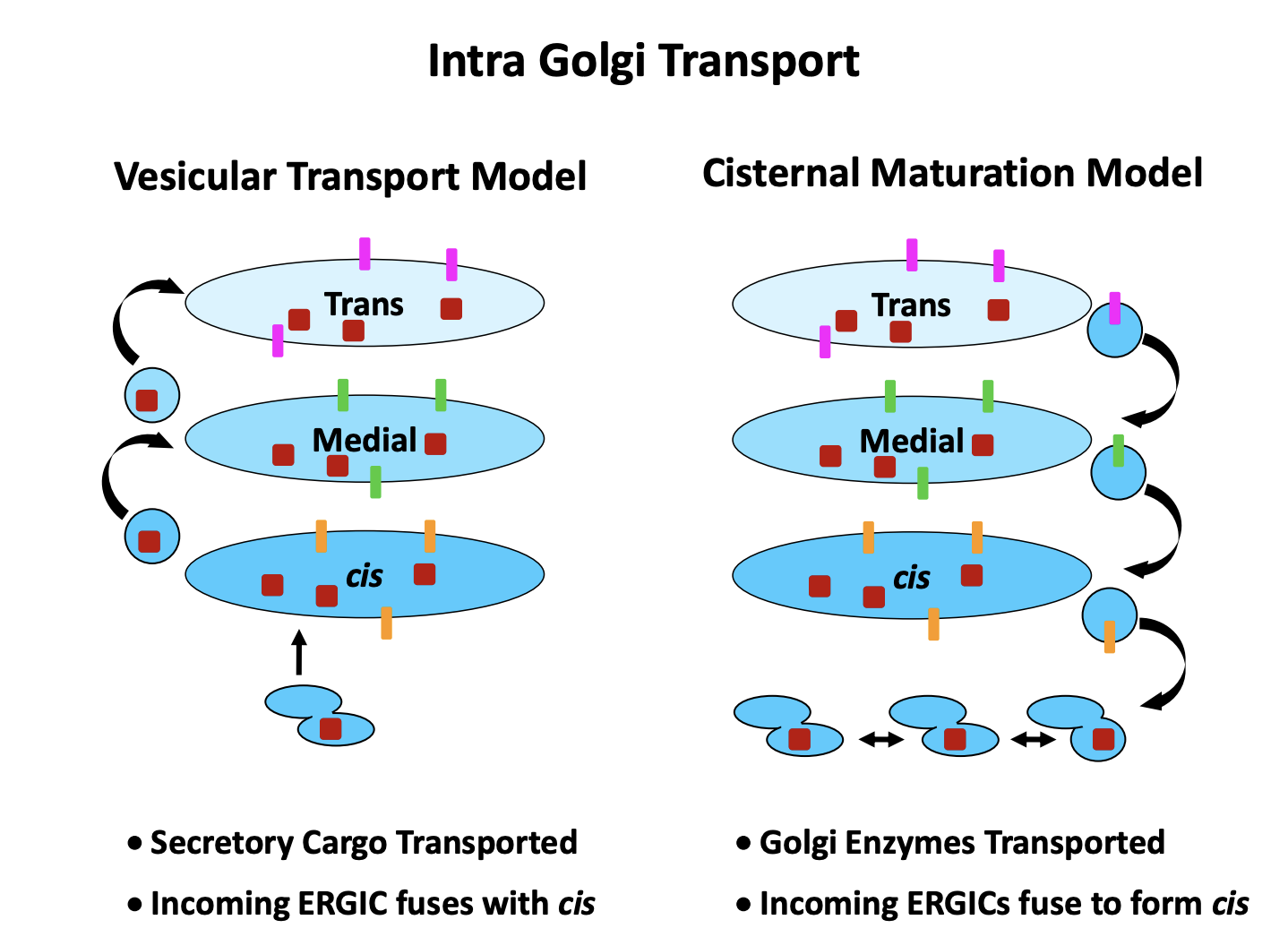
Two models for intra Golgi Transport
Vesicular transport model
vesicles bud from each compartment to the next
cargo: cis→ trans
Cisternal Maturation Model
Proteins arrive in tubular cluster
fuse together
make the newest cisternae
Golgi cisternae mature
exchange their protein complements over time
whilst the cargo always stays in the same cisterna
trans cisternae becomes vesicles and proteins taken further on
cargo: trans→ cis
both models assume vesciular tranport between cisternae
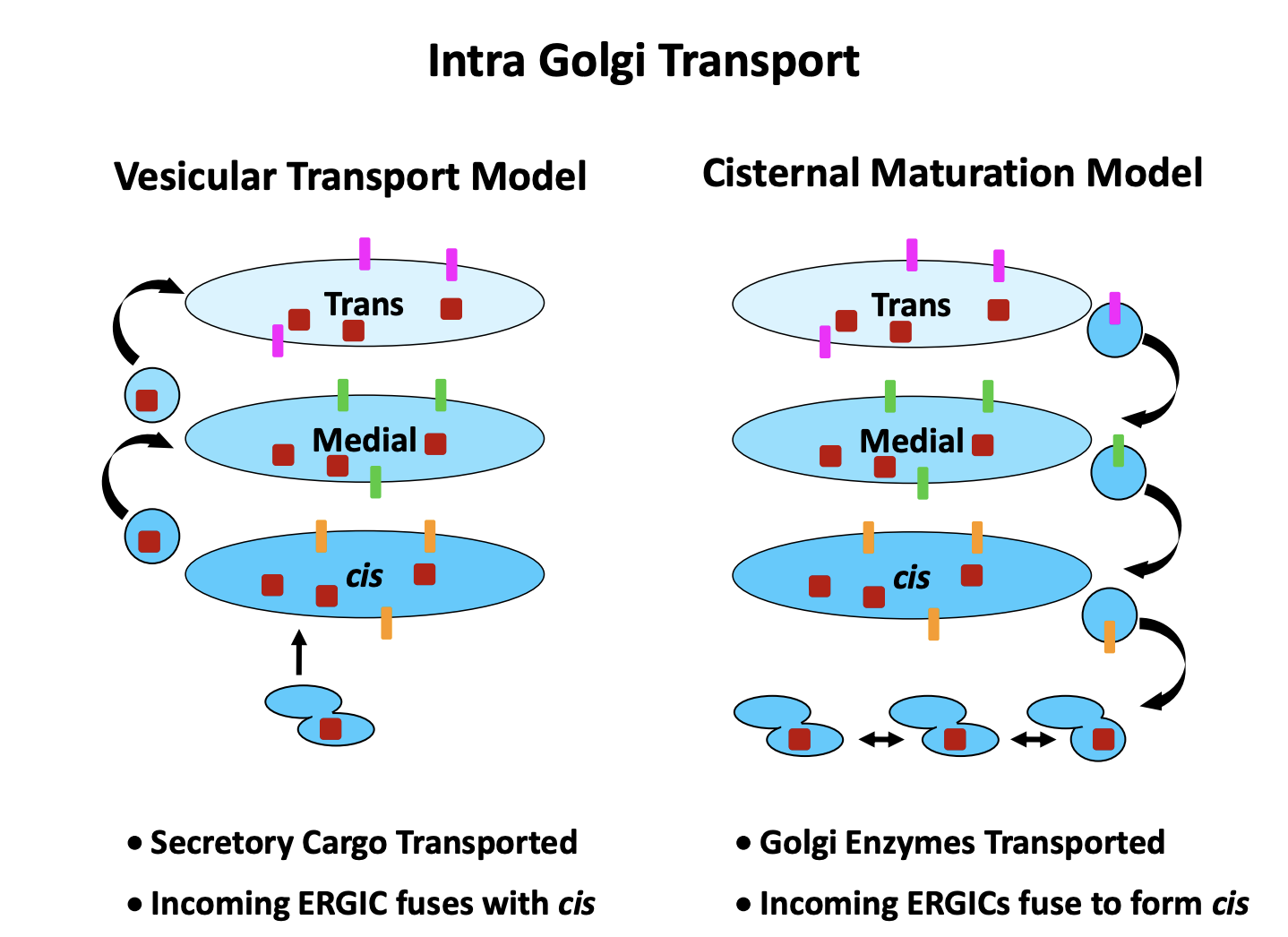
Vesciular tranpsort model EVIDENCE
EM showing vesciles budding from the edges of Golgi cisternae
in vitro transport assays
Cisternae Maturation Model EVIDENCE
Explains how cargo that is too big, e.g procollagen for vesciles can still be transported through
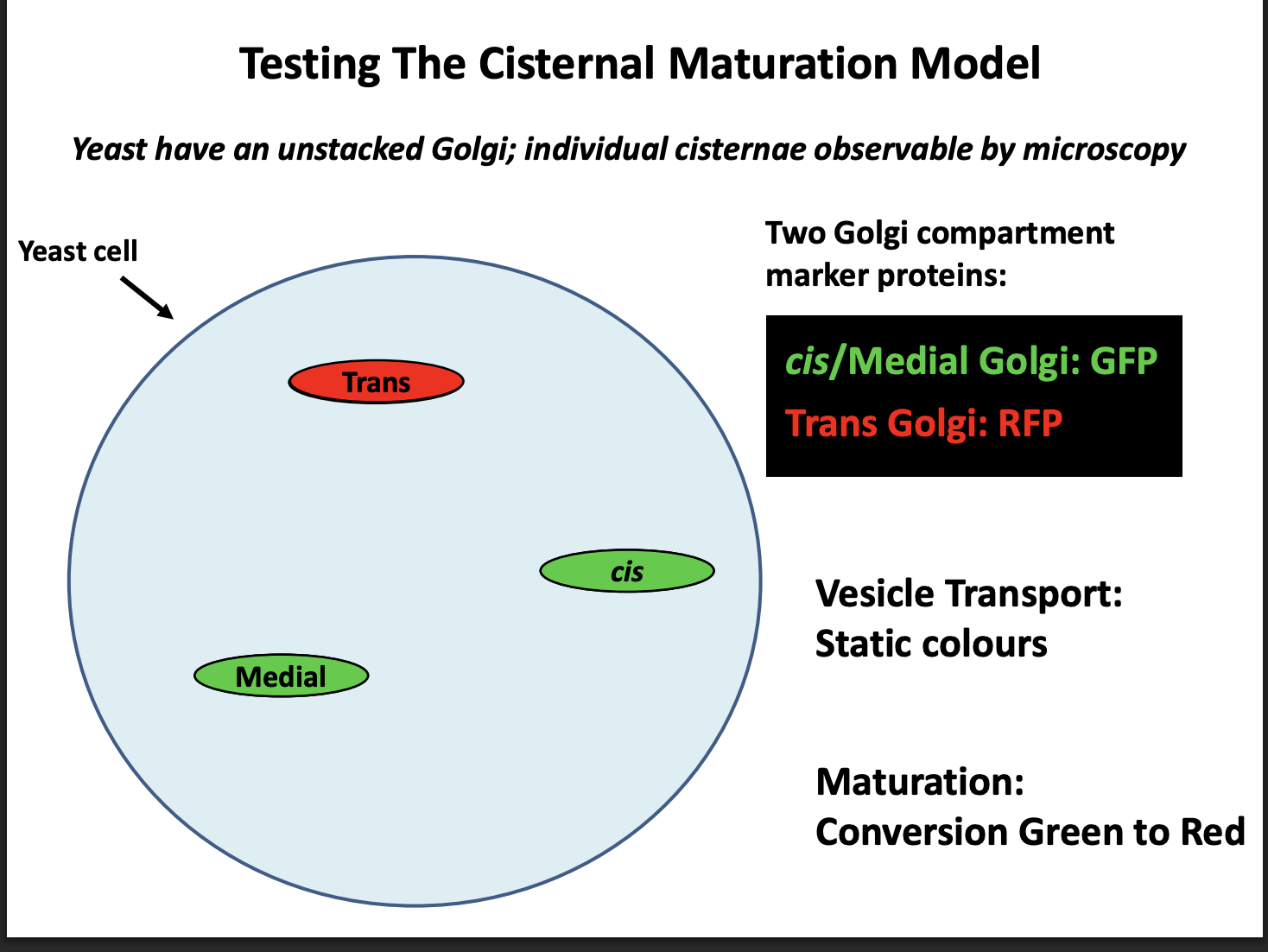
Live cell imaging in yeast
EVIDENCE: PULSE CHASE live cell imaging in yeast→ Why use yeast
Mammalian cells→ golgi compartments are too close together
Yeast→ three compartments are separated far enough as they do not stack
EVIDENCE for cisternal maturation model: PULSE CHASE
Procedure:
Label cis/medial golgi: GFP
Trans Golgi: RFP
Expected observations:
If vesicular transport→ Static colours
If Maturation→ conversion Green to Red
Result:
Conversion green to red
therefore Maturation model is correct
So which model is correct
evidence for the maturation
HOWEVER→ still may be some antereograde traffic still occur
no consesus but probable that both models are partly correct
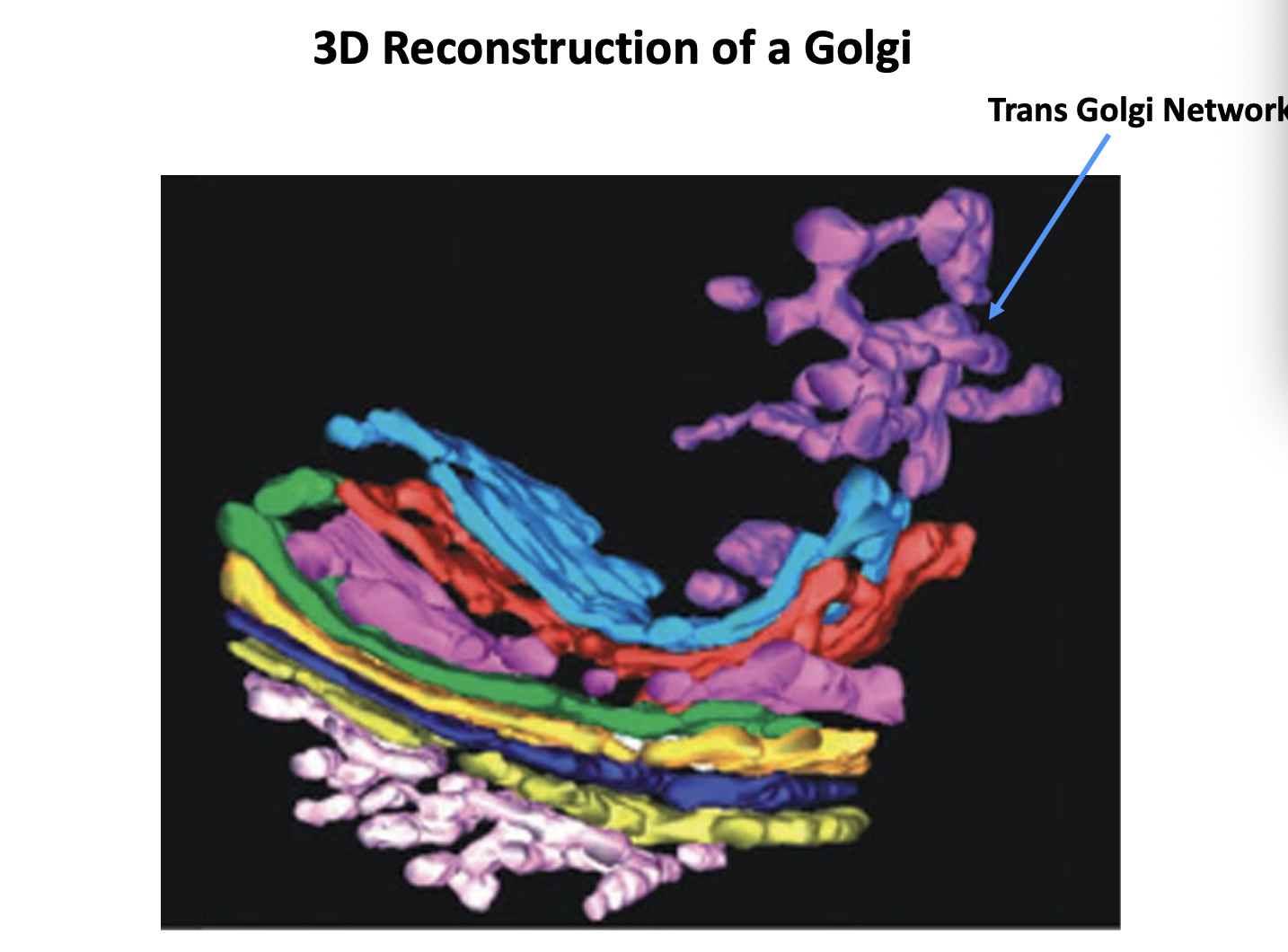
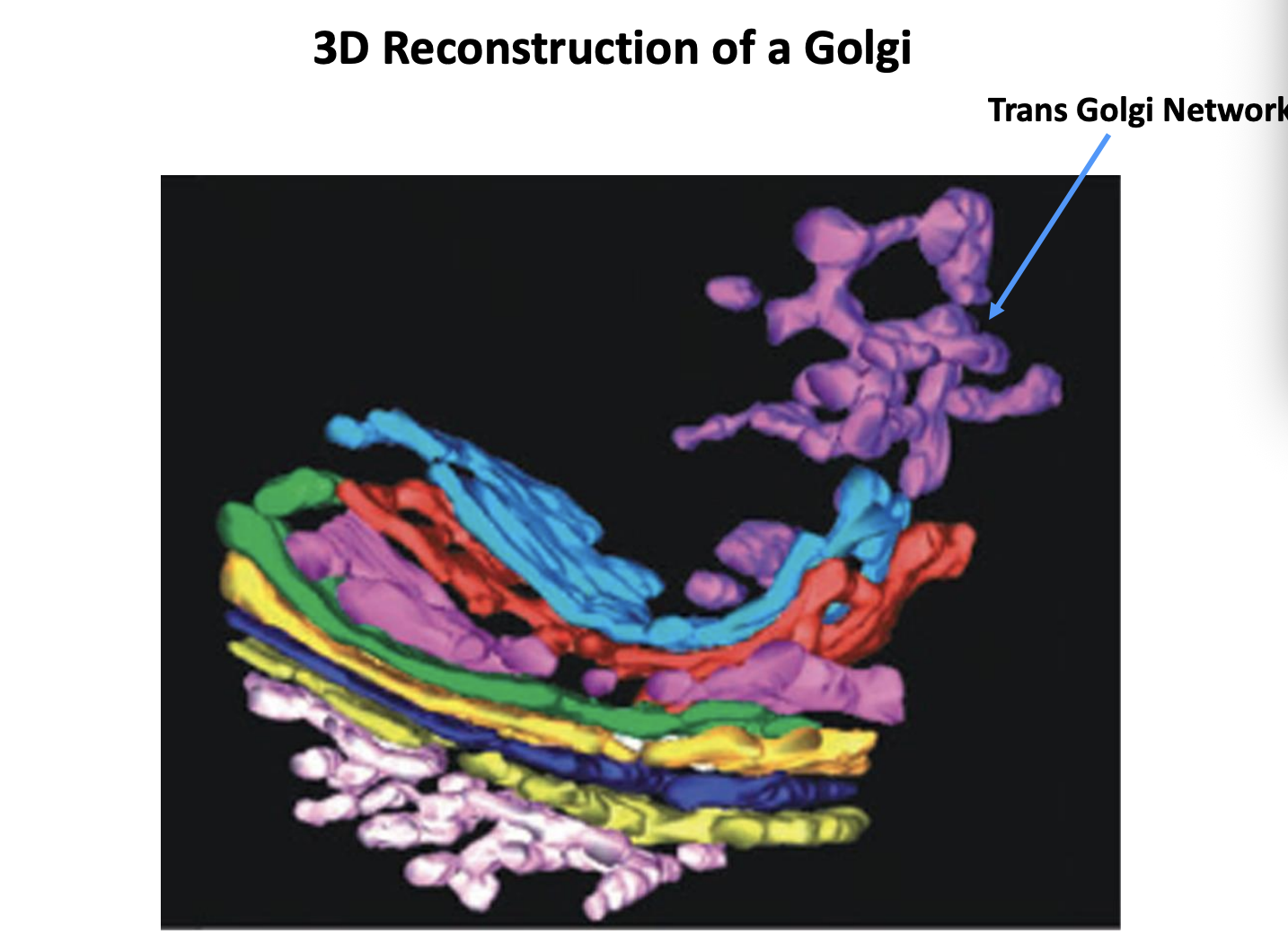
What is the Trans Golgi Network (TGN)
the cluster of tubules and vesicles at the trans-most side of the golgi
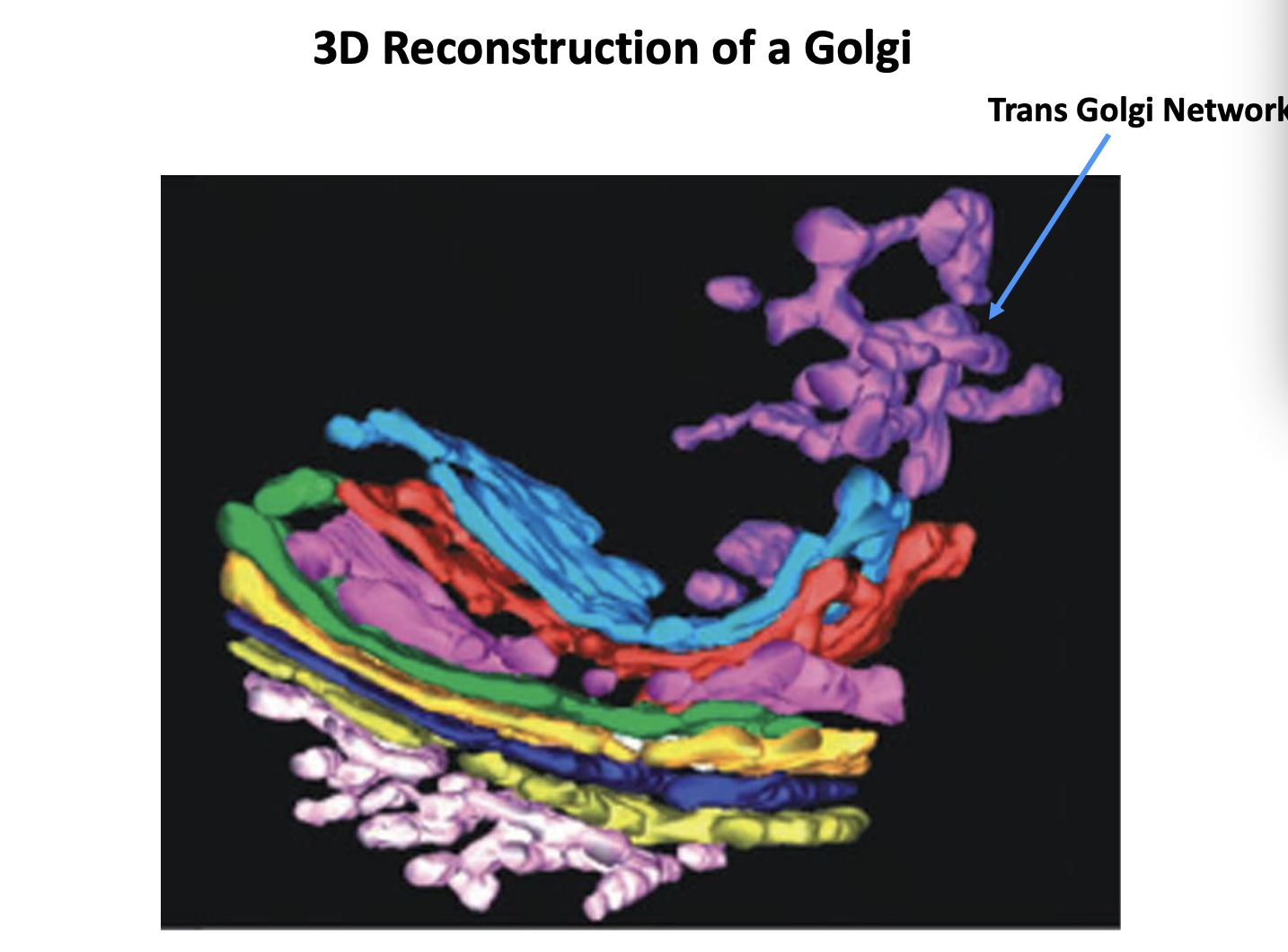
Is it a stable comparment?
Yes→ it has characteristics of a stable comparment
No→ In the cisternal maturation model→ corresponds to a trans-cisterna that is being converted into secretory carriers
What other function does it have in some species?
endosome
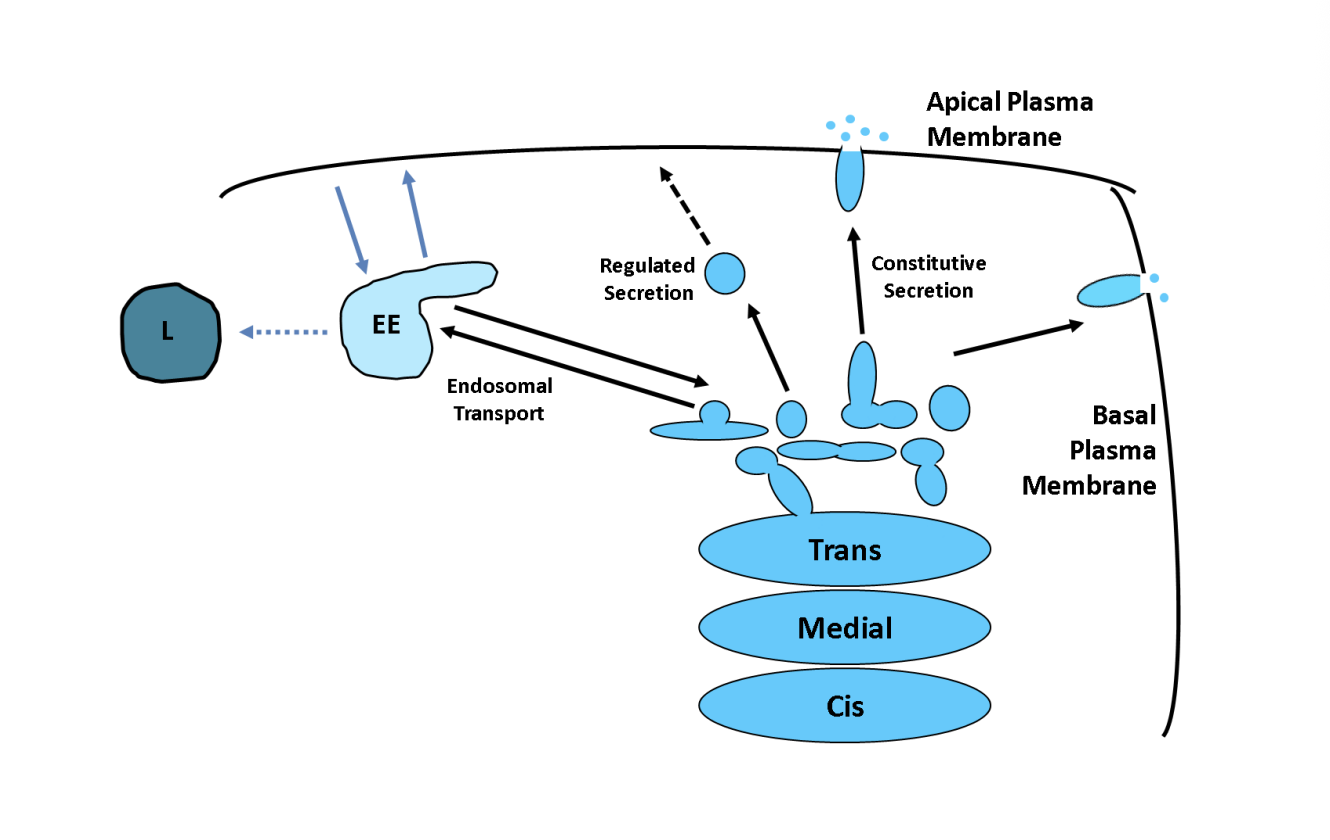
Role of the TGN
Major protein sorting station→ Point at which proteins diverge for the first time
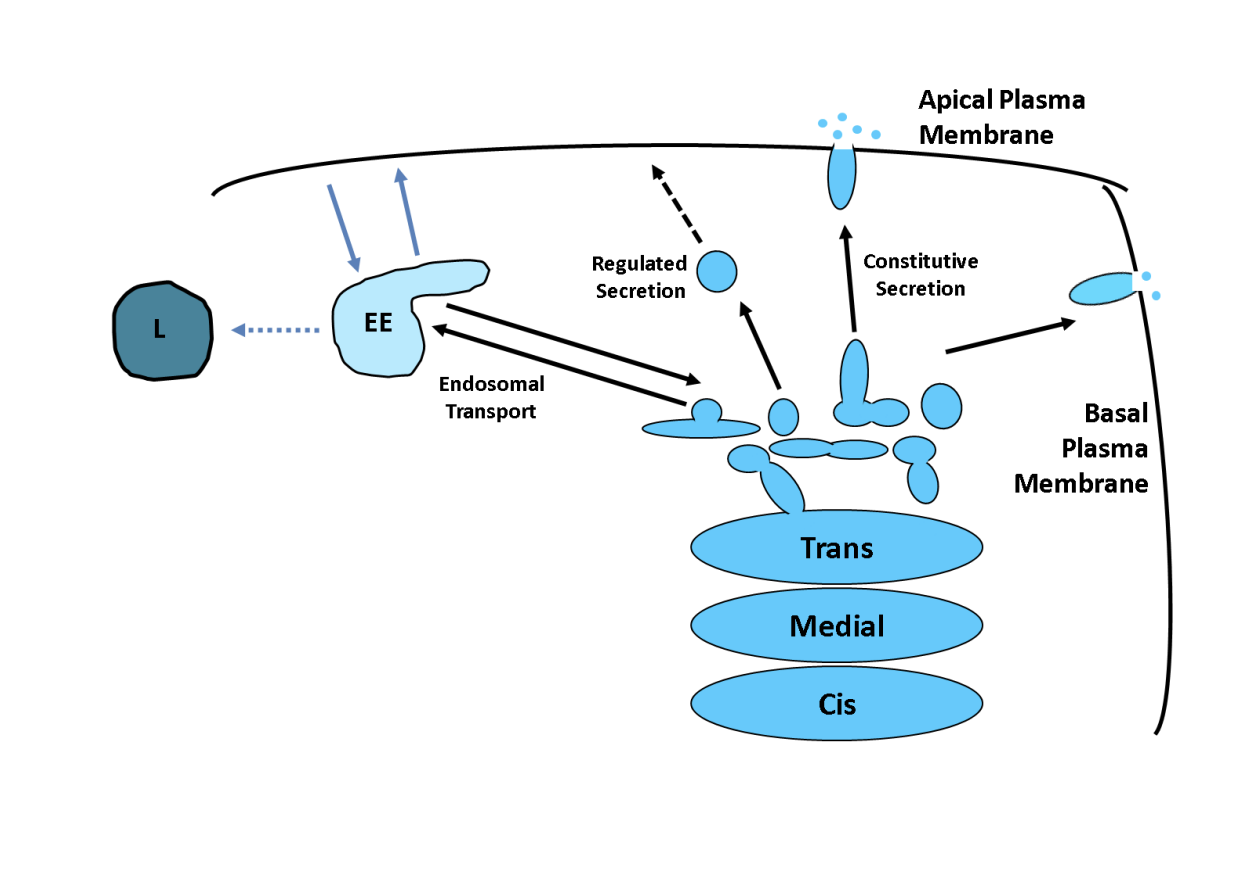
How are proetins sorted by the TGN
Bulk or default→ directly to plasma membrane.
may require no further sorting signals
Sorting into different carriers (direct secretion in polarised cells)
Concentrated in regulated secretory vesicles/ secretory granules
Endosome transport
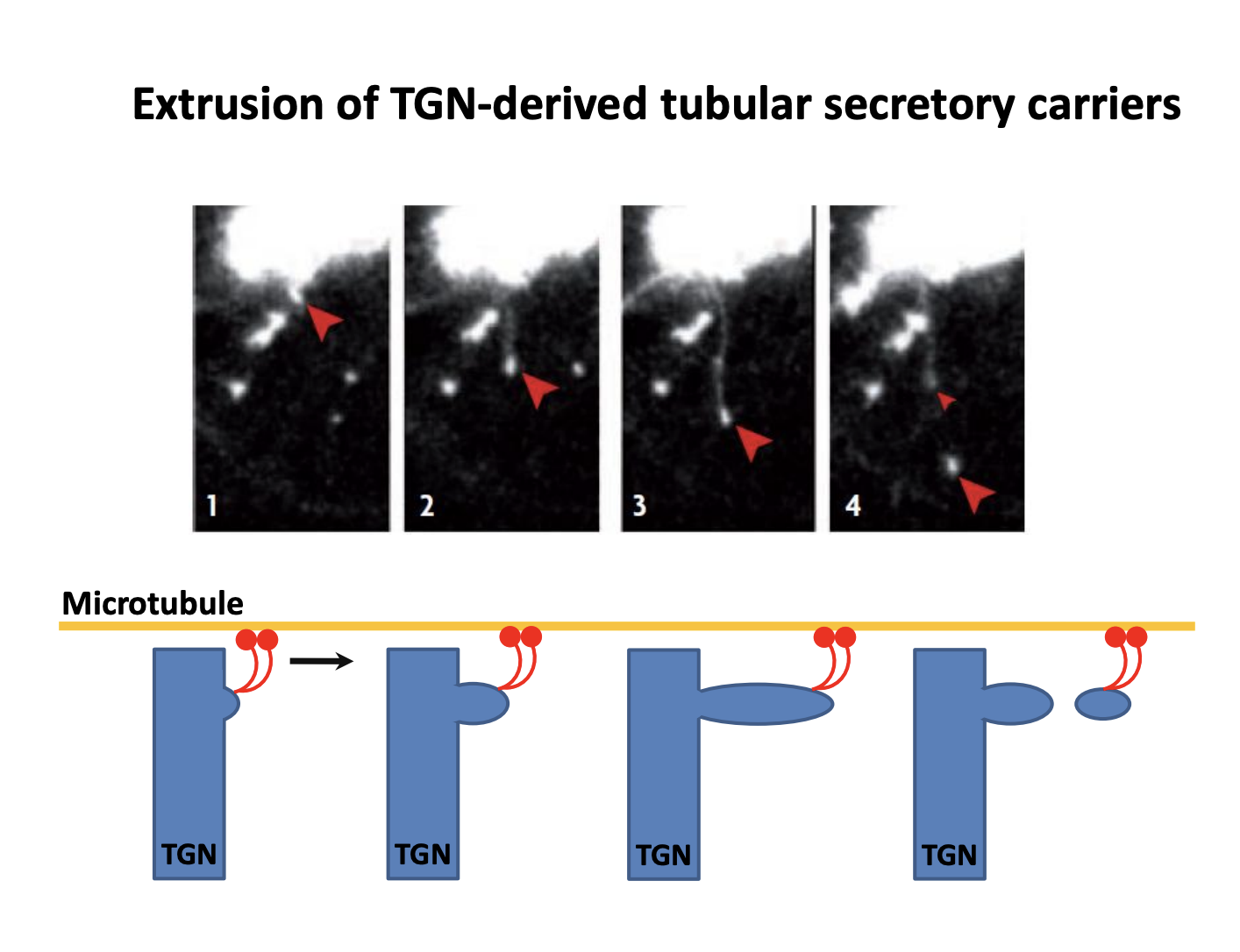
Bulk transport→ live imaging shows how this secretion is mediated
Tubular carriers are pulled out of the TGN by molecular motors on microtubules
do not know about protein coat or other details
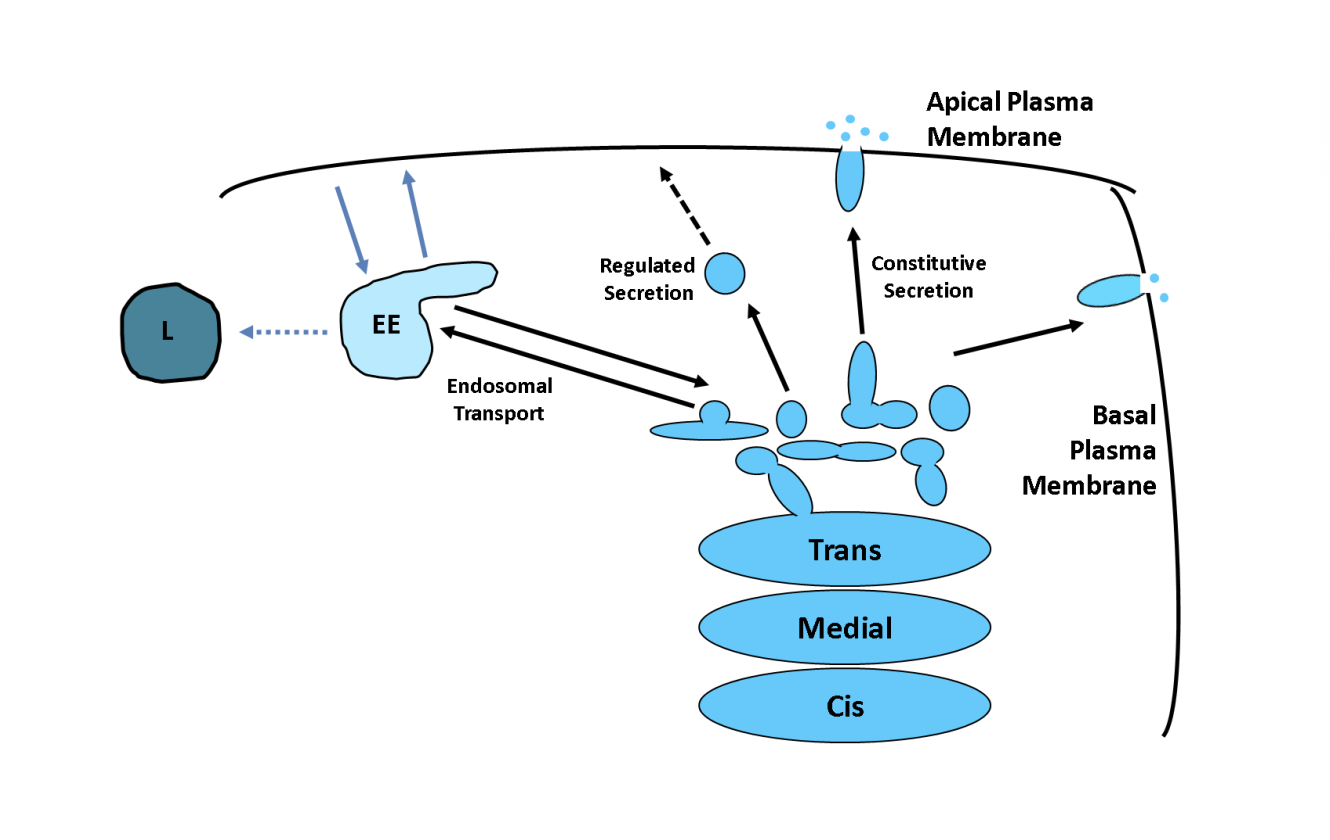
Sorting into different carriers (direct secretion in polarised cells)
Apical vs basal
Axon vs dendrites
have very different protein and lipid compositions
Sort proteins into destined vesicles for certain places
Example of sorting
e.g Vesicular stomatitis virus envelope glycoprotein (VSV-G)
into basal Sorting into different carriers (direct secretion in polarised cells laterally-destined vesicles
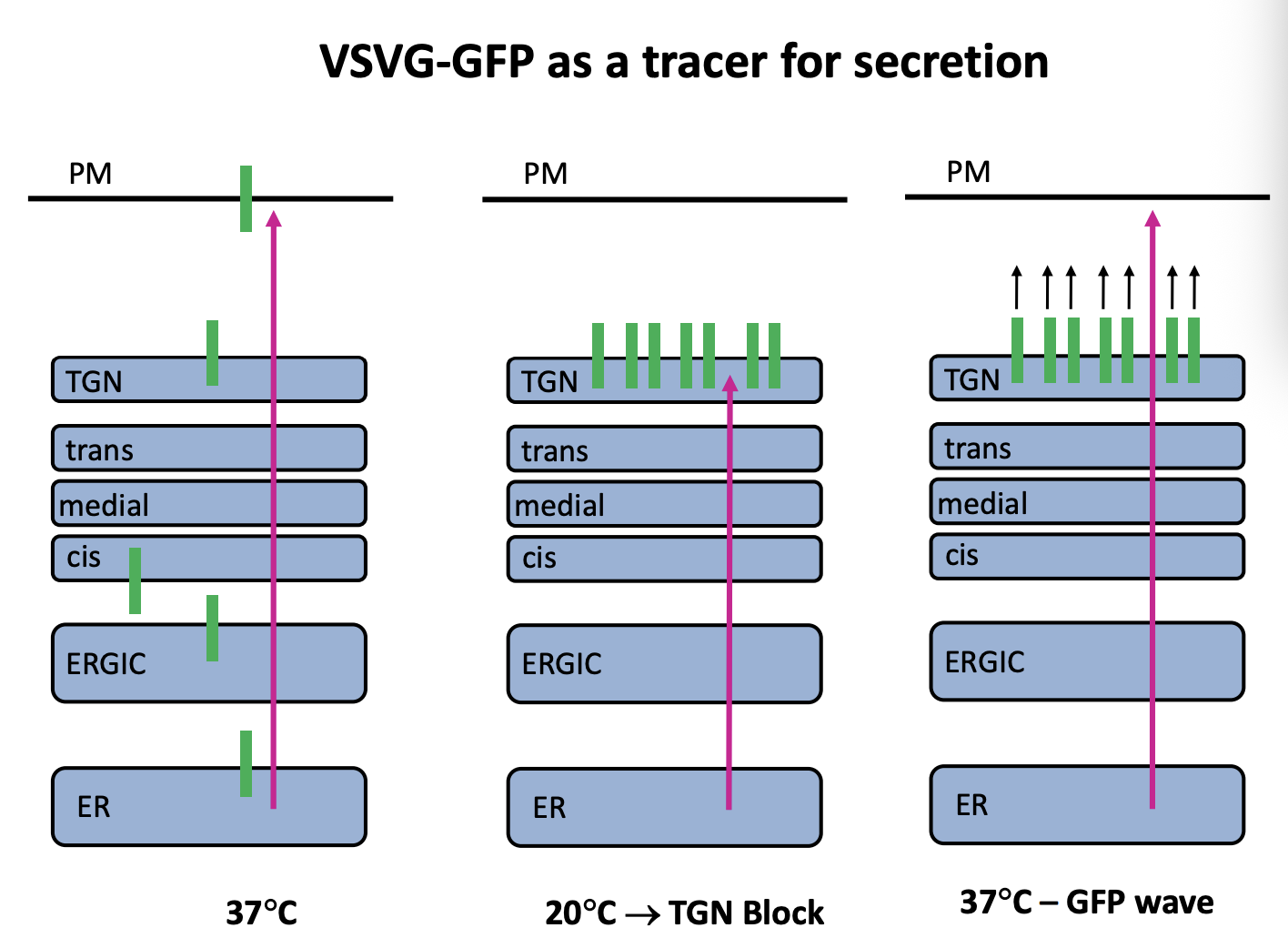
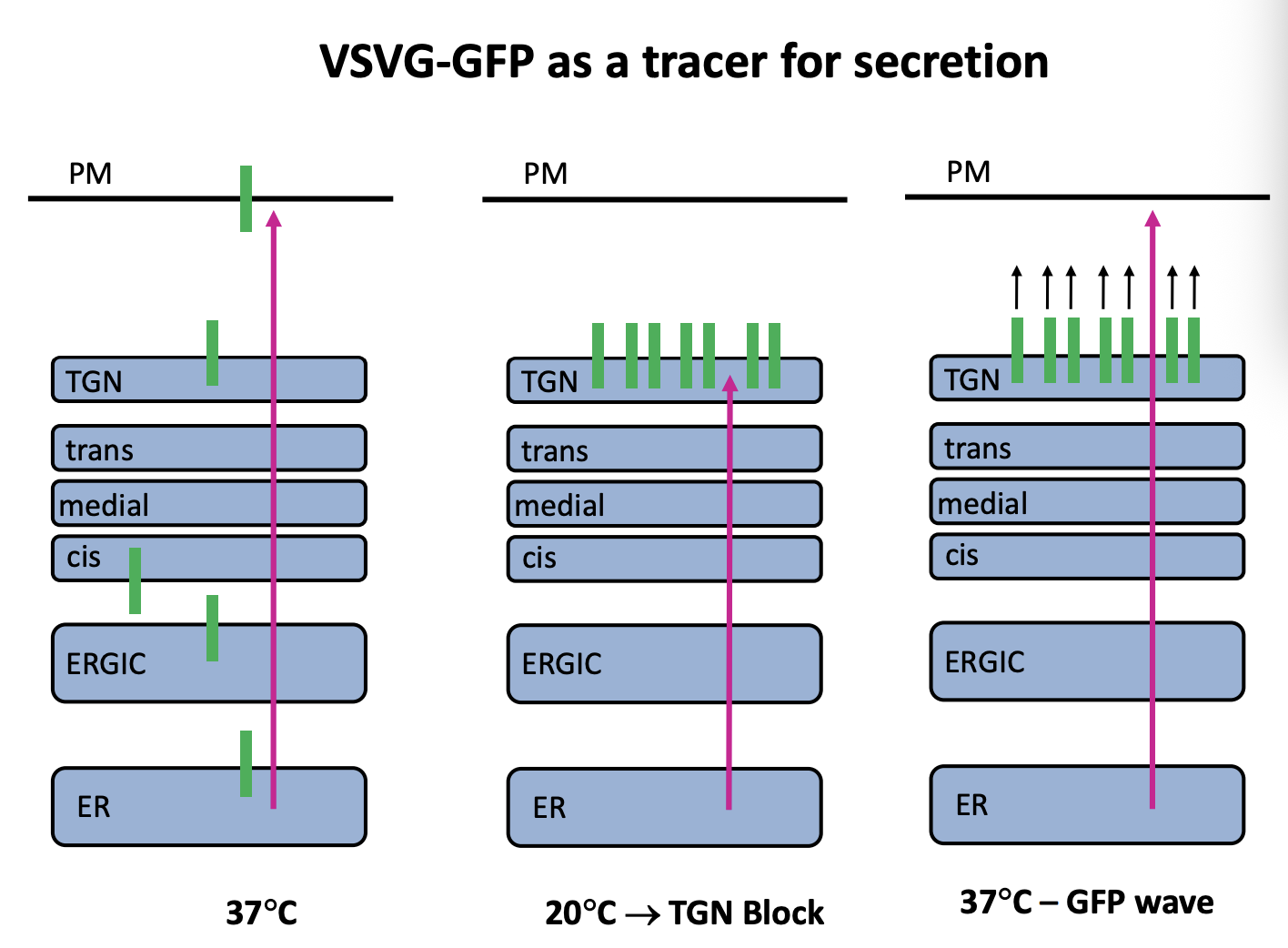
Investigating the secretion of VSVG with GFP
shows that it is temperature dependent
At a low temperature→ there is a TGN block
Regulated secretion and examples
special carriers only fuse with the plasma membrane in response to the appropriate signal
e.g→ insulin release in pancreatic cells
e.g→ NT release in nerve cells
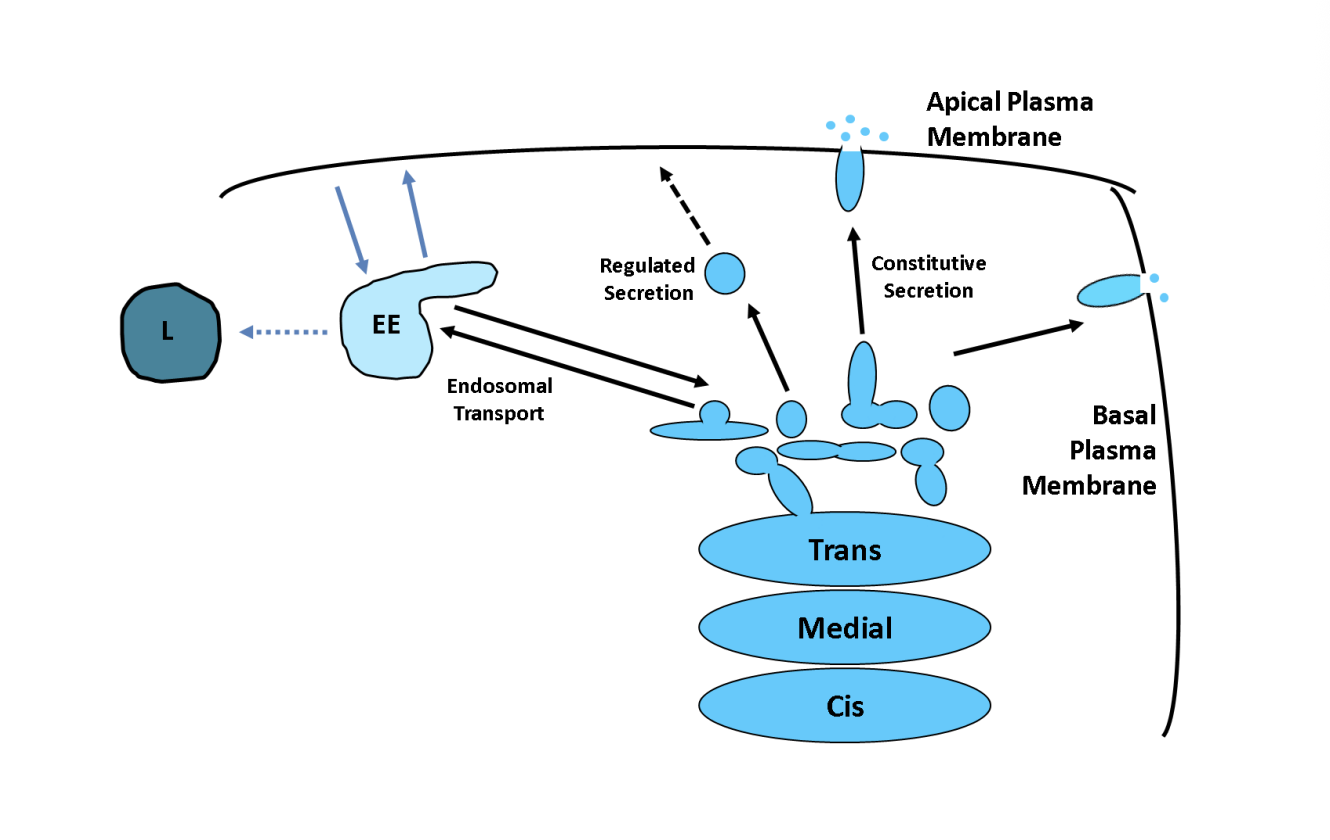
Endosome transport
Clathrin-coated or other types of non-clathrin coated
delivered to endosomes
works an an interface between the secretory and endocytotic pathways