Microbio exam 2
5.0(1)
5.0(1)
Card Sorting
1/201
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
202 Terms
1
New cards
a
Alive/Dead

2
New cards
-sterilization
The process by which all living cells, spores, viruses, and viroids are either destroyed or removed from an object or habitat.
-disinfection
The killing, inhibition, or removal of microorganisms that may cause disease. It usually refers to the treatment of inanimate objects with chemicals.
-disinfectant
An agent, usually chemical, that disinfects; normally, it is employed only with inanimate objects. (disinfectant
An agent, usually chemical, that disinfects; normally, it is employed only with inanimate objects.)
-sanitization
Reduction of the microbial population on an inanimate object to levels judged safe by public health standards. (example, sanitizers are used to clean eating utensils in restaurants.)
-antisepsis
The prevention of infection or sepsis.
-antiseptic
Chemical agent applied to tissue to prevent infection by killing or inhibiting pathogens. (they also reduce the total microbial population. Because they must not cause too much harm to the host, antiseptics are generally not as toxic as disinfectants)
The process by which all living cells, spores, viruses, and viroids are either destroyed or removed from an object or habitat.
-disinfection
The killing, inhibition, or removal of microorganisms that may cause disease. It usually refers to the treatment of inanimate objects with chemicals.
-disinfectant
An agent, usually chemical, that disinfects; normally, it is employed only with inanimate objects. (disinfectant
An agent, usually chemical, that disinfects; normally, it is employed only with inanimate objects.)
-sanitization
Reduction of the microbial population on an inanimate object to levels judged safe by public health standards. (example, sanitizers are used to clean eating utensils in restaurants.)
-antisepsis
The prevention of infection or sepsis.
-antiseptic
Chemical agent applied to tissue to prevent infection by killing or inhibiting pathogens. (they also reduce the total microbial population. Because they must not cause too much harm to the host, antiseptics are generally not as toxic as disinfectants)
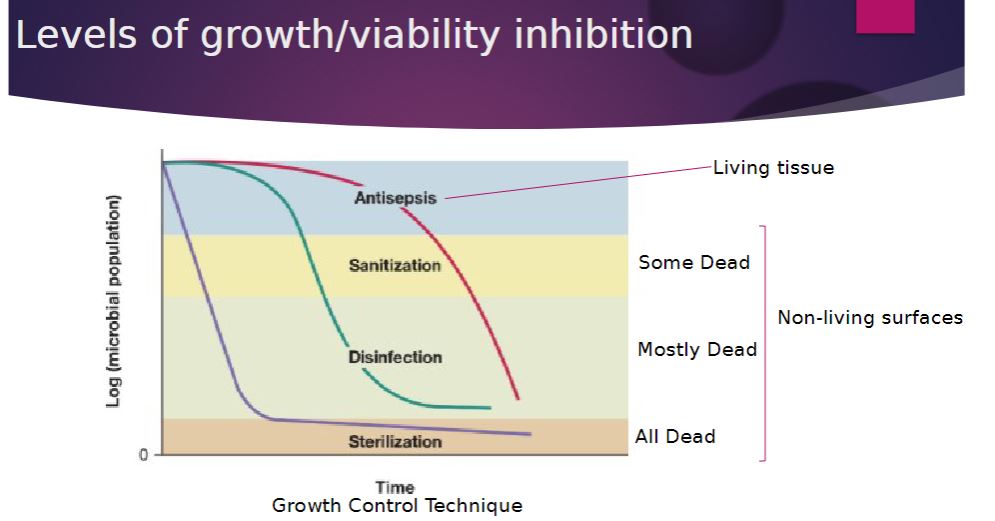
Levels of growth
3
New cards
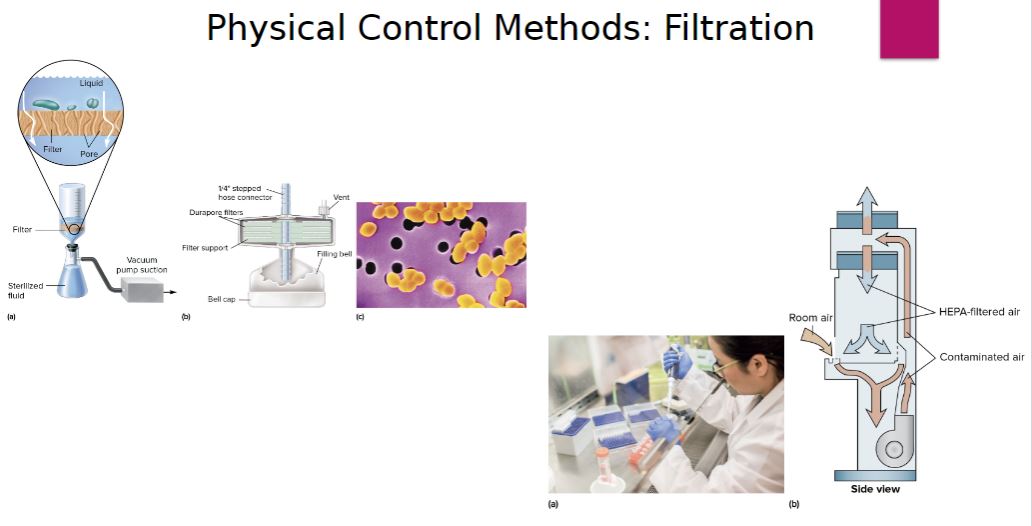
-Filtration is an excellent way to reduce the microbial population in solutions of heat-sensitive material and can be used to sterilize liquids and gases (including air). Rather than directly destroying contaminating microorganisms, the filter simply acts as a barrier to retain them.
There are two types of filters:
-depth filters-
Filters composed of fibrous or granular materials that are used to decrease microbial load and sometimes to sterilize solutions.
-membrane filter-
Porous material that retains microorganisms as the suspending liquid passes through the pores. (replaced depth filters for many purposes.)
Image below-
Membrane Filter Sterilization. (a) Diagram of a membrane filtration setup that uses a vacuum pump to force liquid through the filter. The inset shows a cross-section of the filter and its pores, which retain microbes. (b) Cross section of a membrane filtration unit. Several membranes are used to increase its capacity. (c) Enterococcus faecalis on a membrane filter with 0.4-µm pores.
(a) A technician pipetting potentially hazardous material in a safety cabinet. (b) A schematic diagram showing the airflow pattern within a class II safety cabinet.
There are two types of filters:
-depth filters-
Filters composed of fibrous or granular materials that are used to decrease microbial load and sometimes to sterilize solutions.
-membrane filter-
Porous material that retains microorganisms as the suspending liquid passes through the pores. (replaced depth filters for many purposes.)
Image below-
Membrane Filter Sterilization. (a) Diagram of a membrane filtration setup that uses a vacuum pump to force liquid through the filter. The inset shows a cross-section of the filter and its pores, which retain microbes. (b) Cross section of a membrane filtration unit. Several membranes are used to increase its capacity. (c) Enterococcus faecalis on a membrane filter with 0.4-µm pores.
(a) A technician pipetting potentially hazardous material in a safety cabinet. (b) A schematic diagram showing the airflow pattern within a class II safety cabinet.
Filtration
4
New cards
-Most microorganisms require specific temperatures for normal growth and replication; temperatures in excess damage cellular structures and alter chemical reactions. Moist and dry heat readily destroy viruses, bacteria, and fungi in this way.
Physical control: heat

5
New cards
-Many objects are best sterilized by dry heat. For instance, metal inoculating loops, which are used routinely in the laboratory, can be sterilized in a small, bench-top incinerator. Other metal or glass items are sterilized in an oven at 160° to 170°C for 2 to 3 hours. Microbial death results from the oxidation of cell constituents and protein denaturation. Dry heat is less effective than moist heat. The endospores of Clostridium botulinum, the cause of botulism, are killed in 5 minutes at 121°C by moist heat but only after 2 hours at 160°C by dry heat. However, dry heat has some advantages. It does not corrode glassware and metal instruments as moist heat does, and it can be used to sterilize powders, oils, and similar items. Despite these advantages, dry heat sterilization is slow and not suitable for heat-sensitive materials such as plastic and rubber.
Dry Heat

6
New cards
-Therefore boiling can be used for disinfecting drinking water and objects not harmed by water, but boiling does not sterilize.
-To destroy bacterial endospores, moist heat sterilization must be carried out at temperatures above 100°C, and this requires the use of saturated steam under pressure. Steam sterilization is carried out with an autoclave (figure 27.5), a device somewhat like a pressure cooker.
-To destroy bacterial endospores, moist heat sterilization must be carried out at temperatures above 100°C, and this requires the use of saturated steam under pressure. Steam sterilization is carried out with an autoclave (figure 27.5), a device somewhat like a pressure cooker.
Wet heat

7
New cards
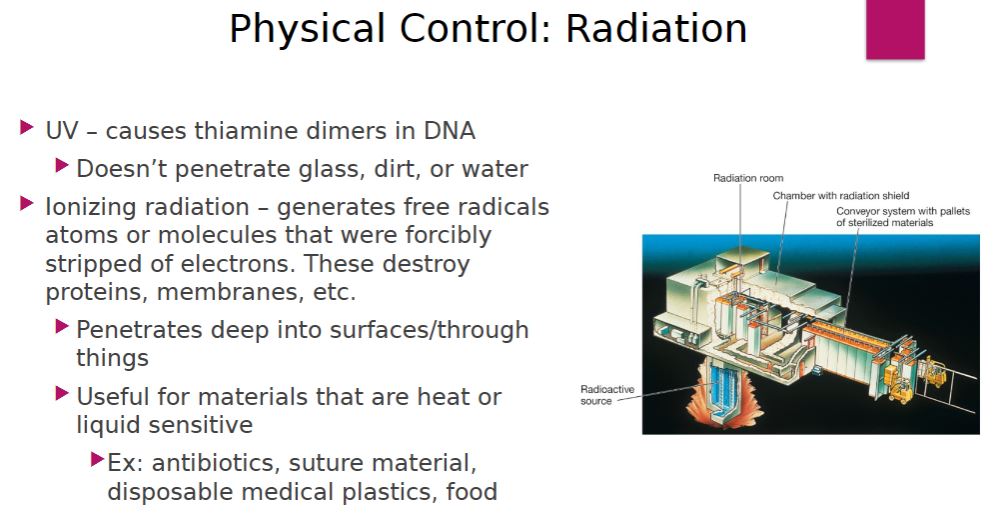
-Ultraviolet (UV) radiation around 260 nm is quite lethal. It causes thymine-thymine dimerization of DNA, preventing replication and transcription. However, UV radiation does not penetrate glass, dirt films, water, and other substances effectively. Therefore, UV radiation is used as a sterilizing agent only in a few situations. UV lamps are sometimes placed on lab ceilings or in biological safety cabinets to sterilize the air and any exposed surfaces. Because UV radiation burns skin and damages eyes, the lamps are off when the areas are in use.
-Ionizing radiation is an excellent sterilizing agent that penetrates deep into objects. Ionizing radiation has sufficient energy to dislodge electrons from atoms, producing chemically reactive free radicals. The free radicals react with nearby matter to weaken or destroy it. Ionizing radiation destroys bacterial endospores and all microbial cells; however, it is not always effective against viruses. Gamma radiation (from a cobalt 60 source) and beta radiation (accelerated electrons from high-voltage electricity) are used in the cold sterilization of antibiotics, hormones, sutures, and plastic disposable supplies such as syringes. Gamma radiation and electron beams have also been used to sterilize and “pasteurize” meat and other foods (figure 27.6). Currently irradiation is used to treat meat, fruits, vegetables, and spices. Various methods are used to control food spoilage (section 29.2)
-Ionizing radiation is an excellent sterilizing agent that penetrates deep into objects. Ionizing radiation has sufficient energy to dislodge electrons from atoms, producing chemically reactive free radicals. The free radicals react with nearby matter to weaken or destroy it. Ionizing radiation destroys bacterial endospores and all microbial cells; however, it is not always effective against viruses. Gamma radiation (from a cobalt 60 source) and beta radiation (accelerated electrons from high-voltage electricity) are used in the cold sterilization of antibiotics, hormones, sutures, and plastic disposable supplies such as syringes. Gamma radiation and electron beams have also been used to sterilize and “pasteurize” meat and other foods (figure 27.6). Currently irradiation is used to treat meat, fruits, vegetables, and spices. Various methods are used to control food spoilage (section 29.2)
Physical control: Radiation
8
New cards
-sterilization-
The process by which all living cells, spores, viruses, and viroids are either destroyed or removed from an object or habitat. When sterilization is achieved by a chemical agent, the chemical is called a sterilant.
-disinfection-
The killing, inhibition, or removal of microorganisms that may cause disease. It usually refers to the treatment of inanimate objects with chemicals. Disinfectants are agents, usually chemical, used to carry out disinfection and normally used only on inanimate objects. A disinfectant does not necessarily sterilize an object because viable spores and a few microorganisms may remain.
-In sanitization-
the microbial population is reduced to levels that are considered safe by public health standards. The inanimate object is usually cleaned as well as partially disinfected. For example, sanitizers are used to clean eating utensils in restaurants.
-Antisepsis-
The prevention of infection or sepsis. Antiseptics are chemical agents applied to tissue to prevent infection by killing or inhibiting pathogen growth; they also reduce the total microbial population. Because they must not cause too much harm to the host, antiseptics are generally not as toxic as disinfectants.
The process by which all living cells, spores, viruses, and viroids are either destroyed or removed from an object or habitat. When sterilization is achieved by a chemical agent, the chemical is called a sterilant.
-disinfection-
The killing, inhibition, or removal of microorganisms that may cause disease. It usually refers to the treatment of inanimate objects with chemicals. Disinfectants are agents, usually chemical, used to carry out disinfection and normally used only on inanimate objects. A disinfectant does not necessarily sterilize an object because viable spores and a few microorganisms may remain.
-In sanitization-
the microbial population is reduced to levels that are considered safe by public health standards. The inanimate object is usually cleaned as well as partially disinfected. For example, sanitizers are used to clean eating utensils in restaurants.
-Antisepsis-
The prevention of infection or sepsis. Antiseptics are chemical agents applied to tissue to prevent infection by killing or inhibiting pathogen growth; they also reduce the total microbial population. Because they must not cause too much harm to the host, antiseptics are generally not as toxic as disinfectants.
Chemical used to inhibit growth

9
New cards
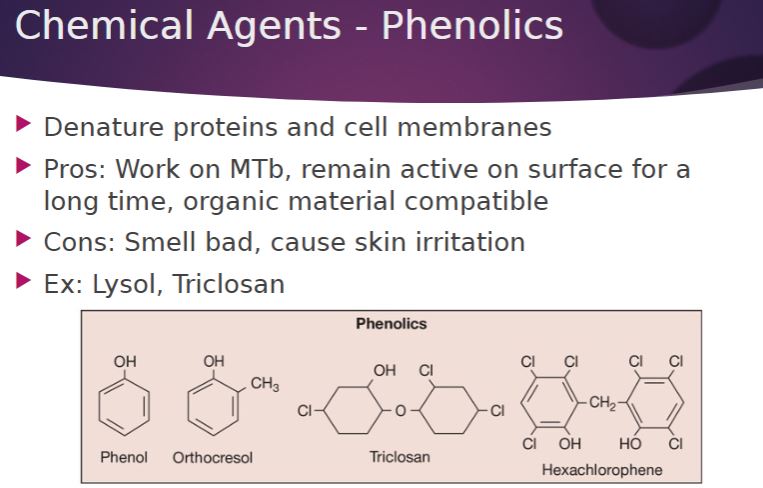
-Phenol was the first widely used antiseptic and disinfectant. In 1867 Joseph Lister employed it to reduce the risk of infection during surgery. Today phenol and phenol derivatives (phenolics) are used as disinfectants in laboratories and hospitals. The commercial disinfectant Lysol is a mixture of phenolics. Phenolics denature proteins and disrupt cell membranes. They have some important advantages as disinfectants: Phenolics are tuberculocidal, effective in the presence of organic material, and remain active on surfaces long after application. Chlorhexidine and chloroxylenol are widely used antiseptics in antimicrobial soaps and preoperative washes.
Chemical agents: Phenolic
10
New cards
-Alcohols are among the most widely used disinfectants, antiseptics, and sanitizers. They are bactericidal and fungicidal but not sporicidal; some enveloped viruses are also destroyed. The two most popular alcohol germicides are ethanol and isopropanol, usually used at 60 to 80% concentration. They act by denaturing proteins and possibly by dissolving membrane lipids. A 10 to 15-minute soaking is sufficient to disinfect small instruments, while rubbing hands with specially formulated alcohol products sanitizes by killing many pathogens.
Chemical agents: Alcohol

11
New cards
-The halogens iodine and chlorine are important antimicrobial agents. Iodine is an antiseptic and kills by oxidizing cell constituents and iodinating proteins. At higher concentrations, it may even kill some endospores. Iodine often is applied as tincture of iodine, 2% or more iodine in a water-ethanol solution of potassium iodide. Although it is an effective antiseptic, skin may be damaged, a stain remains, and iodine allergies can result. Iodine can be complexed with an organic carrier to form an iodophor. Iodophors are water soluble, stable, nonstaining, and release iodine slowly to minimize skin irritation. They are used in hospitals for cleansing preoperative skin and in hospitals and laboratories for disinfecting.
-Chlorine is the usual disinfectant for municipal water supplies and swimming pools, and is also employed in the dairy and food industries. note is the fact that household bleach (diluted to 10% in water, 10-minute contact time) can be used to disinfect surfaces contaminated by human body fluids and that it is made more effective by the addition of household vinegar.
-Chlorine is the usual disinfectant for municipal water supplies and swimming pools, and is also employed in the dairy and food industries. note is the fact that household bleach (diluted to 10% in water, 10-minute contact time) can be used to disinfect surfaces contaminated by human body fluids and that it is made more effective by the addition of household vinegar.
Chemical agents: Halogens

12
New cards
a
a

13
New cards
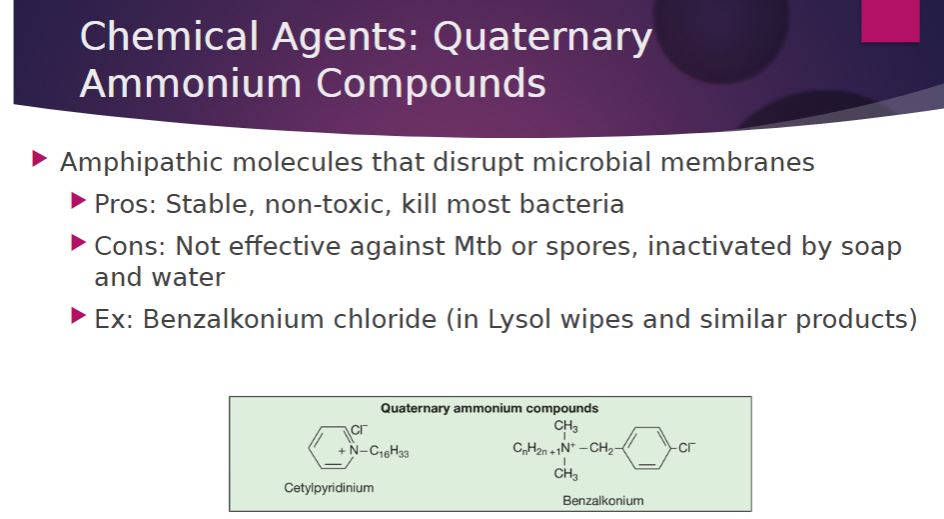
-Quaternary ammonium compounds (QACs) are detergents that have broad spectrum antimicrobial activity and are effective disinfectants used for decontamination purposes. Detergents (Latin detergere, to wipe away) are organic cleansing agents that are amphipathic, having both polar hydrophilic and nonpolar hydrophobic components. The hydrophilic portion of a QAC is a positively charged quaternary nitrogen; thus QACs are cationic detergents. Their antimicrobial activity is the result of their ability to disrupt microbial membranes; they may also denature proteins. QACs such as benzalkonium chloride and cetylpyridinium chloride kill most bacteria but not M. tuberculosis or endospores. They have the advantages of being stable and nontoxic, but they are inactivated by hard water and soap. QACs are often used as disinfectants for food utensils and small instruments, and as antiseptics.
Chemical agents: QAC
14
New cards
-Both of the commonly used aldehydes, formaldehyde and glutaraldehyde, are highly reactive molecules that inactivate nucleic acids and proteins, probably by cross-linking and alkylating molecules. They are sporicidal and can be used as chemical sterilant. Formaldehyde is usually dissolved in water or alcohol before use. A 2% buffered solution of glutaraldehyde is an effective disinfectant. It is less irritating than formaldehyde and is used to disinfect hospital and laboratory equipment. Glutaraldehyde usually disinfects objects within about 10 minutes but may require as long as 12 hours to destroy all endospores.
Chemical Agents: Aldehydes

15
New cards
- Many heat-sensitive items such as plastic Petri dishes, heart-lung machine components, sutures, and catheters are sterilized with ethylene oxide gas. Ethylene oxide (EO) is both microbicidal and sporicidal. It is a strong alkylating agent that kills by reacting with DNA and proteins to block replication and enzymatic activity. It is a particularly effective sterilizing agent because it rapidly penetrates packing materials, even plastic wraps. Sterilization is carried out in an EO sterilizer, which resembles an autoclave in appearance. It controls the EO concentration, temperature, and humidity. Because pure EO is explosive, it is usually supplied in a 10 to 20% concentration mixed with either CO2 or dichlorodifluoromethane. The EO concentration, humidity, and temperature influence the rate of sterilization. Because it is so toxic to humans, extensive aeration of the sterilized materials is necessary to remove residual EO.
Chemical Agents: Gases

16
New cards
Considerations:
- types of microbe
- what kind of surface/object it is
- living vs. non-living
- Risk of treatment (exposure risk)
- environmental condition
- Pop. size/concentration
- cost
-Resistance
- types of microbe
- what kind of surface/object it is
- living vs. non-living
- Risk of treatment (exposure risk)
- environmental condition
- Pop. size/concentration
- cost
-Resistance
Measuring efficacy

17
New cards
a
Measuring efficacy

18
New cards
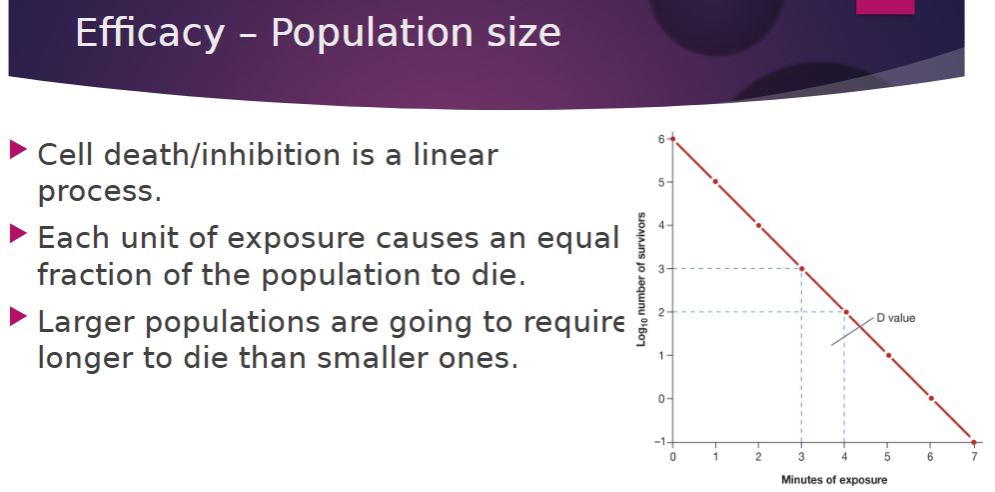
-decimal reduction time (D or D value)-
The time required to kill 90% of the microorganisms or endospores in a sample at a specified temperature.
-Because an equal fraction of a microbial population is killed during each interval, a larger population requires a longer time to die than does a smaller one
The time required to kill 90% of the microorganisms or endospores in a sample at a specified temperature.
-Because an equal fraction of a microbial population is killed during each interval, a larger population requires a longer time to die than does a smaller one
Population size
19
New cards
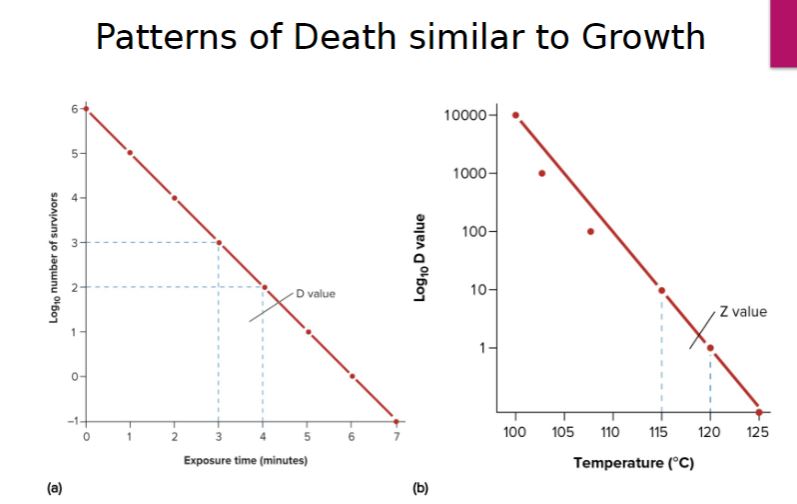
-If the logarithm of the population number remaining is plotted against the time of exposure of the microorganism to the agent, a straight-line plot will result (figure 27.2). When the population has been greatly reduced, the rate of killing may slow due to the survival of a more resistant strain of the microorganism.
-decimal reduction time (D or D value)-
The time required to kill 90% of the microorganisms or endospores in a sample at a specified temperature.
-The time required to kill 90% of the microorganisms or endospores in a sample at a specified temperature.
One measure of an agent’s killing efficiency is the decimal reduction time (D) or D value. The decimal reduction time is the time required to kill 90% of the microorganisms in a sample under specified conditions. For example, in a semilogarithmic plot of the viable population versus the time of heating, the D value is the time required for the line to drop by one log cycle or 10-fold (figure 27.2a). It is also possible to determine the temperature change at a given D value that decreases the microbial population by one log cycle (90%). This temperature change is referred to as the Z value and is predicted from a semilogarithmic plot of D values versus temperature
-decimal reduction time (D or D value)-
The time required to kill 90% of the microorganisms or endospores in a sample at a specified temperature.
-The time required to kill 90% of the microorganisms or endospores in a sample at a specified temperature.
One measure of an agent’s killing efficiency is the decimal reduction time (D) or D value. The decimal reduction time is the time required to kill 90% of the microorganisms in a sample under specified conditions. For example, in a semilogarithmic plot of the viable population versus the time of heating, the D value is the time required for the line to drop by one log cycle or 10-fold (figure 27.2a). It is also possible to determine the temperature change at a given D value that decreases the microbial population by one log cycle (90%). This temperature change is referred to as the Z value and is predicted from a semilogarithmic plot of D values versus temperature
Patterns of death similar to growth
20
New cards
-The effectiveness of an agent varies greatly with the nature of the organisms being treated because microorganisms differ markedly in susceptibility. Bacterial endospores are much more resistant to most antimicrobial agents than vegetative cells, and younger cells are usually more readily destroyed than mature organisms. Some species are able to withstand adverse conditions better than others. For instance, M. tuberculosis is much more resistant to antimicrobial agents than most other bacteria.
Population composition
21
New cards
-Often, but not always, the more concentrated a chemical agent or intense a physical agent, the more rapidly microorganisms are destroyed. However, agent effectiveness usually is not directly related to concentration or intensity. Over a short range, a small increase in concentration leads to an exponential rise in effectiveness; beyond a certain point, increases may not raise the killing rate much at all. Sometimes an agent is more effective at lower concentrations. For example, 70% ethanol is more bactericidal than 95% ethanol because the activity of ethanol is enhanced by the presence of water.
Concentration/intensity of an antimicrobial agent

22
New cards
-The longer a population is exposed to a microbicidal agent, the more organisms are killed. To achieve sterilization, contact time should be long enough to reduce the probability of survival by at least 6 logs.
Contact time
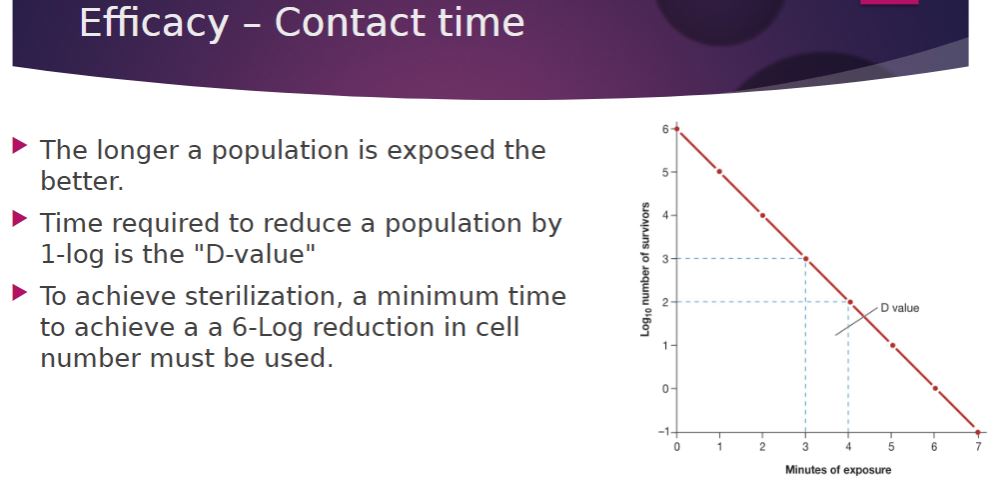
23
New cards
-An increase in the temperature at which a chemical is used often enhances its activity. Frequently a lower concentration of disinfectant or sterilizing agent can be used at a higher temperature.
Temperature
24
New cards
-The population to be controlled is not isolated but surrounded by environmental factors that may either offer protection or aid in its destruction. For example, because heat kills more readily at an acidic pH, acidic foods and beverages such as fruits and tomatoes are easier to pasteurize than more alkaline foods such as milk. A second important environmental factor is organic matter, which can protect microorganisms against physical and chemical disinfecting agents. Biofilms are a good example. The organic matter in a biofilm protects the biofilm’s residents. Furthermore, bacteria in biofilms are altered physiologically, and this makes them less susceptible to many antimicrobial agents. Because of the impact of organic matter, it may be necessary to clean objects, especially medical and dental equipment, before they are disinfected or sterilized.
Local environment

25
New cards
a
How to choose what cleaning method

26
New cards
-The emerging field of biological control of microorganisms holds great promise. Scientists are learning to exploit natural control processes such as predation of one microorganism on another, viral-mediated lysis, and toxin-mediated killing. While these mechanisms occur in nature, their use by humans is relatively new. Studies evaluating control of the human intestinal pathogens Salmonella spp., Shigella spp., and E. coli by Gram-negative predators such as Bdellovibrio spp. suggest that poultry farms may be sprayed with a predatory bacterium to reduce potential contamination.
Biological Control Mechanisms

27
New cards
-Bactericidal Action of Penicillin. The Penicillium mold colony secretes penicillin that kills the Staphylococcus aureus streaked nearby.
Antibiotics
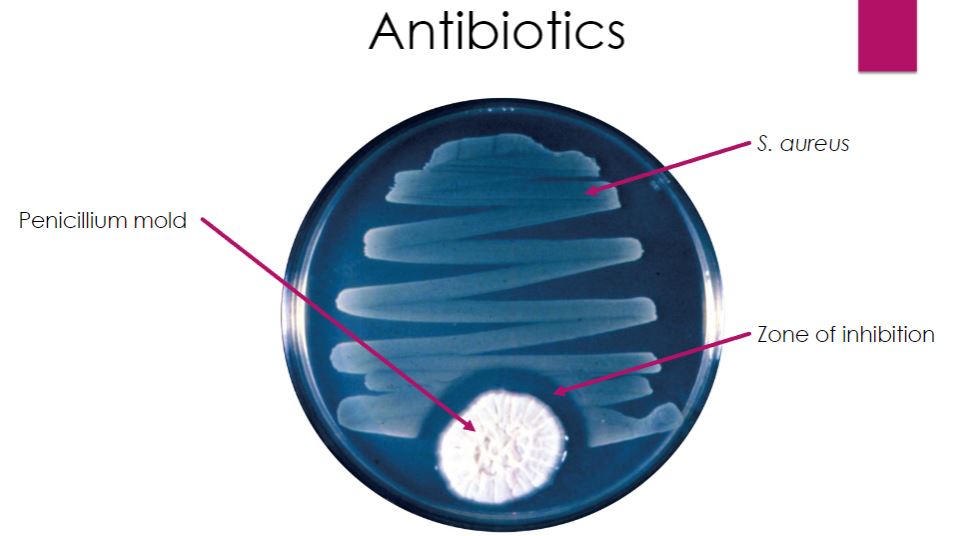
28
New cards
a
Antibiotic definition

29
New cards
-The modern era of chemotherapy began with the work of the German physician Paul Ehrlich (1854–1915). Ehrlich was fascinated with dyes that specifically bind to microbial cells. He reasoned that one of the dyes could be a chemical that would selectively destroy pathogens without harming human cells—a “magic bullet.” As mentioned in the chapter opener, Ehrlich and a young Japanese scientist named Sahachiro Hata (1873–1938) developed a treatment for syphilis by testing a variety of arsenic-based chemicals on infected rabbits. They found that arsphenamine was active against the syphilis spirochete. Arsphenamine was made available in 1910 under the trade name Salvarsan. Ehrlich and Hata’s success led to the screening of a vast number of chemicals for other magic bullets. In 1927 Gerhard Domagk (1895–1964) discovered that Prontosil red, a new dye for staining leather, protected mice completely against pathogenic streptococci and staphylococci without apparent toxicity. Jacques (1897–1977) and Therese (1892–1978) Trefouel later showed that the body metabolized the dye to sulfanilamide. Domagk received the 1939 Nobel Prize in Physiology or Medicine for his discovery of sulfonamides, or sulfa drugs.
Origin of antibiotics

30
New cards
a
Ehrlich and Hata
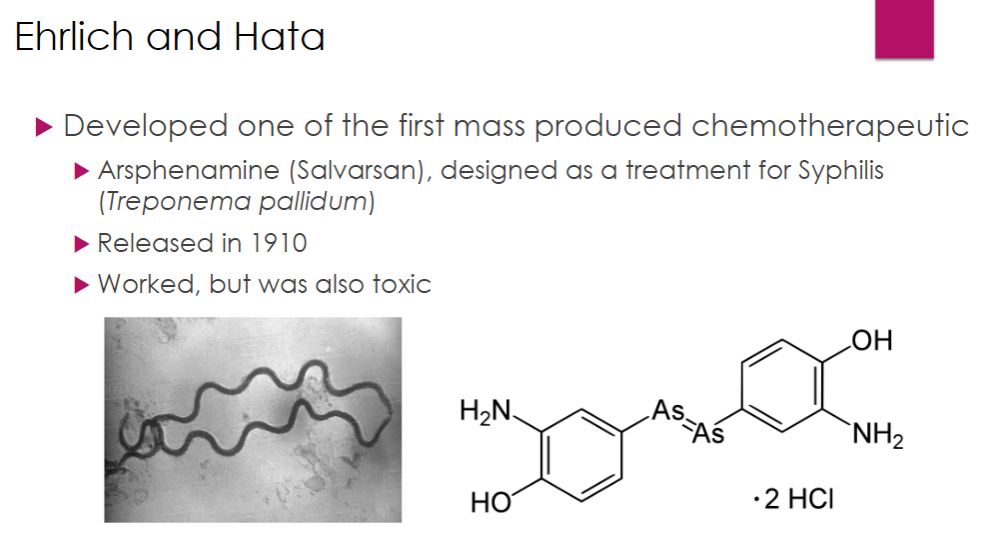
31
New cards
a
Gehard Domagk
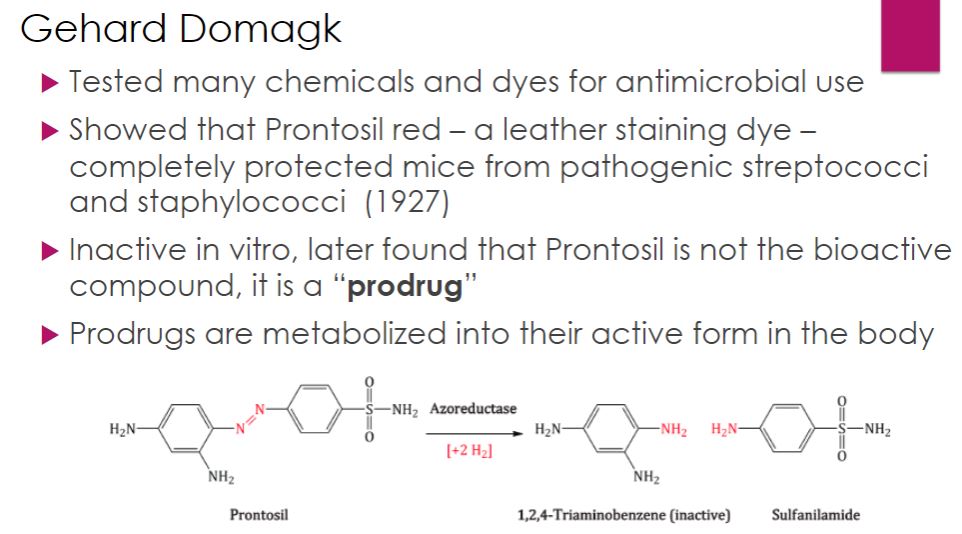
32
New cards
-penicillin-
A β-lactam antibiotic. The first true antibiotic to be discovered and used clinically.
-was first discovered in 1896 by a twenty-one-year-old French medical student named Ernest Duchesne (1874–1912). His work was forgotten until Alexander Fleming (1881–1955) accidentally rediscovered penicillin in 1928. After returning from a weekend vacation, Fleming noticed that a Petri plate of staphylococci also had mold growing on it and there were no bacterial colonies surrounding it (figure 28.1). Although the precise events are still unclear, it has been suggested that a Penicillium notatum spore had contaminated the Petri dish before it had been inoculated with the staphylococci. The mold apparently grew before the bacteria and produced penicillin. The bacteria nearest the fungus were lysed. Fleming correctly deduced that the mold produced a diffusible substance, which he called penicillin. However, Fleming could not demonstrate that penicillin remained active in vivo long enough to destroy pathogens and he dropped the research.
A β-lactam antibiotic. The first true antibiotic to be discovered and used clinically.
-was first discovered in 1896 by a twenty-one-year-old French medical student named Ernest Duchesne (1874–1912). His work was forgotten until Alexander Fleming (1881–1955) accidentally rediscovered penicillin in 1928. After returning from a weekend vacation, Fleming noticed that a Petri plate of staphylococci also had mold growing on it and there were no bacterial colonies surrounding it (figure 28.1). Although the precise events are still unclear, it has been suggested that a Penicillium notatum spore had contaminated the Petri dish before it had been inoculated with the staphylococci. The mold apparently grew before the bacteria and produced penicillin. The bacteria nearest the fungus were lysed. Fleming correctly deduced that the mold produced a diffusible substance, which he called penicillin. However, Fleming could not demonstrate that penicillin remained active in vivo long enough to destroy pathogens and he dropped the research.
Penicillin
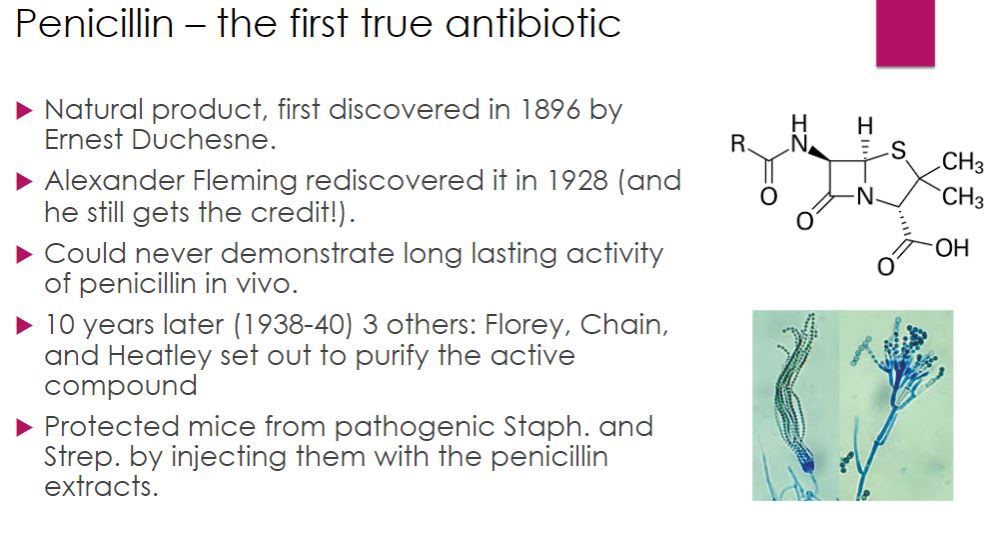
33
New cards
a
Penicillin demand

34
New cards
a
Penicillin continued

35
New cards
a
The antibiotic era

36
New cards
a
Streptomyces antibiotic powerhouses
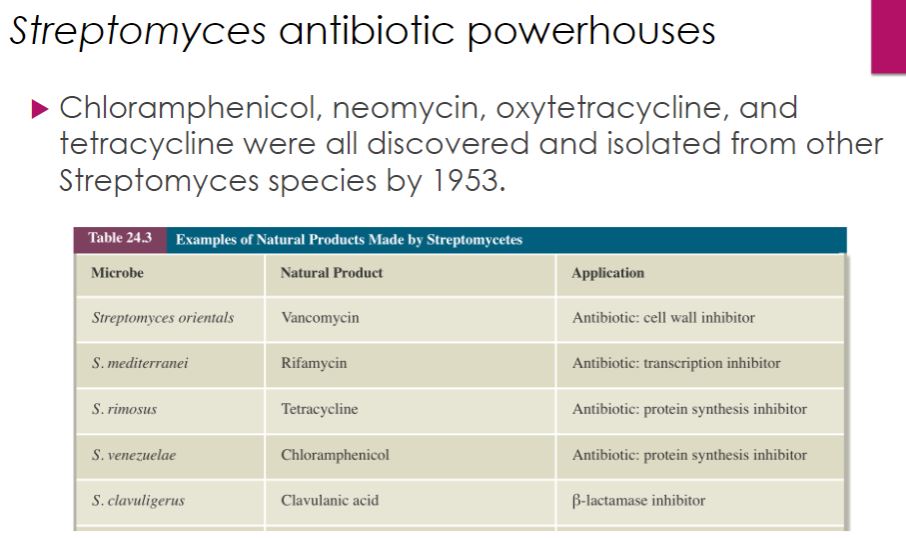
37
New cards
-selective toxicity-
The ability of a chemotherapeutic agent to kill or inhibit a microbial pathogen while damaging the host as little as possible.
The degree of selective toxicity may be expressed in terms of: (1) the therapeutic dose—the drug level required for treatment of a particular infection, and
(2) the toxic dose—the drug level at which the agent becomes too toxic for the host.
-therapeutic index
The ratio between the toxic dose and the therapeutic dose of a drug, used as a measure of the drug’s relative safety.
-The larger the therapeutic index, the better the chemotherapeutic agent in general.
The ability of a chemotherapeutic agent to kill or inhibit a microbial pathogen while damaging the host as little as possible.
The degree of selective toxicity may be expressed in terms of: (1) the therapeutic dose—the drug level required for treatment of a particular infection, and
(2) the toxic dose—the drug level at which the agent becomes too toxic for the host.
-therapeutic index
The ratio between the toxic dose and the therapeutic dose of a drug, used as a measure of the drug’s relative safety.
-The larger the therapeutic index, the better the chemotherapeutic agent in general.
Properties of good antibiotics

38
New cards
-Antimicrobial agents are often classified as either narrow-spectrum drugs—that is, they are effective only against a limited variety of pathogens—or broad-spectrum drugs that attack many different kinds of bacteria. Drugs may also be classified based on the general microbial group they act against: antibacterial, antifungal, antiprotozoan, and antiviral. A few agents can be used against more than one group; for example, sulfonamides are active against bacteria and some protozoa.
Properties of good antibiotics

39
New cards
a
Antibiotic Derivative
40
New cards
-chemotherapeutic agents can be either cidal or static (table 28.1). Static agents reversibly inhibit growth; if the agent is removed, the microorganisms will recover and grow again. Although a cidal agent kills the target pathogen, it may be static at low levels. The effect of an agent also varies with the target species; that is, an agent may be cidal for one species and static for another. Because static agents do not directly destroy the pathogen, elimination of the infection depends on the host’s own immunity mechanisms. A static agent may not be effective if the host is immunosuppressed.
Mechanistic categories of antibiotics

41
New cards
An estimate of the effectiveness of a chemotherapeutic agent against a pathogen can be obtained by determining the minimal inhibitory concentration (MIC; referred to as “mick”). The MIC is the lowest concentration of a drug that prevents growth of a particular pathogen. The minimal lethal concentration (MLC) is the lowest drug concentration that kills the pathogen. A cidal drug generally kills pathogens at levels two to four times higher than the MIC, whereas a static agent kills at much greater concentrations, if at all.
Effectiveness

42
New cards
-Dilution susceptibility tests can be used to determine MIC and MLC values. Antibiotic dilution tests can be done in both agar and broth. Media called Mueller-Hinton broth or agar is used because all conditions must be standardized. In the broth dilution test, tubes containing broth with antibiotic concentrations in the range of 0.1 to 128 mg per milliliter (twofold dilutions) are inoculated with a standard density of the test organism. The lowest concentration of the antibiotic resulting in no growth after 16 to 20 hours of incubation is the MIC. The MLC can be ascertained if the tubes showing no growth are then cultured into fresh medium lacking antibiotic. The lowest antibiotic concentration from which the microorganisms do not subsequently grow is the MLC. The agar dilution test is similar to the broth dilution test. Plates containing Mueller-Hinton agar and various amounts of antibiotic are inoculated and examined for growth.
Measuring effectiveness

43
New cards
-In many situations, such as in analyzing rapidly growing pathogens, a disk diffusion technique is used. This assay is fairly simple. Mueller-Hinton agar is inoculated with the bacterium isolated from the clinical sample. Small paper disks, each impregnated with a different antibiotic, are placed on the inoculated agar. When the antibiotic diffuses radially outward through the agar, it produces a concentration gradient. The antibiotic is present at high concentrations near the disk and affects even minimally susceptible microorganisms. On the other hand, resistant organisms will grow close to the disk. As the distance from the disk increases, the antibiotic concentration decreases and only more susceptible pathogens are harmed. A clear zone or ring forms around an antibiotic disk after incubation if the agent inhibits bacterial growth. The wider this zone of inhibition, the more susceptible the pathogen is. Zone width also is a function of the antibiotic’s initial concentration, its solubility, and its diffusion rate through agar. Thus zone widths around different antibiotic discs cannot be used to compare their effectiveness.
-Kirby-Bauer test
-Kirby-Bauer test
Disk Diffusion Tests

44
New cards
-The Etest from bioMérieux S.A. is often used in sensitivity testing. Unlike the Kirby-Bauer test, the MIC is also determined by the Etest. In general, each bacterial isolate to be tested for antimicrobial sensitivities is inoculated on the surface of an agar medium and Etest strips are then placed on the surface . Each strip contains a gradient of an antibiotic and is labeled with a scale of values. After 24 to 48 hours of incubation, an elliptical zone of inhibition appears. As shown in figure 28.4, MICs are determined from the point of intersection between the zone of inhibition and the strip’s scale of values.
Etest

45
New cards
-The most selective antibiotics are those that interfere with bacterial cell wall synthesis. Drugs such as penicillins, cephalosporins, and vancomycin have a high therapeutic index because they target structures and functions not found in eukaryotic cells.
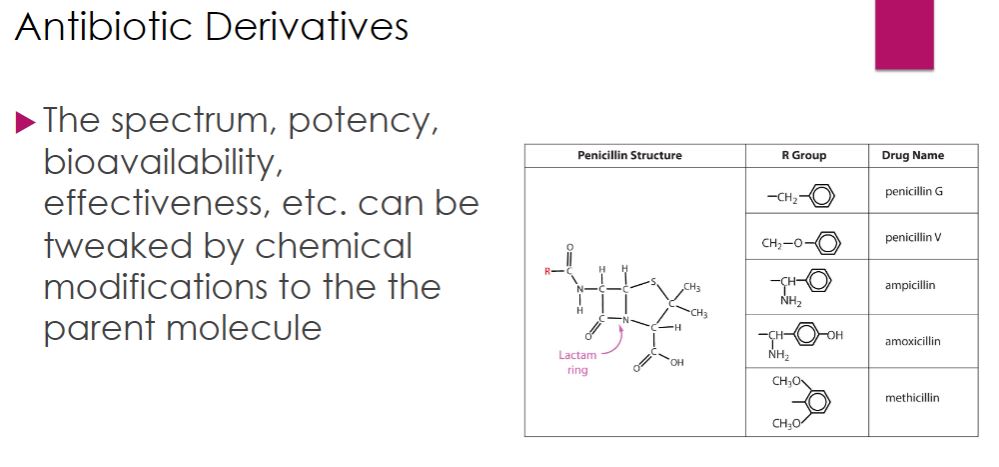
-Penicillins differ in several ways. The two naturally occurring penicillins, penicillin G and penicillin V, are narrow-spectrum drugs, targeting Gram-positive bacteria (figure 28.5). Penicillin G must be administered by injection (parenterally) because it is destroyed by stomach acid. Penicillin V can be given orally because it is more resistant to stomach acid. Almost immediately after penicillin was introduced, bacterial resistance was discovered. Bacteria were found to produce enzymes called penicillinases, which cleave the β-lactam ring thereby inactivating the drug (figure 28.5). For this reason, they are also called β-lactamases. To fight this and broaden the spectrum of activity, semisynthetic penicillins were developed. Among the first semisynthetic penicillins were the antistaphylococcal drugs nafcillin, oxacillin, dicloxacillin, and methicillin. These compounds have bulkier side chains than natural penicillin, making them more difficult for penicillinase enzymes to degrade. Next came the aminopenicillins, which are more hydrophilic allowing passage through porins in Gram-negative outer membranes. While the antistaphylococcal penicillins retain the narrow Gram-positive spectrum of the parent compound, aminopenicillins such as ampicillin and amoxicillin have broader coverage that includes many Gram negatives. Later, extended-spectrum penicillins, such as piperacillin (figure 28.5) were developed to specifically resist penicillinase-producing bacteria. Unfortunately, but not surprising, these newer drugs are not immune from bacterial resistance.
-Penicillins differ in several ways. The two naturally occurring penicillins, penicillin G and penicillin V, are narrow-spectrum drugs, targeting Gram-positive bacteria (figure 28.5). Penicillin G must be administered by injection (parenterally) because it is destroyed by stomach acid. Penicillin V can be given orally because it is more resistant to stomach acid. Almost immediately after penicillin was introduced, bacterial resistance was discovered. Bacteria were found to produce enzymes called penicillinases, which cleave the β-lactam ring thereby inactivating the drug (figure 28.5). For this reason, they are also called β-lactamases. To fight this and broaden the spectrum of activity, semisynthetic penicillins were developed. Among the first semisynthetic penicillins were the antistaphylococcal drugs nafcillin, oxacillin, dicloxacillin, and methicillin. These compounds have bulkier side chains than natural penicillin, making them more difficult for penicillinase enzymes to degrade. Next came the aminopenicillins, which are more hydrophilic allowing passage through porins in Gram-negative outer membranes. While the antistaphylococcal penicillins retain the narrow Gram-positive spectrum of the parent compound, aminopenicillins such as ampicillin and amoxicillin have broader coverage that includes many Gram negatives. Later, extended-spectrum penicillins, such as piperacillin (figure 28.5) were developed to specifically resist penicillinase-producing bacteria. Unfortunately, but not surprising, these newer drugs are not immune from bacterial resistance.
Cell wall biosynthesis inhibitor
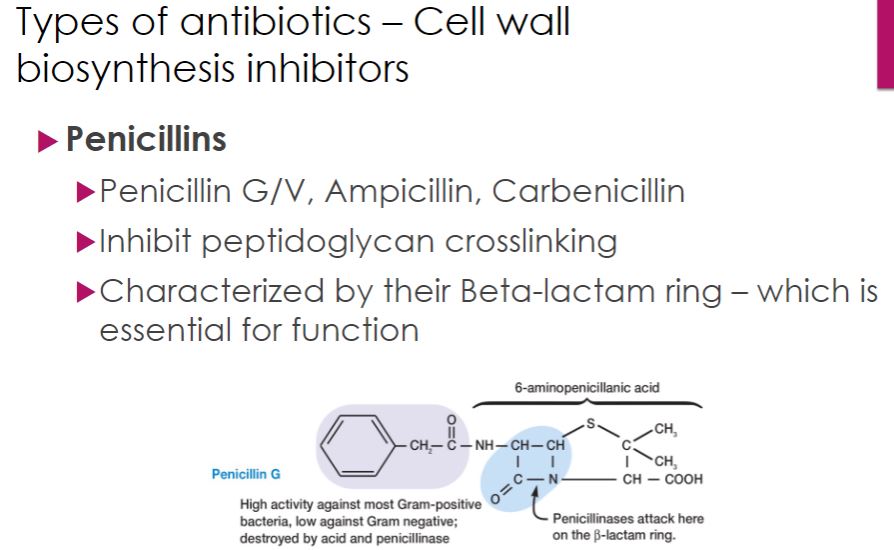
46
New cards
Recall that when new peptidoglycan units are added to a growing cell wall, the peptide side chain on one peptidoglycan subunit must be linked to the peptide side chain on another subunit in a process known as transpeptidation (figure 28.6; also see figure 3.20). The structure of the penicillins resembles the two amino acids at the end of the side chain, called the terminal d-alanyl-d-alanine. This structural similarity blocks the transpeptidation reaction that forms the peptidoglycan cross-links because the transpeptidation enzyme binds penicillin instead of its true substrate. In fact, these transpeptidases are commonly called penicillin-binding proteins (PBPs). Because cell wall formation is blocked, osmotic lysis occurs. Penicillins also destroy bacteria by activating their own autolytic enzymes. Thus penicillins act only on growing bacteria actively synthesizing new peptidoglycan.
cell wall biosynthesis inhibitor continue
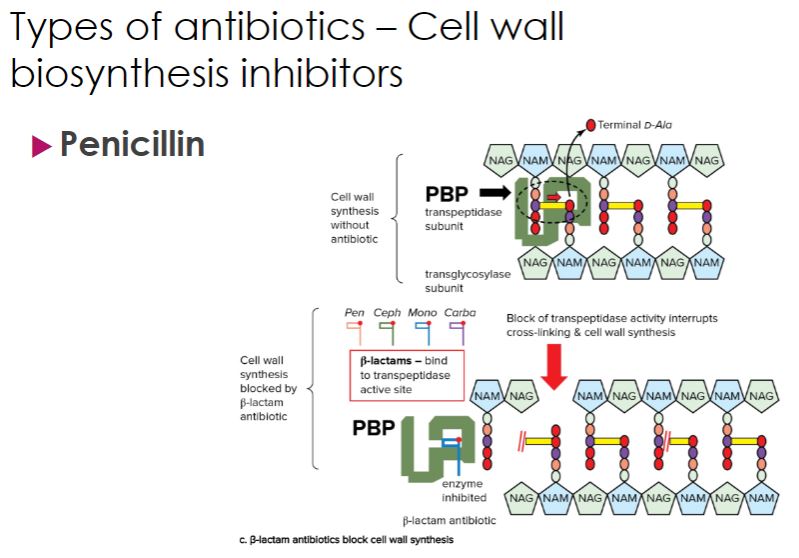
47
New cards
-Cephalosporins are a family of antibiotics originally isolated in 1948 from the fungus Cephalosporium. They contain a β-lactam structure similar to that of the penicillins but were found to be more resistant to β-lactamase attack. As might be expected from their structural similarities to penicillins, cephalosporins also inhibit the transpeptidation reaction during peptidoglycan synthesis. The earlier generation cephalosporins have better Gram-positive coverage than later generations, which are more effective against a wider range of Gram negatives.
cephalosporin
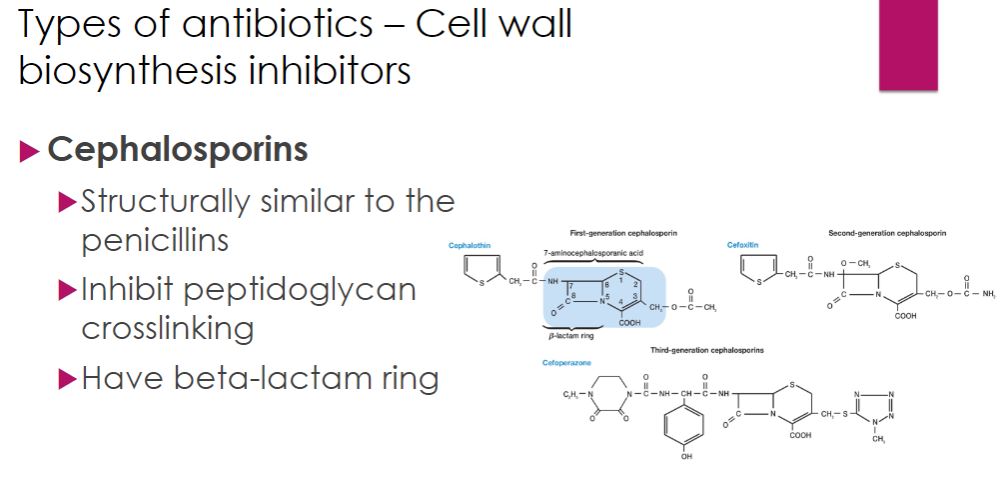
48
New cards
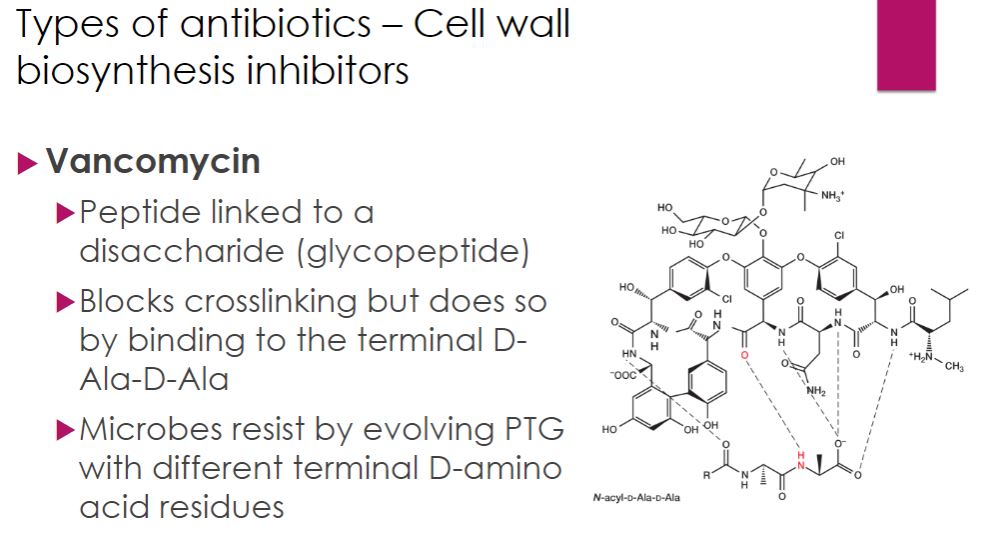
-Vancomycin is a glycopeptide antibiotic produced by the bacterium Streptomyces orientalis. It binds specifically to the d-alanyl-d-alanine terminal sequence on the pentapeptide portion of peptidoglycan. Thus like β-lactam antibiotics, vancomycin blocks transpeptidation, but it binds to the substrate rather than the enzyme itself.
-Vancomycin is bactericidal only for Gram-positive bacteria. It is most often given intravenously because it is not absorbed from the gastrointestinal tract. For this reason, it is only prescribed orally for Clostridioides difficile infection, which causes debilitating diarrhea (see chapter 24 opening story). It is an important drug in the treatment of antibiotic-resistant staphylococcal and enterococcal infections. However, vancomycin-resistant strains of Enterococcus have emerged, and a few cases of resistant Staphylococcus aureus have been reported. Vancomycin resistance poses a serious public health threat; vancomycin is one of the “drugs of last resort” in cases of methicillin-resistant Staphylococcus aureus (MRSA), which are resistant to all β-lactam drugs
-Vancomycin is bactericidal only for Gram-positive bacteria. It is most often given intravenously because it is not absorbed from the gastrointestinal tract. For this reason, it is only prescribed orally for Clostridioides difficile infection, which causes debilitating diarrhea (see chapter 24 opening story). It is an important drug in the treatment of antibiotic-resistant staphylococcal and enterococcal infections. However, vancomycin-resistant strains of Enterococcus have emerged, and a few cases of resistant Staphylococcus aureus have been reported. Vancomycin resistance poses a serious public health threat; vancomycin is one of the “drugs of last resort” in cases of methicillin-resistant Staphylococcus aureus (MRSA), which are resistant to all β-lactam drugs
Vancomycin
49
New cards
-Many antibiotics inhibit protein synthesis by binding bacterial ribosomal proteins or rRNA. Because these drugs discriminate between bacterial and eukaryotic ribosomes, their therapeutic index is fairly high but not as high as that of cell wall synthesis inhibitors. Several different steps in protein synthesis can be affected by drugs in this category.
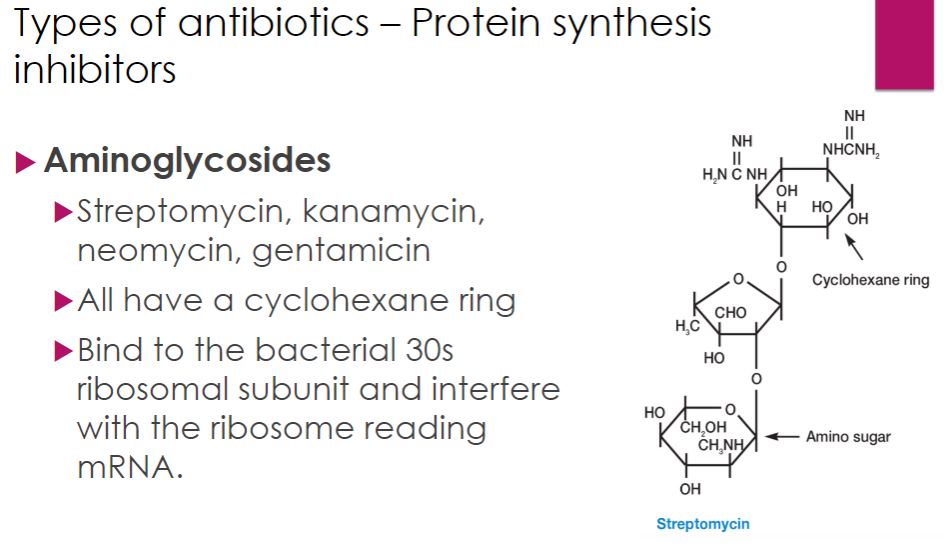
-Aminoglycosides Although considerable variation in structure occurs among aminoglycoside antibiotics, all contain a cyclohexane ring and amino sugars (figure 28.9). Streptomycin, kanamycin, neomycin, and tobramycin are synthesized by different species of the bacterial genus Streptomyces, whereas gentamicin comes from a related bacterium, Micromonospora purpurea. Aminoglycosides are generally used to treat infections caused by Gram-negative bacteria. However, because they can be quite toxic they are used sparingly.
-All aminoglycoside antibiotics disrupt peptide elongation during translation. This occurs as aminoglycosides bind to ribosomal RNA of the bacterial 30S ribosomal subunit, interfering with mRNA reading and/or causing early termination of peptide synthesis. Aminoglycoside side effects are thought to be due to their ability to bind to host mitochondrial ribosomes, which share the same 30S binding site as their bacterial ancestors.
-Aminoglycosides Although considerable variation in structure occurs among aminoglycoside antibiotics, all contain a cyclohexane ring and amino sugars (figure 28.9). Streptomycin, kanamycin, neomycin, and tobramycin are synthesized by different species of the bacterial genus Streptomyces, whereas gentamicin comes from a related bacterium, Micromonospora purpurea. Aminoglycosides are generally used to treat infections caused by Gram-negative bacteria. However, because they can be quite toxic they are used sparingly.
-All aminoglycoside antibiotics disrupt peptide elongation during translation. This occurs as aminoglycosides bind to ribosomal RNA of the bacterial 30S ribosomal subunit, interfering with mRNA reading and/or causing early termination of peptide synthesis. Aminoglycoside side effects are thought to be due to their ability to bind to host mitochondrial ribosomes, which share the same 30S binding site as their bacterial ancestors.
Protein synthesis inhibitor
50
New cards
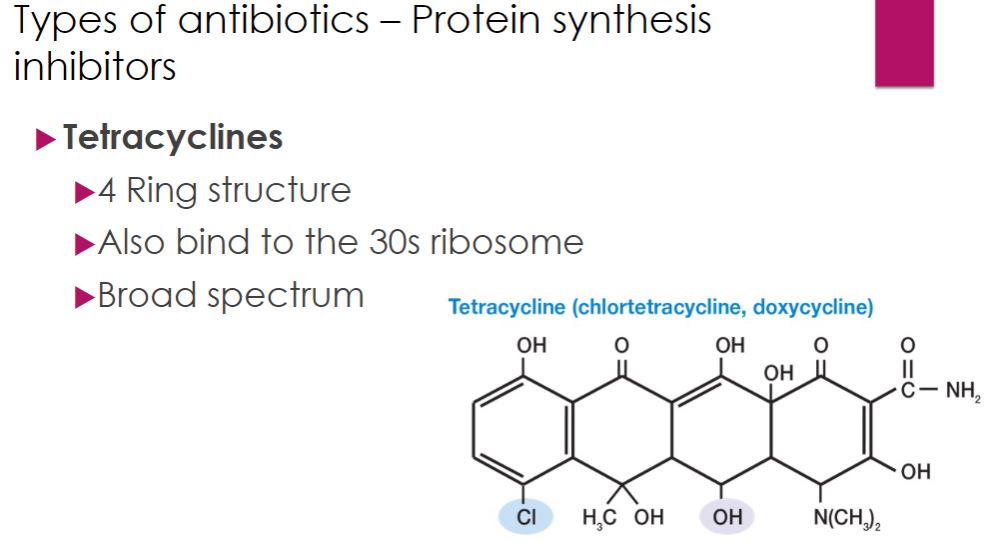
-The tetracyclines are a family of antibiotics with a common four-ring structure to which a variety of side chains are attached (figure 28.10). Oxytetracycline and chlortetracycline are produced naturally by Streptomyces spp., whereas other tetracyclines are semisynthetic. These antibiotics are similar to aminoglycosides in that they target the 30S subunit of the ribosome, inhibiting protein synthesis. Tetracyclines are broad-spectrum antibiotics active against most bacteria, including the intracellular pathogens rickettsias, chlamydiae, and mycoplasmas.
tetracycline
51
New cards
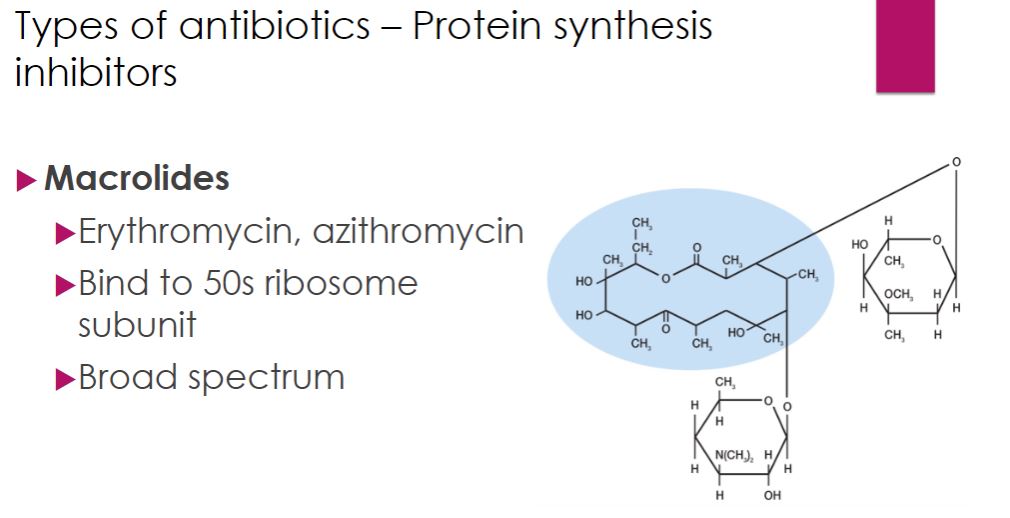
-The macrolide antibiotics contain a ring structure consisting of 12 to 22 carbons called a lactone ring. The lactone ring is linked to one or more sugars (figure 28.11). Erythromycin binds to the 50S ribosomal subunit to inhibit bacterial protein elongation. Erythromycin is a relatively broad-spectrum antibiotic effective against Gram-positive bacteria, mycoplasmas, and some Gram-negative bacteria. It is used with patients who are allergic to penicillins and in the treatment of diarrhea caused by Campylobacter, and pneumonia from Legionella or Mycoplasma infections. Azithromycin has surpassed erythromycin in use. Ketolides are semisynthetic macrolide derivatives with a similar mechanism of action. They benefit from better acid stability and bioavailability (the amount of a drug that reaches the circulation). Telithromycin is a ketolide used to treat community-acquired pneumonia.
Macrolides
52
New cards
-Several antimicrobial drugs act as antimetabolites, as they antagonize, or block, the functioning of metabolic pathways. Antimetabolites are structurally similar to substrates of key enzymes and compete with the metabolites for the binding site of these enzymes. However, once bound to the enzyme, the antimetabolites are different enough to block enzyme activity and further progression of the pathway. By preventing metabolism, they are broad spectrum
Antimetabolites

53
New cards
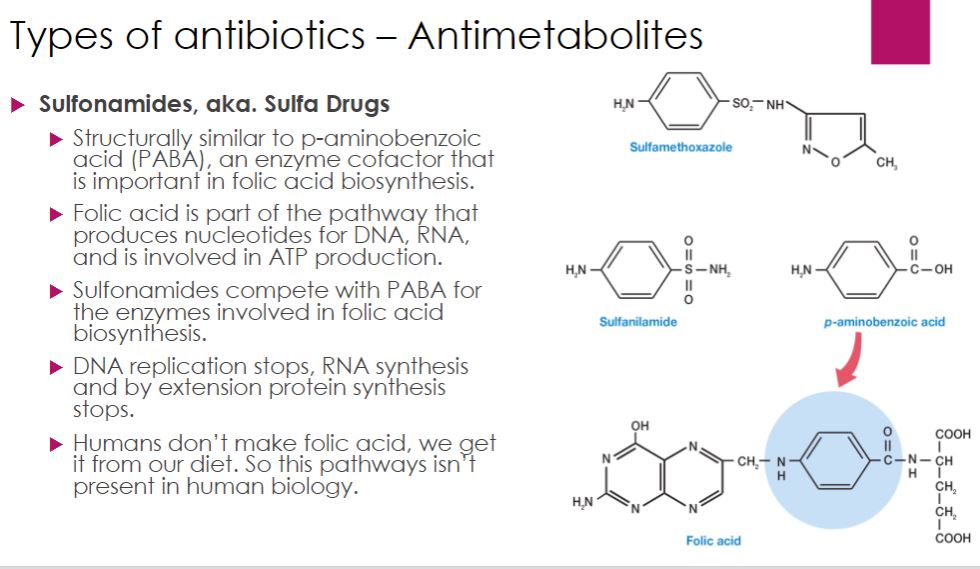
Sulfonamides, or sulfa drugs, are structural analogues of p-aminobenzoic acid, or PABA (figure 28.12). PABA is needed for folic acid (folate) synthesis. Folic acid is required for the synthesis of purines. Thus by depleting the pool of folate in the cell, sulfonamides prevent synthesis of DNA, RNA, proteins, and other important cell constituents (e.g., ATP). When a sulfa drug enters a bacterial cell, it competes with PABA for the active site of an enzyme (dihydropteroate synthase) involved in one of the first steps in folic acid synthesis (see figure 6.14). Sulfonamides are selectively toxic for many bacteria and protozoa because these microbes manufacture their own folate and cannot effectively take up this cofactor, whereas humans do not synthesize folate; instead, we must obtain it in our diet. Sulfonamides thus have a high therapeutic index.
Sulfonamides or Sulfa drugs
54
New cards
-The most commonly used antibacterial drugs that inhibit nucleic acid synthesis function by inhibiting (1) DNA gyrase and topoisomerases (fluoroquinolones) or (2) RNA polymerase (rifamycins). Neither are as selectively toxic as other antibacterial agents because bacteria and eukaryotes do not differ greatly with respect to nucleic acid synthesis. The most commonly used drugs in this category are the fluoroquinolones.
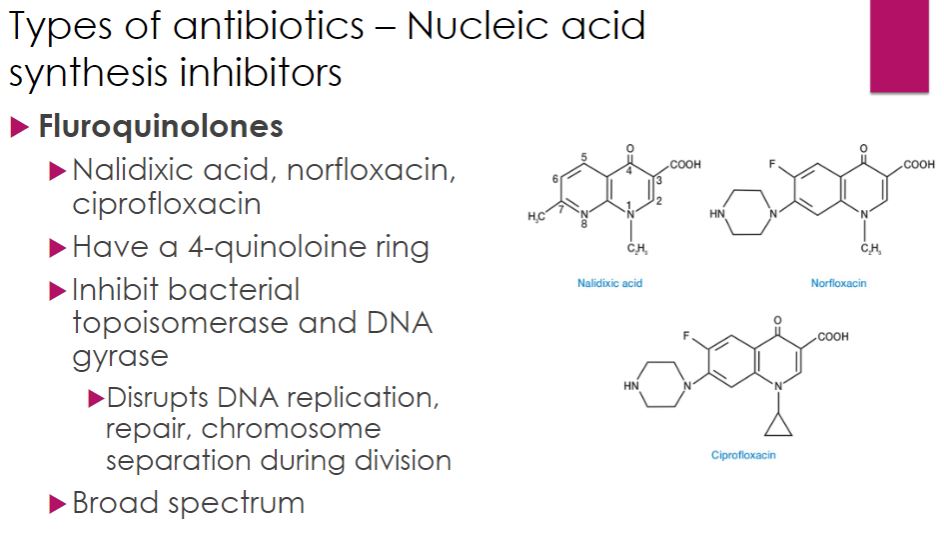
-Fluoroquinolones are synthetic drugs that feature a fluorinated four-quinolone ring (figure 28.14). They act by binding to the bacterial topoisomerases DNA gyrase and topoisomerase IV. Inhibition of these enzymes disrupts DNA replication and repair, bacterial chromosome separation during division, and other processes involving DNA.
-The fluoroquinolones have been used to treat a wide variety of infections. This success has led to their overuse and fluoroquinolone resistance is now problematic. In addition, because they can cause disabling damage to tendons and nerves, the U.S. Food and Drug Administration has deemed them too dangerous to use except for specific infections.
-Fluoroquinolones are synthetic drugs that feature a fluorinated four-quinolone ring (figure 28.14). They act by binding to the bacterial topoisomerases DNA gyrase and topoisomerase IV. Inhibition of these enzymes disrupts DNA replication and repair, bacterial chromosome separation during division, and other processes involving DNA.
-The fluoroquinolones have been used to treat a wide variety of infections. This success has led to their overuse and fluoroquinolone resistance is now problematic. In addition, because they can cause disabling damage to tendons and nerves, the U.S. Food and Drug Administration has deemed them too dangerous to use except for specific infections.
Nucleic Acid: fluoroquinolones
55
New cards
-Treatment of fungal infections generally has been less successful than that of bacterial infections largely because as eukaryotes, fungal cells are much more similar to human cells than are bacterial cells. Many drugs that inhibit or kill fungi therefore have a low therapeutic index. Nonetheless, a few drugs are useful in treating many major fungal diseases.
Antifungals

56
New cards
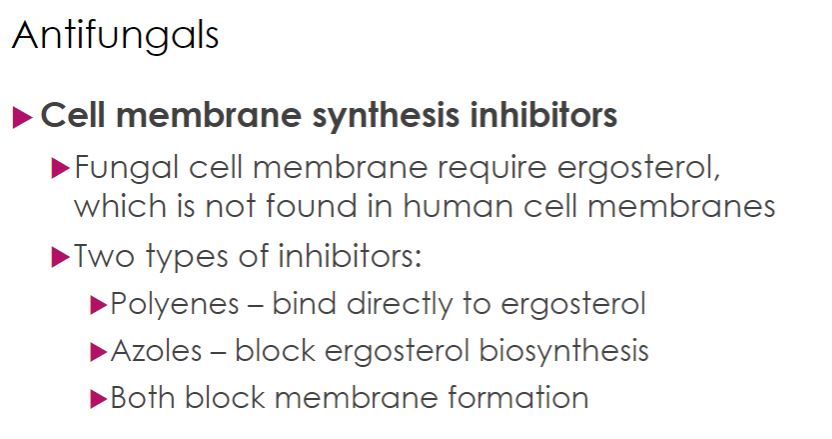
-There are two major classes of antifungal drugs for severe and systemic infections: polyenes and azoles; both block fungal cell membrane synthesis. This is a good drug target because fungal membranes require the sterol ergosterol, not found in human membranes. Polyenes bind directly to ergosterol, and azoles block the last step in ergosterol biosynthesis. Thus both drugs disrupt fungal membrane formation.
Cell membrane synthesis inhibitors
57
New cards
-As discussed in chapter 17, fungal infections are often subdivided into infections called superficial mycoses, subcutaneous mycoses, systemic mycoses, and opportunistic mycoses. Treatment for each type of mycosis differs. Because superficial and cutaneous mycoses are limited to the skin and hair, these are treated with topical forms of azole drugs. Superficial mycoses of the nails are often treated with the agent griseofulvin, which is taken orally. This agent disrupts fungal mitotic spindles thereby preventing cell division. Nystatin, a polyene antibiotic from Streptomyces noursei, is used to control opportunistic Candida infections of the skin, vagina, or mouth and esophagus. It is too toxic to be taken systemically; when used as a “swish and swallow” agent for thrush, it is not absorbed from the gastrointestinal tract.
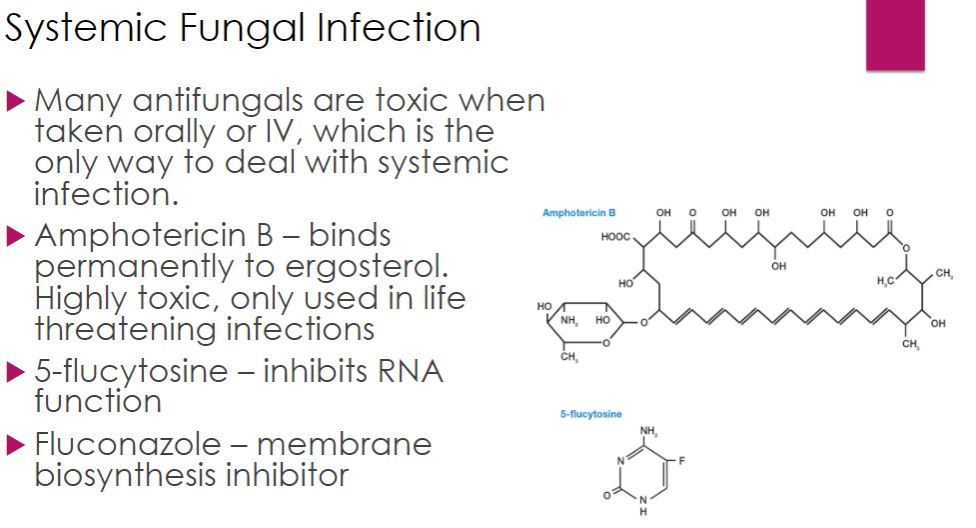
-Systemic fungal infections are very difficult to control and can be fatal. Drugs commonly used against systemic mycoses include amphotericin B, 5-flucytosine, and azoles such as fluconazole, itraconazole, and voriconazole. Amphotericin B, a polyene from Streptomyces spp., is quite toxic to humans earning it the nickname “ampho-terrible.” It is used only for serious, life-threatening infections.
-Systemic fungal infections are very difficult to control and can be fatal. Drugs commonly used against systemic mycoses include amphotericin B, 5-flucytosine, and azoles such as fluconazole, itraconazole, and voriconazole. Amphotericin B, a polyene from Streptomyces spp., is quite toxic to humans earning it the nickname “ampho-terrible.” It is used only for serious, life-threatening infections.
Types of fungal infection

58
New cards
a
Systemic fungal infection
59
New cards
-The discovery of small molecules that inhibit virus-specific enzymes and replication cycle processes has led to the development of antiviral drugs. However, antiviral drugs have had mixed success and the vast majority of viral infections cannot be cured. Some antiviral drugs simply limit the duration of the illness
Antiviral

60
New cards
-Like fungi, protozoa are eukaryotes so the potential for drug action on host cells and tissues is greater than it is when targeting bacteria. Most of the drugs used to treat protozoan infections have significant side effects; nonetheless, the side effects are usually acceptable when weighed against the parasitic burden. The number of antiprotozoan drugs is relatively small, and the mechanism of action for most is not completely understood. The drugs described here appear to act on protozoan nucleic acids or some metabolic process.
Antiprotozoan

61
New cards
a
antimicrobial resistance

62
New cards
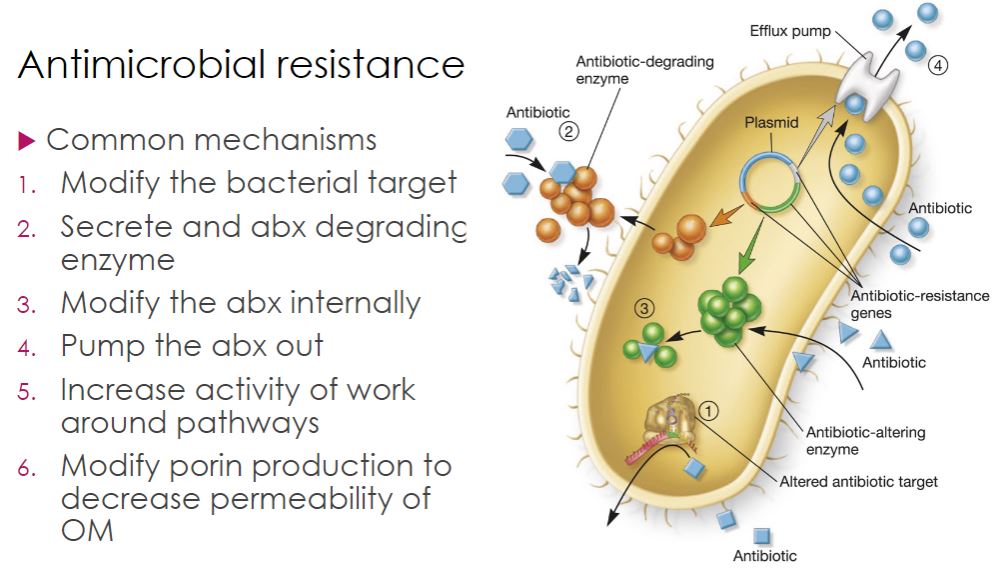
-Bacteria have evolved several resistance mechanisms (figure 28.18). One is to modify the target of the antibiotic. For example, MRSA is resistant to all β-lactam antibiotics because it has replaced its transpeptidase (penicillin-binding protein [PBP]) with another version of this enzyme (figure 28.6c). This altered PBP still functions as a transpeptidase, but has no binding site for β-lactam antibiotics. This explains why MRSA is resistant to all members of this class of antibiotics but remains sensitive to vancomycin. Recall that vancomycin binds a different target—the terminal d-ala of the pentapeptide on the peptidoglycan subunit (figure 28.6a). Resistance to this drug among some enterococci is another example of changing the target. Vancomycin resistance occurs when bacteria gain the capacity to change the terminal d-ala to a d-lactate or d-serine residue to which the drug cannot bind.
-A second resistance strategy is drug inactivation. The best-known example is hydrolysis of the β-lactam ring of penicillins by penicillinase and other β-lactamase enzymes. Drugs also are inactivated by the addition of chemical groups. For example, aminoglycosides can be modified and inactivated by acetylation of amino groups and phosphorylation or adenylylation of hydroxyl groups present on the aminoglycoside. Because these resistance strategies involve gaining enzymes that catalyze the inactivating reaction, the new function is acquired when bacteria share genes by horizontal gene transfer.
-The third resistance strategy minimizes the concentration of the antibiotic in the cell. This can be accomplished by altering membrane structure, particularly the outer membrane for Gram-negative bacteria, so that less antibiotic enters the cell. Another approach is to pump the drug out of the cell after it has entered, using translocases, called efflux pumps, that expel drugs. Efflux pumps are relatively nonspecific and pump many different drugs; therefore, they often confer multidrug resistance. Many efflux pumps are drug/proton antiporters; that is, protons enter the cell as the drug leaves. - Finally, resistant bacteria may either use an alternate pathway to bypass the biochemical reaction inhibited by the agent or increase the production of the target metabolite. For example, some bacteria are resistant to sulfonamides simply because they use preformed folic acid from their surroundings, rather than synthesize it themselves. Other strains increase their rate of folic acid production and thus counteract sulfonamide inhibition. Again, these changes are most often mediated by horizontal gene transfer.
-A second resistance strategy is drug inactivation. The best-known example is hydrolysis of the β-lactam ring of penicillins by penicillinase and other β-lactamase enzymes. Drugs also are inactivated by the addition of chemical groups. For example, aminoglycosides can be modified and inactivated by acetylation of amino groups and phosphorylation or adenylylation of hydroxyl groups present on the aminoglycoside. Because these resistance strategies involve gaining enzymes that catalyze the inactivating reaction, the new function is acquired when bacteria share genes by horizontal gene transfer.
-The third resistance strategy minimizes the concentration of the antibiotic in the cell. This can be accomplished by altering membrane structure, particularly the outer membrane for Gram-negative bacteria, so that less antibiotic enters the cell. Another approach is to pump the drug out of the cell after it has entered, using translocases, called efflux pumps, that expel drugs. Efflux pumps are relatively nonspecific and pump many different drugs; therefore, they often confer multidrug resistance. Many efflux pumps are drug/proton antiporters; that is, protons enter the cell as the drug leaves. - Finally, resistant bacteria may either use an alternate pathway to bypass the biochemical reaction inhibited by the agent or increase the production of the target metabolite. For example, some bacteria are resistant to sulfonamides simply because they use preformed folic acid from their surroundings, rather than synthesize it themselves. Other strains increase their rate of folic acid production and thus counteract sulfonamide inhibition. Again, these changes are most often mediated by horizontal gene transfer.
a
63
New cards
-Microbes, like all organisms, use their vast repertoire of chemical reactions to survive and reproduce. Indeed, metabolism is central to all life. Recall that metabolism consists of two parts: catabolism—the breaking down of nutrients and other molecules for energy and electrons, and anabolism—the use of carbon to build cell material. Despite the diversity of metabolic processes that have evolved over billions of years, there are several aspects that are common to all organisms. Here we list the common features, noting where they are discussed in this chapter.
-Life obeys the laws of thermodynamics (this section). Organisms must obtain energy from their environment; they cannot “make” energy, it must be captured or conserved.
-The energy cells obtain from their environment is most often conserved as ATP (section 6.2). ATP supplies the energy needed for certain chemical reactions to proceed.
-Oxidation-reduction (redox) reactions play a critical role in energy conservation (section 6.3). Many of these redox reactions occur in electron transport chains (section 6.4).
-The chemical reactions that occur in cells are organized into pathways (section 6.5). The product of one reaction is the substrate for the next reaction in the pathway.
-Each reaction of a pathway is catalyzed by an enzyme or a ribozyme (section 6.6). Enzymes and ribozymes are critical to all life as they speed up the reactions that enable life to exist.
-The functioning of biochemical pathways is regulated (section 6.7). This ensures that their products are made at the correct time.
-Life obeys the laws of thermodynamics (this section). Organisms must obtain energy from their environment; they cannot “make” energy, it must be captured or conserved.
-The energy cells obtain from their environment is most often conserved as ATP (section 6.2). ATP supplies the energy needed for certain chemical reactions to proceed.
-Oxidation-reduction (redox) reactions play a critical role in energy conservation (section 6.3). Many of these redox reactions occur in electron transport chains (section 6.4).
-The chemical reactions that occur in cells are organized into pathways (section 6.5). The product of one reaction is the substrate for the next reaction in the pathway.
-Each reaction of a pathway is catalyzed by an enzyme or a ribozyme (section 6.6). Enzymes and ribozymes are critical to all life as they speed up the reactions that enable life to exist.
-The functioning of biochemical pathways is regulated (section 6.7). This ensures that their products are made at the correct time.
Energy Needs

64
New cards
-Examine the common features of metabolism just described. It should be clear that cells must do work to survive and reproduce. Cells carry out three major types of work. Chemical work involves the synthesis of complex biological molecules from much simpler precursors (i.e., anabolism); energy is needed to increase the molecular complexity of a cell. Transport work requires energy to take up nutrients, eliminate wastes, and maintain ion balances. Energy input is needed because molecules and ions often must be transported across cell membranes against concentration and electrochemical gradients. The third type of work is mechanical work. Energy is required for cell motility and the movement of structures within cells, such as partitioning chromosomes during cell division. As just indicated, cells need energy to do work. Indeed, energy may be defined most simply as the capacity to do work. This is because all physical and chemical processes are the result of the application or movement of energy. Organisms obtain the energy they need from an energy source present in their environment. They convert the energy it provides into a useful form. Cellular energy is most commonly stored in the nucleotide triphosphate ATP. To understand how energy is conserved in ATP and how ATP is used to do cellular work, some knowledge of the basic principles of thermodynamics is required. The science of thermodynamics analyzes energy changes in a collection of matter (e.g., a cell or a plant) called a system. Thermodynamics focuses on the energy differences between the initial state and the final state of a system. It is not concerned with the rate of the process. For instance, if a pan of water is heated to boiling, only the condition of the water at the start and at boiling is important in thermodynamics, not how fast it is heated.
Where does energy come from?

65
New cards
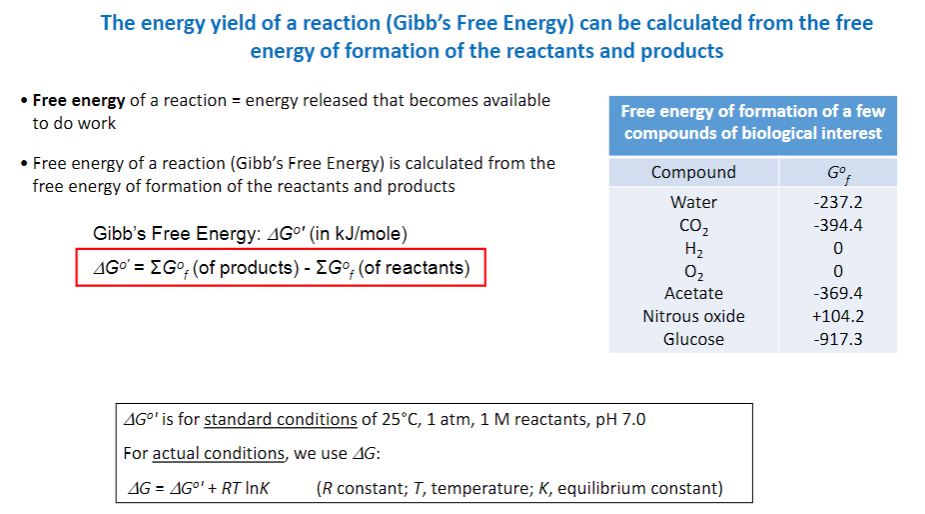
-The first and second laws of thermodynamics can be combined in a useful equation, relating the changes in energy that can occur in chemical reactions and other processes. ΔG = ΔH − T ΔS ΔG is the change in free energy, ΔH is the change in enthalpy, T is the temperature in Kelvin (°C + 273), and ΔS is the change in entropy occurring during the reaction. The change in enthalpy is the change in heat content. Cellular reactions occur under conditions of constant pressure and volume. Thus the change in enthalpy is about the same as the change in total energy during the reaction. The free energy change is the amount of energy in a system (or cell) available to do useful work at constant temperature and pressure. Therefore the change in entropy (ΔS) is a measure of the proportion of the total energy change that the system cannot use in performing work. Free energy and entropy changes do not depend on how the system gets from start to finish. A reaction will occur spontaneously—that is, without any external cause—if the free energy of the system decreases during the reaction or, in other words, if ΔG is negative. It follows from the equation that a reaction with a large positive change in entropy will normally tend to have a negative ΔG value and therefore occur spontaneously. A decrease in entropy will tend to make ΔG more positive and the reaction less favorable.
-R is the gas constant (1.9872 cal/mole-degree or 8.3145 J/mole-degree), and T is the absolute temperature. Inspection of this equation shows that when ΔG°′ is negative, the equilibrium constant is greater than one and the reaction goes to completion as written. It is said to be an exergonic reaction (figure 6.1). In an endergonic reaction, ΔG°′ is positive and the equilibrium constant is less than one. That is, the reaction is not favorable, and little product will be formed at equilibrium under standard conditions. Keep in mind that the ΔG°′ value shows only where the reaction lies at equilibrium, not how fast the reaction reaches equilibrium.
-R is the gas constant (1.9872 cal/mole-degree or 8.3145 J/mole-degree), and T is the absolute temperature. Inspection of this equation shows that when ΔG°′ is negative, the equilibrium constant is greater than one and the reaction goes to completion as written. It is said to be an exergonic reaction (figure 6.1). In an endergonic reaction, ΔG°′ is positive and the equilibrium constant is less than one. That is, the reaction is not favorable, and little product will be formed at equilibrium under standard conditions. Keep in mind that the ΔG°′ value shows only where the reaction lies at equilibrium, not how fast the reaction reaches equilibrium.
Free Energy
66
New cards
a
Gibbs free energy
67
New cards
a
Oxidation reduction (look into it and identify)
68
New cards
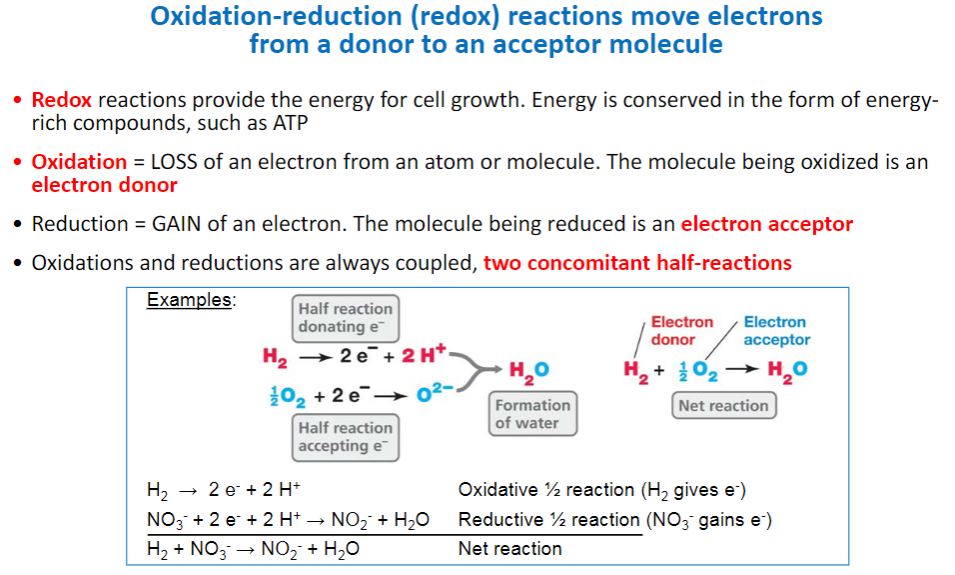
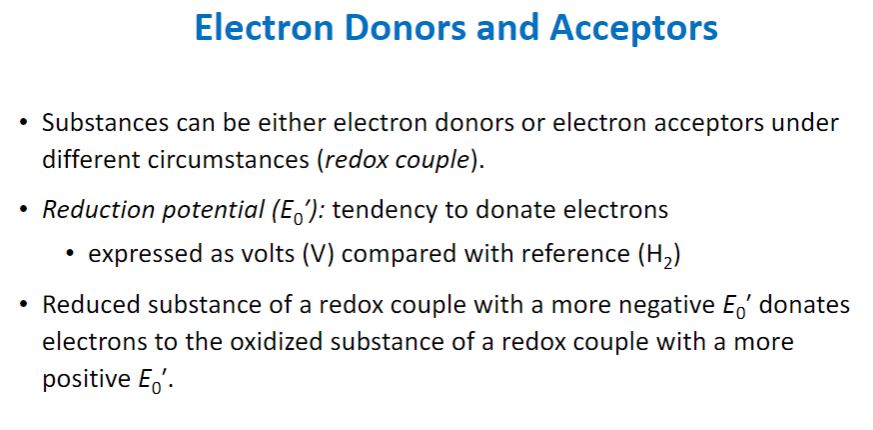
-Free energy changes are related to the equilibria of all chemical reactions, including the equilibria of oxidation-reduction reactions. The release of energy from an energy source normally involves oxidation-reduction reactions. Oxidation-reduction (redox) reactions are those in which electrons move from an electron donor to an electron acceptor.1 As electrons transfer from donor to acceptor, the donor becomes less energy rich and the acceptor becomes more energy rich. Thus electrons can be thought of as packets of energy. The more electrons a molecule has and is able to donate in a redox reaction, the more energy rich the molecule is. This explains why molecules such as glucose, which can donate up to 24 electrons in redox reactions, are such excellent sources of energy. Each redox reaction consists of two half reactions. One half reaction functions as the electron-donating half (i.e., an oxidation reaction), and the other functions as the electron-accepting half (i.e., the reduction). By convention, half reactions are written as reductions. As shown in table 6.2, the left side of each half reaction consists of a molecule that can accept electrons and the number (n) of electrons (e−) it accepts, while the right side includes the molecule it becomes after accepting the electrons. This molecule is referred to as a donor, because it has electrons it can give up. The acceptor and donor of a half reaction are referred to as a conjugate redox pair.
Electron donors and acceptors
69
New cards
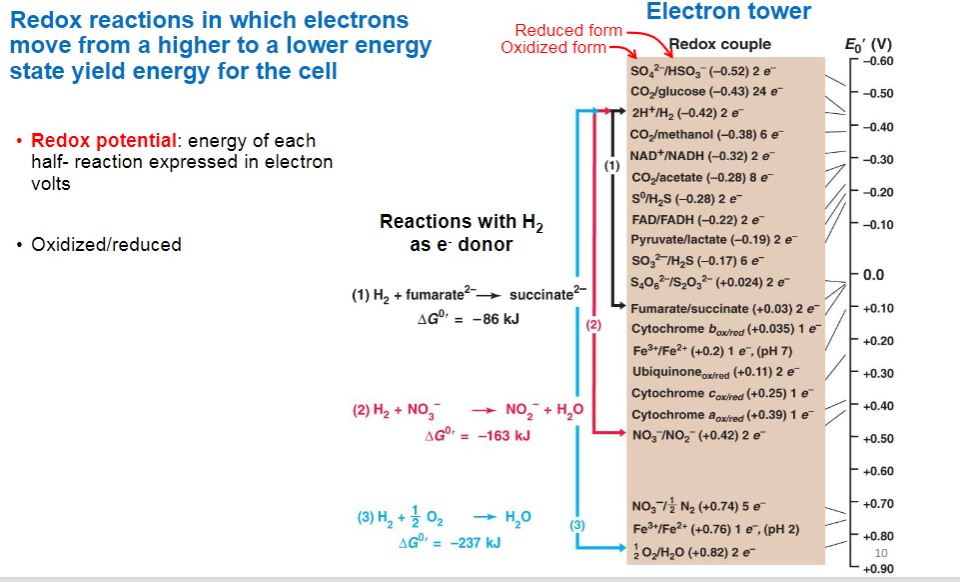
-The equilibrium constant for a redox half reaction is called the standard reduction potential (E0) and is a measure of the tendency of the donor of a half reaction to lose electrons. By convention, the standard reduction potentials for half reactions, such as those in table 6.2, are determined at pH 7 and are represented by E0′. Standard reduction potentials are measured in volts, a unit of electrical potential or electromotive force. Therefore conjugate redox pairs are a potential source of energy. The reduction potential has a concrete meaning. Conjugate redox pairs with more negative reduction potentials will spontaneously donate electrons to pairs with more positive potentials and greater affinity for electrons. Thus electrons tend to move from donors at the top of the list in table 6.2 to acceptors at the bottom because the latter have more positive potentials.
Redox reduction
70
New cards
a
Capturing/Coupling energy
71
New cards
a
High energy bonds and energy rich compounds
72
New cards
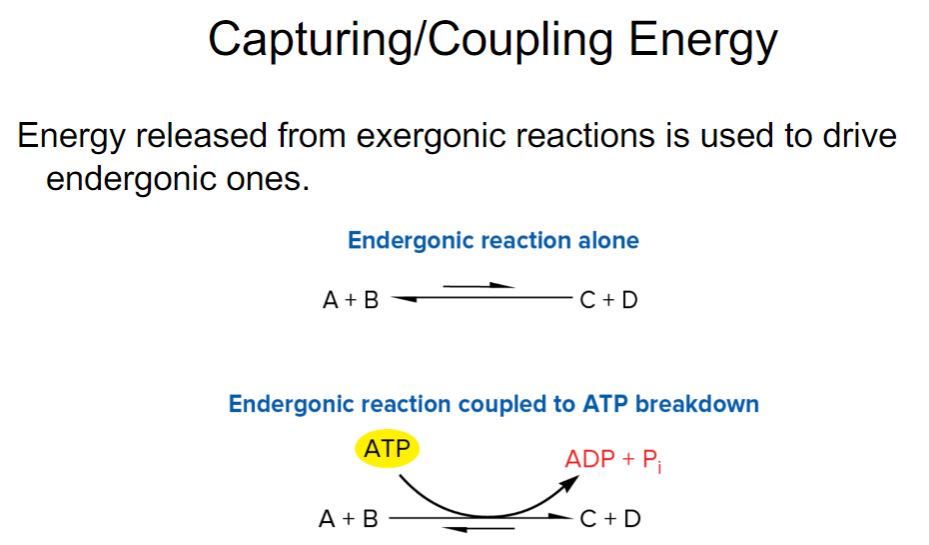
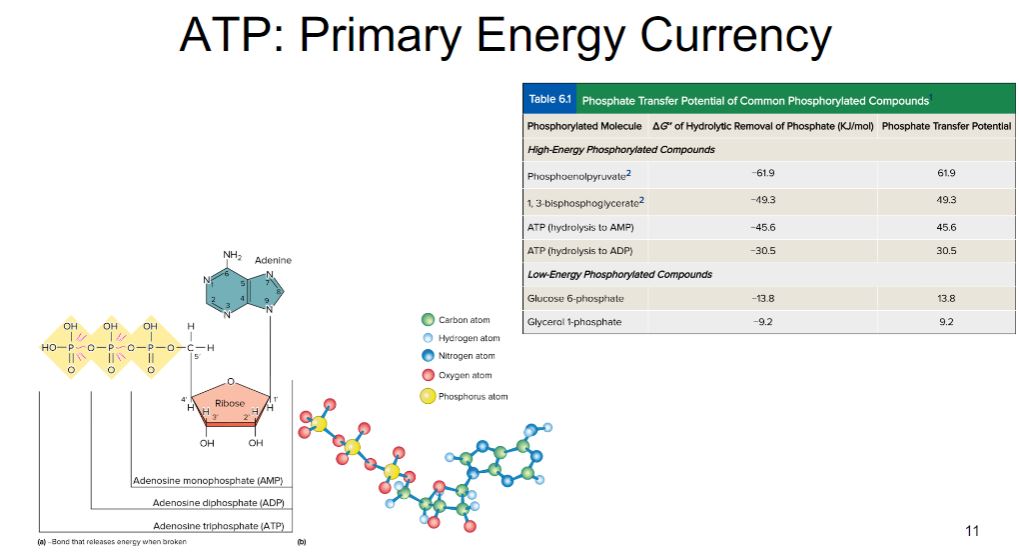
-Energy is released from a cell’s energy source in exergonic reactions (i.e., those reactions with a negative ΔG). Rather than wasting this energy, much of it is trapped in a practical form that allows it to be used to do work. Cells carry out endergonic reactions (e.g., anabolism), and the energy captured by the cell is used to drive these reactions to completion. In living organisms, the most commonly used practical form of energy is the nucleotide adenosine 5′-triphosphate (ATP; figure 6.2). In a sense, cells carry out certain processes so that they can “earn” ATP, which they “spend” in carrying out other processes. Thus ATP is often referred to as the cell’s energy currency. In the cell’s economy, ATP serves as the link between exergonic reactions and endergonic reactions
-image below-
Adenosine Triphosphate. (a) Structure of ATP, ADP, and AMP. The two red bonds (∼) are more easily broken and release considerable energy that can be used in endergonic reactions. The pyrimidine ring atoms have been numbered, as have the carbon atoms in ribose. (b) A stick-and-ball model of ATP. For simplification, some H atoms are not shown.
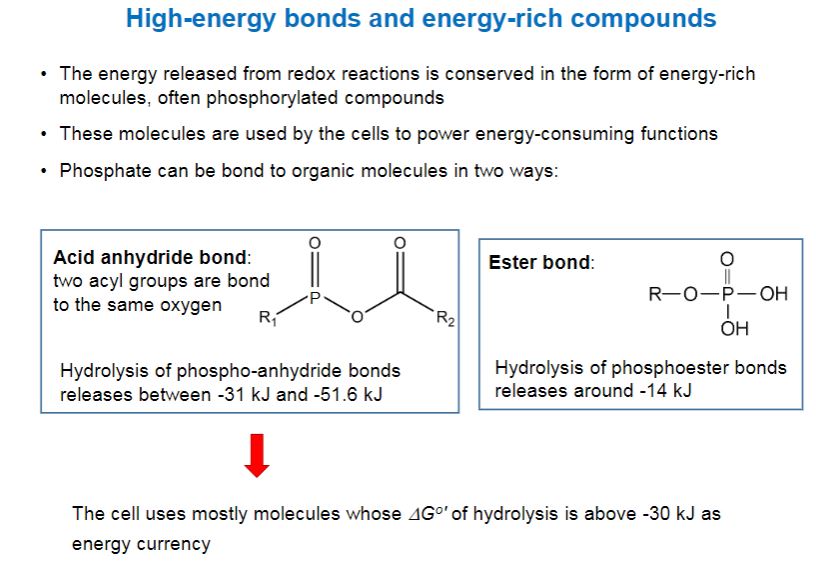
- What makes ATP suited for its role as energy currency? ATP is a high-energy molecule. That is, it is hydrolyzed almost completely to the products adenosine diphosphate (ADP) and orthophosphate (Pi), with a strongly exergonic ΔG°′ of −7.3 kcal/mole (−30.5 kJ/mole).
-The very negative ΔG°′ of ATP hydrolysis is related to another important characteristic of ATP: its ability to transfer a phosphoryl group to another molecule. ATP is said to have a high phosphate transfer potential because it readily donates a phosphoryl group to other molecules. Note in table 6.1 that there are other phosphorylated compounds found in cells. These molecules are generated during catabolism of organic molecules such as glucose. Some of these molecules have even higher phosphate transfer potentials than ATP. The fact that ATP does not have the highest phosphate transfer potential means that it can easily be made by cells from ADP, using molecules such as phosphoenolpyruvate (PEP) as the source of the phosphoryl group. This mechanism for making ATP is called substrate-level phosphorylation.
-image below-
Adenosine Triphosphate. (a) Structure of ATP, ADP, and AMP. The two red bonds (∼) are more easily broken and release considerable energy that can be used in endergonic reactions. The pyrimidine ring atoms have been numbered, as have the carbon atoms in ribose. (b) A stick-and-ball model of ATP. For simplification, some H atoms are not shown.
- What makes ATP suited for its role as energy currency? ATP is a high-energy molecule. That is, it is hydrolyzed almost completely to the products adenosine diphosphate (ADP) and orthophosphate (Pi), with a strongly exergonic ΔG°′ of −7.3 kcal/mole (−30.5 kJ/mole).
-The very negative ΔG°′ of ATP hydrolysis is related to another important characteristic of ATP: its ability to transfer a phosphoryl group to another molecule. ATP is said to have a high phosphate transfer potential because it readily donates a phosphoryl group to other molecules. Note in table 6.1 that there are other phosphorylated compounds found in cells. These molecules are generated during catabolism of organic molecules such as glucose. Some of these molecules have even higher phosphate transfer potentials than ATP. The fact that ATP does not have the highest phosphate transfer potential means that it can easily be made by cells from ADP, using molecules such as phosphoenolpyruvate (PEP) as the source of the phosphoryl group. This mechanism for making ATP is called substrate-level phosphorylation.
ATP
73
New cards
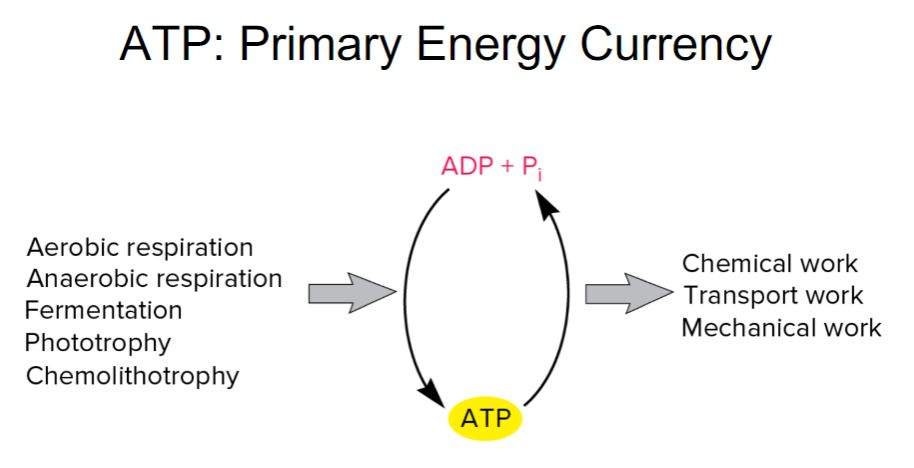
-ATP, ADP, and Pi form an energy cycle. As seen in figure 6.4, the energy released from an energy source is used to synthesize ATP from ADP and Pi. When ATP is hydrolyzed, the energy released drives endergonic processes such as anabolism, transport, and mechanical work. The mechanisms for synthesizing ATP are described in more detail in.
-ATP is the major energy currency for cells, but it is not the only energy currency. Other nucleoside triphosphates (NTPs) have major roles in metabolism. Guanosine 5′-triphosphate (GTP) supplies some of the energy used during protein synthesis. Cytidine 5′-triphosphate (CTP) is used during lipid synthesis, and uridine 5′-triphosphate (UTP) is used for the synthesis of peptidoglycan and other polysaccharides
-ATP is the major energy currency for cells, but it is not the only energy currency. Other nucleoside triphosphates (NTPs) have major roles in metabolism. Guanosine 5′-triphosphate (GTP) supplies some of the energy used during protein synthesis. Cytidine 5′-triphosphate (CTP) is used during lipid synthesis, and uridine 5′-triphosphate (UTP) is used for the synthesis of peptidoglycan and other polysaccharides
Primary energy currency
74
New cards
a
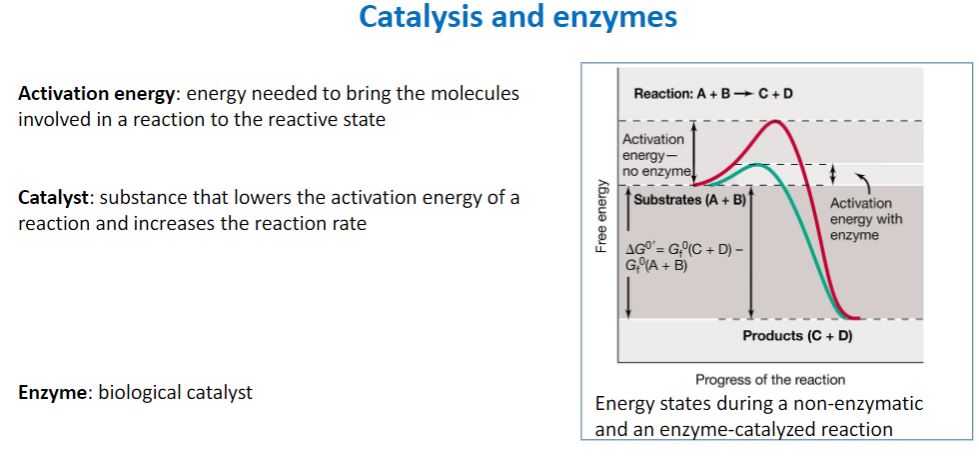
Catalyst and Enzymes
75
New cards
a
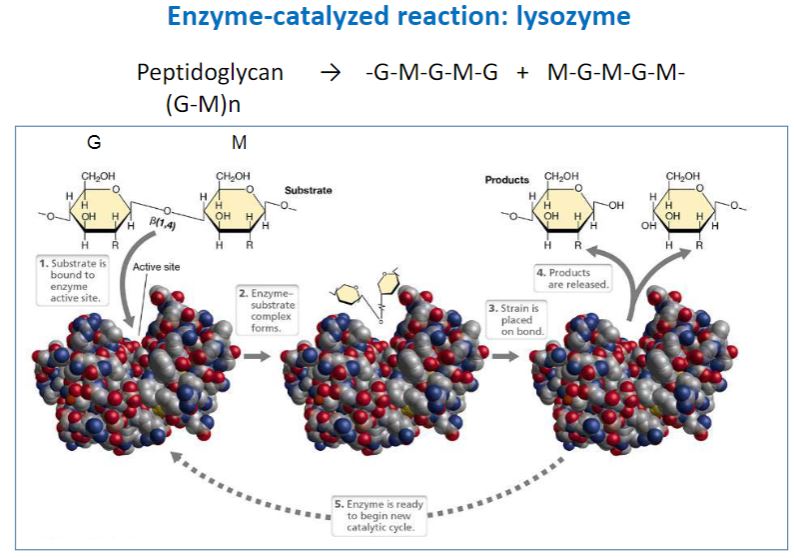
lysozyme
76
New cards
a
Catalyst and enzymes
77
New cards
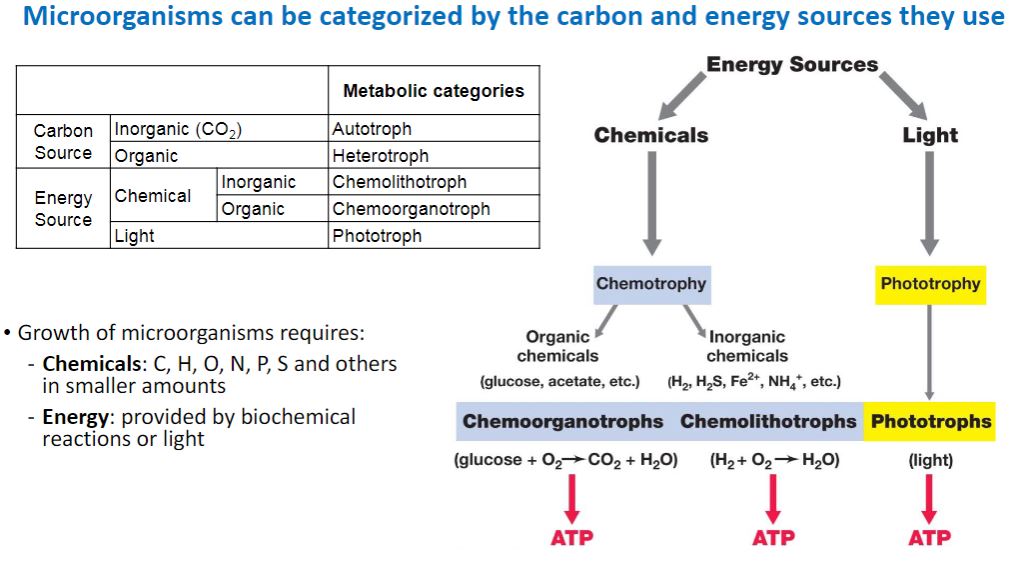
-For each requirement—energy, electrons, and carbon—a dichotomy exists for how an organism fulfills that need. Only two sources of energy are available to organisms: light and certain chemical molecules. Phototrophs use light as their energy source (table 7.1); chemotrophs obtain energy from the oxidation of chemical compounds (either organic or inorganic). Likewise, organisms have only two sources for electrons. Lithotrophs (i.e., “rock-eaters”) use reduced inorganic substances as their electron source, whereas organotrophs extract electrons from reduced organic compounds. Finally, organisms may be heterotrophs—organisms that use reduced, preformed organic molecules as their carbon source for growth, or they may be autotrophs—organisms that use carbon dioxide (CO2) as their sole or principal source of carbon.
microorganism can be categorized by the carbon and energy sources they use
78
New cards
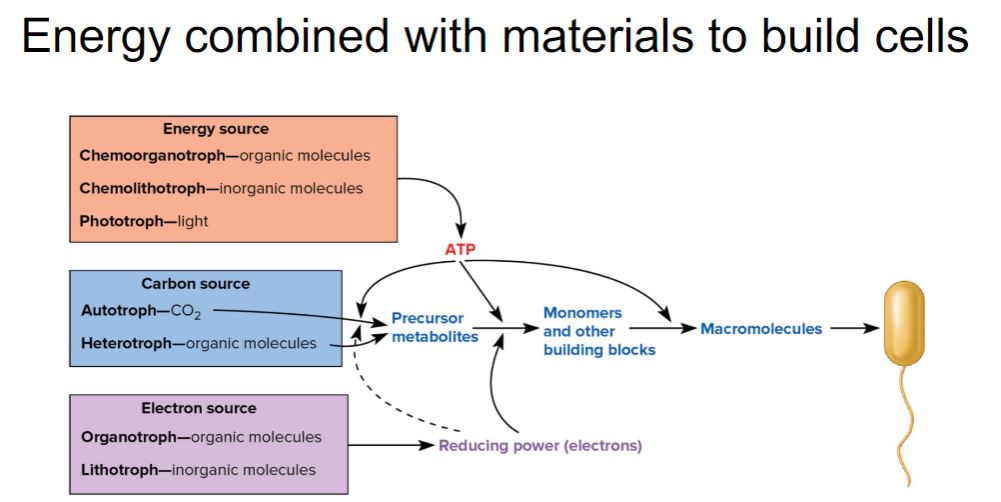
-Despite the diversity of energy, electron, and carbon sources used by organisms, all are used to generate three main products: ATP, the primary molecule used to conserve the energy supplied by an energy source; reducing power, molecules that serve as a ready supply of electrons for a variety of chemical reactions; and precursor metabolites, small organic molecules that provide the carbon skeletons needed for biosynthesis of important chemical building blocks (monomers) such as amino acids (figure 7.1). The processes that supply ATP, reducing power, and precursor metabolites are called fueling reactions. With the exception of CO2 fixation, the fueling reactions are part of catabolism. As seen in figure 7.1, the generation of monomers using the products of the fueling reactions paves the way for anabolic reactions that synthesize macromolecules (e.g., polymers such as proteins). The macromolecules are used to construct cellular structures such as ribosomes and flagella.
-image below-
-The Fueling Reactions Convert an Organism’s Carbon, Energy, and Electron Sources into Precursor Metabolites, ATP, and Reducing Power. In autotrophs, the precursor metabolites arise from CO2-fixation and related pathways, which are discussed in chapter 8. In heterotrophs, the precursor metabolites arise from reactions of the central metabolic pathways (see figure 8.2). ATP and reducing power are used in anabolic reactions that convert precursor metabolites into monomers and other building blocks. Monomers are used to generate macromolecules such as proteins, which are assembled into various cell structures.
-image below-
-The Fueling Reactions Convert an Organism’s Carbon, Energy, and Electron Sources into Precursor Metabolites, ATP, and Reducing Power. In autotrophs, the precursor metabolites arise from CO2-fixation and related pathways, which are discussed in chapter 8. In heterotrophs, the precursor metabolites arise from reactions of the central metabolic pathways (see figure 8.2). ATP and reducing power are used in anabolic reactions that convert precursor metabolites into monomers and other building blocks. Monomers are used to generate macromolecules such as proteins, which are assembled into various cell structures.
Energy combined to build cells
79
New cards
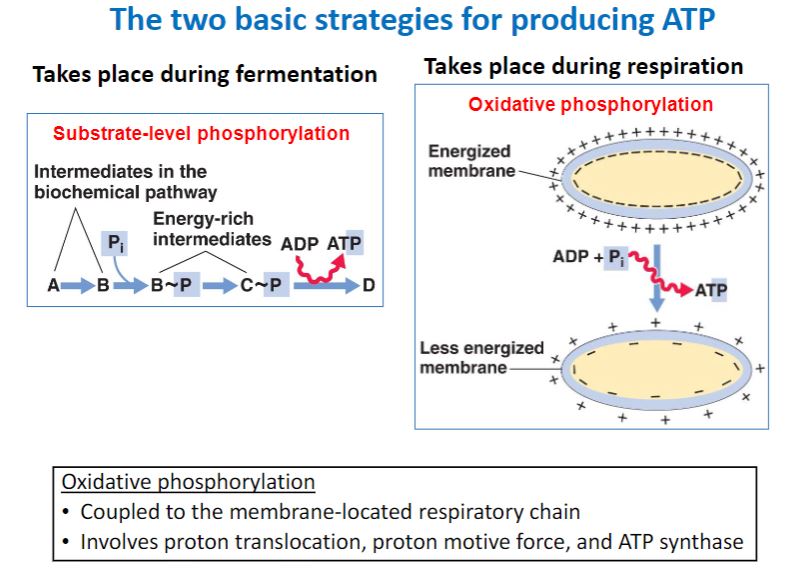
a
Two basic strategies for producing ATP
80
New cards
-As illustrated in figure 7.1, chemoorganotrophs use reduced organic compounds for catabolic and anabolic processes. When an organic compound is oxidized to release energy (catabolism), it also provides the carbon and electrons needed for anabolism. A chemoorganotroph may use aerobic respiration, anaerobic respiration, or fermentation to catabolize its energy source. When the organic energy source is oxidized, the electrons released must be accepted by electron carriers such as NAD+ and FAD. When these reduced electron carriers (e.g., NADH, FADH2) in turn donate the electrons to an electron transport chain, the metabolic process is called respiration and may be either aerobic or anaerobic (figure 7.2). In aerobic respiration, the final electron acceptor is oxygen, whereas the terminal acceptor in anaerobic respiration is a different oxidized molecule such as NO3−, SO42−, CO2, Fe3+, or SeO42−. Organic acceptors such as fumarate and humic acids also may be used. During respiration, electrons pass through the electron transport chain to the final electron acceptor generating a type of potential energy called the proton motive force (PMF). The PMF is used to synthesize ATP from ADP and phosphate (Pi). In contrast, fermentation (Latin fermentare, to cause to rise) uses an electron acceptor that is endogenous (from within the cell) and does not involve an electron transport chain. The endogenous electron acceptor is usually an intermediate (e.g., pyruvate) of the catabolic pathway used to degrade and oxidize the organic energy source. During fermentation, ATP is synthesized almost exclusively by substrate-level phosphorylation, a process in which a phosphate is transferred to ADP from a high-energy molecule (e.g., phosphoenolpyruvate) generated by catabolism of the energy source.
-image below-
Chemoorganotrophic Fueling Processes. Organic molecules serve as energy and electron sources for all three fueling processes used by chemoorganotrophs. In aerobic respiration and anaerobic respiration, the electrons pass through an electron transport chain. This generates a proton motive force (PMF), which is used to synthesize most of the cellular ATP by a mechanism called oxidative phosphorylation (ox phos); a small amount of ATP is made by a process called substrate-level phosphorylation (SLP). In aerobic respiration, O2 is the terminal electron acceptor, whereas in anaerobic respiration, exogenous molecules other than O2 serve as electron acceptors. During fermentation, endogenous organic molecules act as electron acceptors, electron transport chains do not function, and most organisms synthesize ATP only by substrate-level phosphorylation.
-image below-
Chemoorganotrophic Fueling Processes. Organic molecules serve as energy and electron sources for all three fueling processes used by chemoorganotrophs. In aerobic respiration and anaerobic respiration, the electrons pass through an electron transport chain. This generates a proton motive force (PMF), which is used to synthesize most of the cellular ATP by a mechanism called oxidative phosphorylation (ox phos); a small amount of ATP is made by a process called substrate-level phosphorylation (SLP). In aerobic respiration, O2 is the terminal electron acceptor, whereas in anaerobic respiration, exogenous molecules other than O2 serve as electron acceptors. During fermentation, endogenous organic molecules act as electron acceptors, electron transport chains do not function, and most organisms synthesize ATP only by substrate-level phosphorylation.
Chemoorganotrophic energy producing Reactions
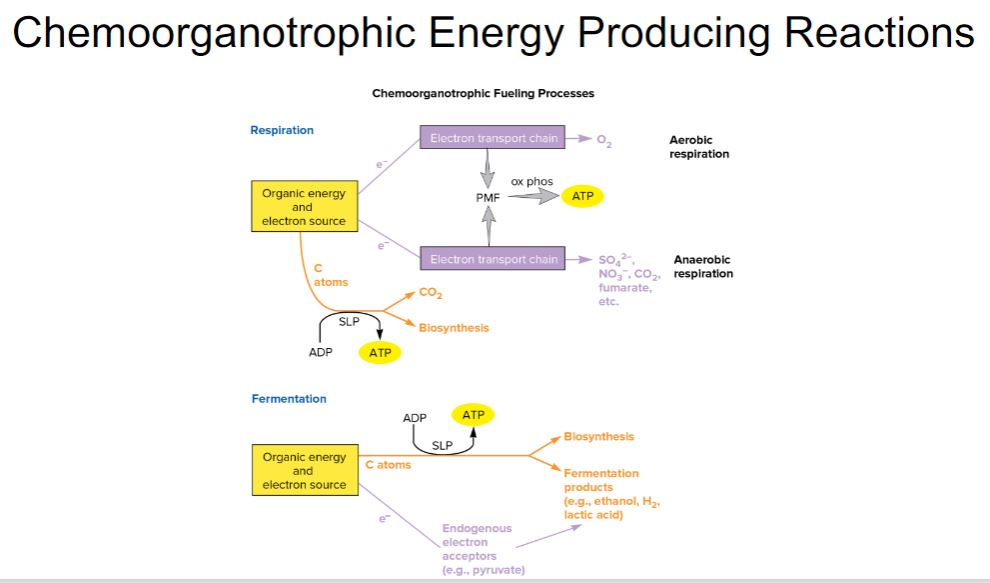
81
New cards
-By convention, aerobic respiration, anaerobic respiration, and fermentation are usually described with glucose as the energy source. This is done for several reasons. One is that glucose is used by many chemoorganotrophs as an energy source. But perhaps more important is the way catabolic pathways are organized. Most chemoorganotrophs use a wide variety of organic molecules as energy sources (figure 7.3). They are degraded by pathways that either generate glucose or intermediates of the pathways used in glucose catabolism. Thus nutrient molecules are funneled into ever fewer metabolic intermediates. Indeed, a common pathway often degrades many similar molecules (e.g., several different sugars). The existence of a few metabolic pathways that each break down many nutrients greatly increases metabolic efficiency by avoiding the need for a large number of less metabolically flexible pathways.
-image below-
Pathways Used by Chemoorganotrophs to Catabolize Organic Energy Sources. Notice that these pathways funnel metabolites into the glycolytic pathways and the tricarboxylic acid cycle, thus increasing metabolic efficiency and flexibility.
-image below-
Pathways Used by Chemoorganotrophs to Catabolize Organic Energy Sources. Notice that these pathways funnel metabolites into the glycolytic pathways and the tricarboxylic acid cycle, thus increasing metabolic efficiency and flexibility.
Carbon is not the only fuel source
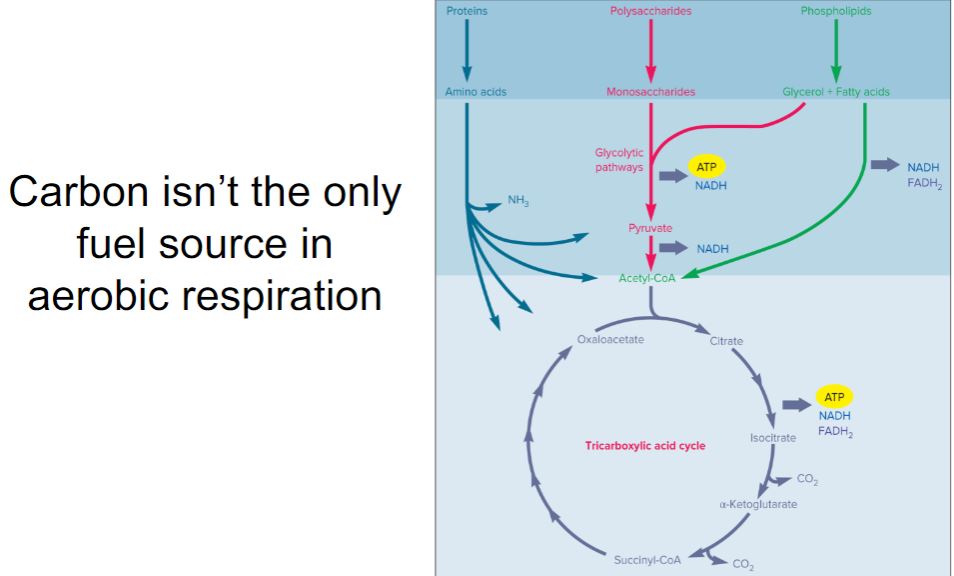
82
New cards
-Because a carbon source can function as an energy source or for anabolic purposes, some pathways include enzymes that function both catabolically and anabolically. These are called amphibolic pathways (Greek amphi, on both sides). For instance, many of the enzymes of the Embden-Meyerhof pathway (a glycolytic pathway) are freely reversible. They function catabolically during glycolysis but anabolically during gluconeogenesis, a pathway that generates glucose from pyruvate and other small molecules (figure 7.4). Other reactions in glycolysis are not reversible. During glycolysis, an enzyme catalyzes the reaction in a catabolic direction, and during gluconeogenesis, a different enzyme catalyzes the reverse, anabolic reaction. Thus glycolysis and gluconeogenesis are two distinct pathways.
-Our discussion of chemoorganotrophy begins with aerobic respiration and introduces the glycolytic pathways, TCA cycle, and other important processes. Many of these also occur during anaerobic respiration. Fermentation is quite distinct from respiration. It involves only a subset of the reactions that function during respiration and only partially catabolizes the energy source.
-image below-
The Embden-Meyerhof Pathway Is an Example of an Amphibolic Pathway. Many reactions of this pathway are catalyzed by enzymes that function in glycolysis and in an anabolic pathway called gluconeogenesis. Gluconeogenesis reverses the glycolytic process and allows cells to synthesize glucose from smaller molecules such as pyruvate. Note that some glycolytic reactions are catalyzed by enzymes unique to that pathway. Likewise, some gluconeogenic reactions are catalyzed by enzymes unique to gluconeogenesis. Therefore, although they share some enzymes, the two pathways are distinct.
-Our discussion of chemoorganotrophy begins with aerobic respiration and introduces the glycolytic pathways, TCA cycle, and other important processes. Many of these also occur during anaerobic respiration. Fermentation is quite distinct from respiration. It involves only a subset of the reactions that function during respiration and only partially catabolizes the energy source.
-image below-
The Embden-Meyerhof Pathway Is an Example of an Amphibolic Pathway. Many reactions of this pathway are catalyzed by enzymes that function in glycolysis and in an anabolic pathway called gluconeogenesis. Gluconeogenesis reverses the glycolytic process and allows cells to synthesize glucose from smaller molecules such as pyruvate. Note that some glycolytic reactions are catalyzed by enzymes unique to that pathway. Likewise, some gluconeogenic reactions are catalyzed by enzymes unique to gluconeogenesis. Therefore, although they share some enzymes, the two pathways are distinct.
Pathways are reversible
83
New cards
three steps in respiration
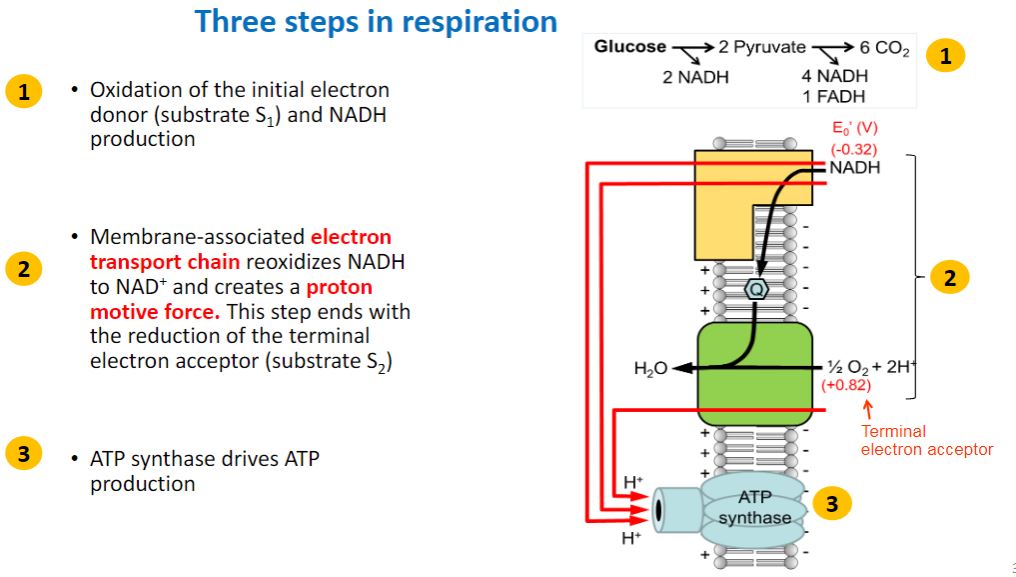
84
New cards
a
Step 1 glucose breakdown
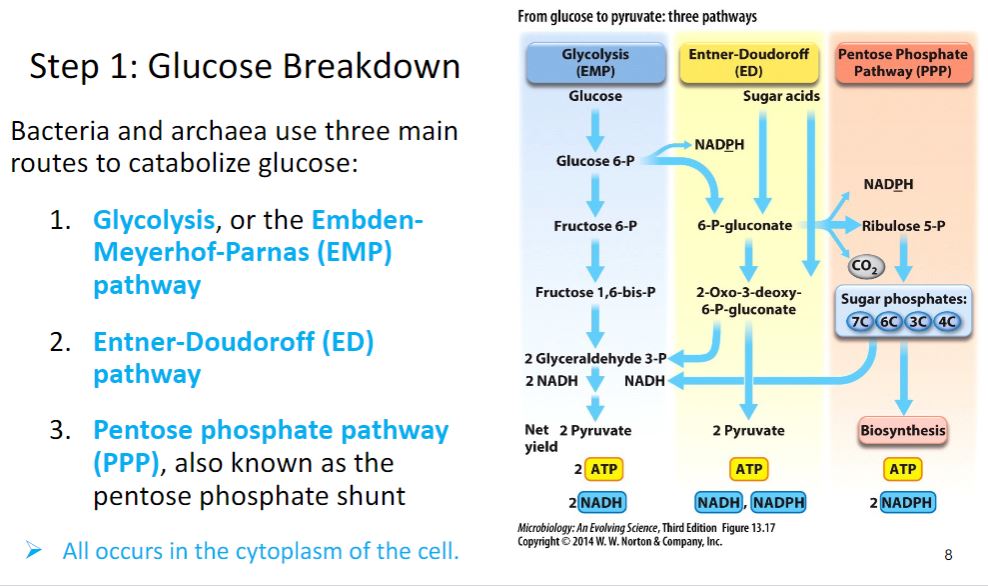
85
New cards
a
glycolysis

86
New cards
a
Glycolysis (EMP)
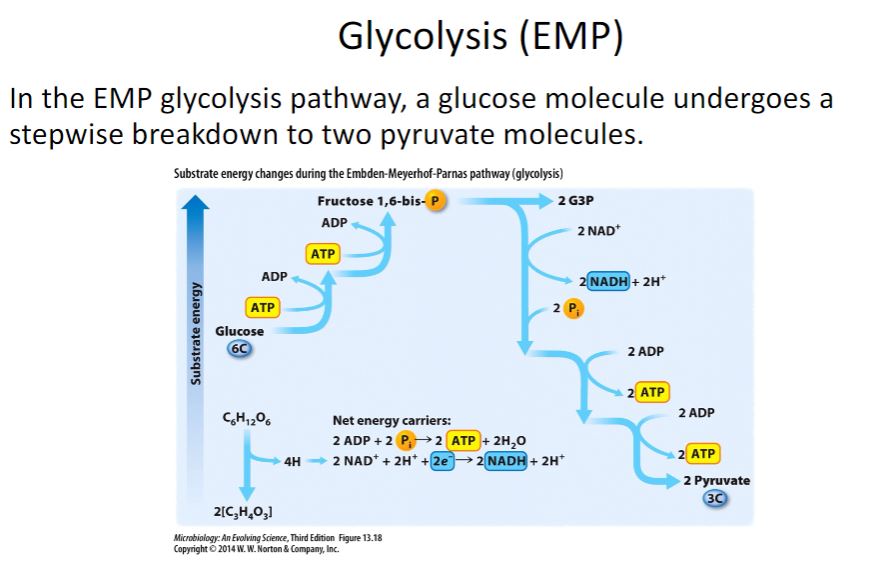
87
New cards
a
Electron carriers move electrons
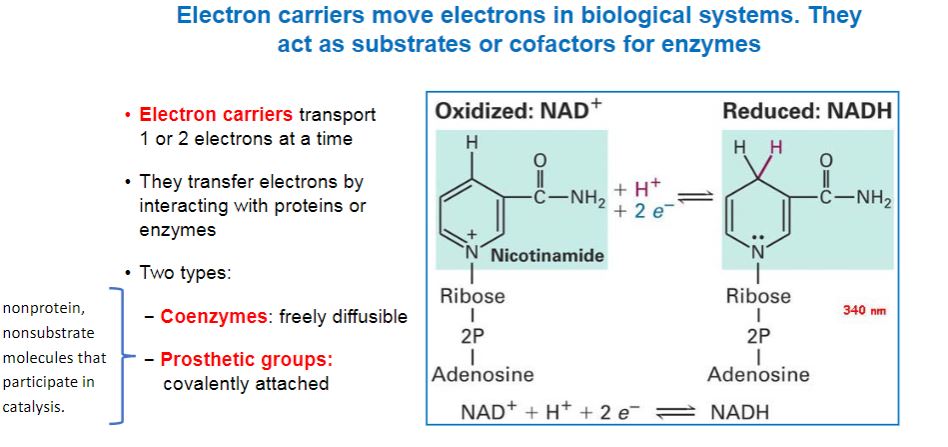
88
New cards
-In the glycolytic pathways, glucose is oxidized to pyruvate. During aerobic respiration, the catabolic process continues by oxidizing pyruvate to three CO2. The first step of this process employs a multienzyme system called the pyruvate dehydrogenase complex (PDH). PDH oxidizes and cleaves pyruvate to form one NADH, one CO2, and the 2-carbon molecule acetyl-coenzyme A (acetyl-CoA) (figure 7.8). The oxidization of a molecule to simultaneously release carbon as CO2 and NADH (or other reduced electron carrier) is called oxidative decarboxylation. Acetyl-CoA is an energy-rich molecule because the bond that links acetic acid to coenzyme A (a thioester bond) has a large negative change in free energy when hydrolyzed.
-image below-
The Tricarboxylic Acid Cycle. The TCA cycle is linked to glycolysis by a connecting reaction catalyzed by the pyruvate dehydrogenase complex. The reaction decarboxylates pyruvate and generates acetyl-CoA. The TCA cycle may be divided into three stages based on the number of carbons present in its intermediates. The three stages are separated from one another by two oxidative decarboxylation reactions. Precursor metabolites are shown in blue. NADH and FADH2 are shown in purple; they can transfer electrons to an electron transport chain (ETC).
-image below-
The Tricarboxylic Acid Cycle. The TCA cycle is linked to glycolysis by a connecting reaction catalyzed by the pyruvate dehydrogenase complex. The reaction decarboxylates pyruvate and generates acetyl-CoA. The TCA cycle may be divided into three stages based on the number of carbons present in its intermediates. The three stages are separated from one another by two oxidative decarboxylation reactions. Precursor metabolites are shown in blue. NADH and FADH2 are shown in purple; they can transfer electrons to an electron transport chain (ETC).
Step 2 citric acid
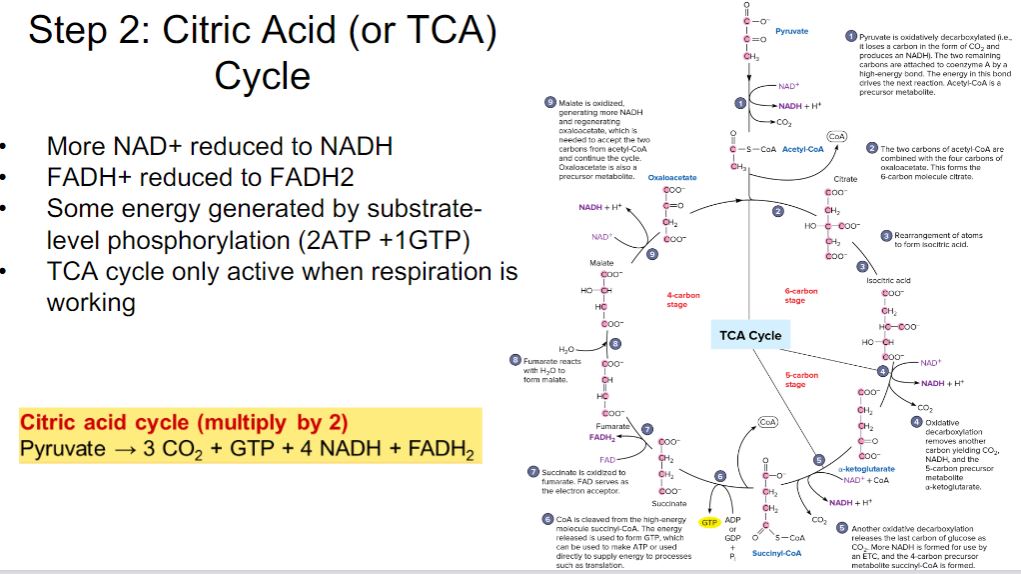
89
New cards
a
FAD
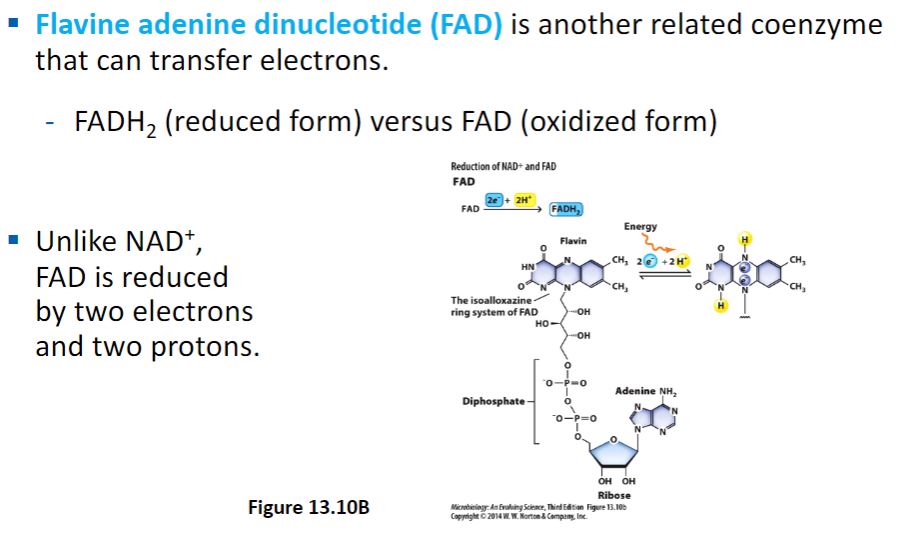
90
New cards
-The mitochondrial electron transport chain (ETC) is composed of a series of electron carriers that operate together to transfer electrons from donors, such as NADH and FADH2, to O2 (figure 7.9). The electrons flow from carriers with more negative reduction potentials to those with more positive potentials and eventually combine with O2 and H+ to form water. Each carrier is reduced when it accepts electrons and then reoxidized when the electrons are transferred to the next carrier in the chain (figure 7.10). The electrons move down this potential gradient much like water flowing down a series of rapids. The difference in reduction potentials between O2 and NADH is large, about 1.14 volts, which makes possible the release of a great deal of energy. However, the ETC breaks up the overall energy release (electron transfer) into small steps, preventing electrons from reducing cellular components outside the ETC. The energy released is sufficient for ATP production. Thus much as the energy from waterfalls can be harnessed by waterwheels and used to generate electricity, so too is the flow of electrons through an ETC harnessed to make ATP.
-image below-
-Figure 7.9 The Reduction Potentials of Carriers in the Mitochondrial Electron Transport Chain. In mitochondria, the electron carriers are organized into four complexes that are linked by the electron carriers coenzyme Q (CoQ) and cytochrome c (Cyt c). Electrons flow from NADH and succinate down the reduction potential gradient to oxygen.
-Figure 7.10 The Electron Tower. Electrons move spontaneously from donors with more negative reduction potentials to acceptors with more positive reduction potentials.
-image below-
-Figure 7.9 The Reduction Potentials of Carriers in the Mitochondrial Electron Transport Chain. In mitochondria, the electron carriers are organized into four complexes that are linked by the electron carriers coenzyme Q (CoQ) and cytochrome c (Cyt c). Electrons flow from NADH and succinate down the reduction potential gradient to oxygen.
-Figure 7.10 The Electron Tower. Electrons move spontaneously from donors with more negative reduction potentials to acceptors with more positive reduction potentials.
Step 3 electron transport chain
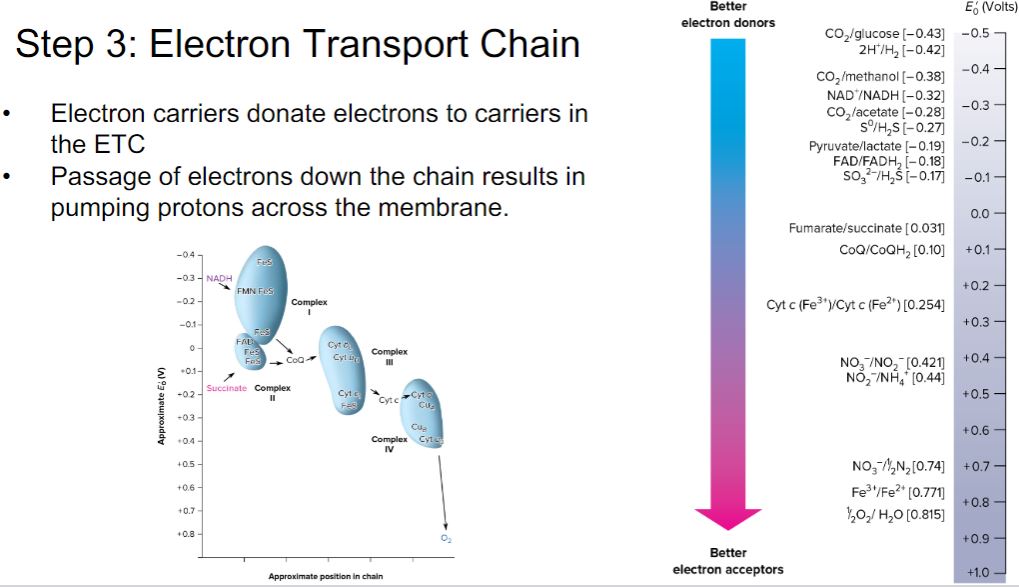
91
New cards
-In eukaryotes, ETC carriers reside within the inner membrane of the mitochondrion. The ETC consists of four large, multiprotein complexes and two additional carriers, coenzyme Q and the small protein cytochrome c, that connect complex I to complex III and complex III to IV, respectively. The final complex (complex IV) is referred to as the terminal oxidase as it is responsible for the final step transferring electrons to O2 (figure 7.11). As electrons move through the ETC, protons are moved across the inner membrane, forming a proton gradient.
-image below-
-Creation of a Proton Gradient by the Mitochondrial Electron Transport Chain and Its Use in Synthesis of ATP. As electrons move along the ETC, protons are transported across the membrane creating a proton gradient. Proton release occurs when electrons are transferred from carriers, such as FMN and coenzyme Q (Q), that carry both electrons and protons to components such as nonheme iron proteins (FeS) and cytochromes (Cyt) that transport only electrons. Complex IV is the terminal oxidase that transfers electrons to oxygen. The number of protons moved across the membrane at each site per pair of electrons transported is still somewhat uncertain and may vary among species; the current consensus is that 10 protons move outward during NADH oxidation. One molecule of ATP is synthesized and released from the enzyme ATP synthase for every 2.5 protons that move back into the matrix by passing through the enzyme.
-image below-
-Creation of a Proton Gradient by the Mitochondrial Electron Transport Chain and Its Use in Synthesis of ATP. As electrons move along the ETC, protons are transported across the membrane creating a proton gradient. Proton release occurs when electrons are transferred from carriers, such as FMN and coenzyme Q (Q), that carry both electrons and protons to components such as nonheme iron proteins (FeS) and cytochromes (Cyt) that transport only electrons. Complex IV is the terminal oxidase that transfers electrons to oxygen. The number of protons moved across the membrane at each site per pair of electrons transported is still somewhat uncertain and may vary among species; the current consensus is that 10 protons move outward during NADH oxidation. One molecule of ATP is synthesized and released from the enzyme ATP synthase for every 2.5 protons that move back into the matrix by passing through the enzyme.
Step 3:
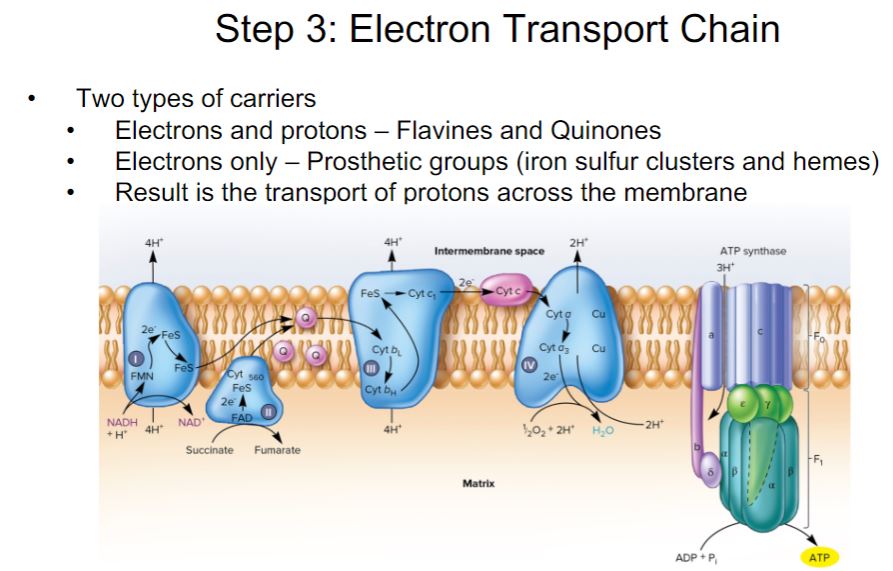
92
New cards
-The use of the PMF for ATP synthesis is catalyzed by ATP synthase. The best-studied ATP synthases are the F1F0 ATP synthases found in mitochondria, chloroplasts, and bacteria. They are also known as F1F0 ATPases because they can catalyze ATP hydrolysis. The mitochondrial F1 component appears as a spherical structure attached to the mitochondrial inner membrane surface by a stalk. It functions much like a rotary engine as described in figure 7.16.
-image below-
-ATP Synthase Structure and Function. (a) The major structural features of ATP synthase. F1 is composed largely of alternating α and β subunits; the three active sites are on the β subunits. The γ subunit extends through the center and can rotate. The stalk (γ and ε subunits) connects the sphere to F0, the membrane embedded complex that serves as a proton channel. F0 contains one a subunit, two b subunits, and nine to twelve c subunits. The stator arm is composed of subunit a, two b subunits, and the δ subunit; it is embedded in the membrane and attached to F1. A ring of c subunits in F0 is connected to the stalk and may act as a rotor and move past the a subunit of the stator. As the c subunit ring turns, it rotates the shaft (γε subunits). The inset is an atomic force micrograph of the F0 rotor. (b) The binding change mechanism is a widely accepted model of ATP synthesis. This simplified drawing of the model shows the three catalytic β subunits and the γ subunit, which is located at the center of the F1 complex. As the γ subunit rotates, it causes conformational changes in each subunit. The βE (empty) conformation is an open conformation, which does not bind nucleotides. When the γ subunit rotates, βE is converted to the βHC (half closed) conformation. Pi and ADP can enter the catalytic site when it is in this conformation. The next rotation by the γ subunit is critical because it brings about three significant conformational changes: (1) βHC to βDP (ADP bound), (2) βDP to βTP (ATP bound), and (3) βTP to βE. Change from βDP to βTP is accompanied by the formation of ATP; change from βTP to βE allows for release of ATP from ATP synthase.
-image below-
-ATP Synthase Structure and Function. (a) The major structural features of ATP synthase. F1 is composed largely of alternating α and β subunits; the three active sites are on the β subunits. The γ subunit extends through the center and can rotate. The stalk (γ and ε subunits) connects the sphere to F0, the membrane embedded complex that serves as a proton channel. F0 contains one a subunit, two b subunits, and nine to twelve c subunits. The stator arm is composed of subunit a, two b subunits, and the δ subunit; it is embedded in the membrane and attached to F1. A ring of c subunits in F0 is connected to the stalk and may act as a rotor and move past the a subunit of the stator. As the c subunit ring turns, it rotates the shaft (γε subunits). The inset is an atomic force micrograph of the F0 rotor. (b) The binding change mechanism is a widely accepted model of ATP synthesis. This simplified drawing of the model shows the three catalytic β subunits and the γ subunit, which is located at the center of the F1 complex. As the γ subunit rotates, it causes conformational changes in each subunit. The βE (empty) conformation is an open conformation, which does not bind nucleotides. When the γ subunit rotates, βE is converted to the βHC (half closed) conformation. Pi and ADP can enter the catalytic site when it is in this conformation. The next rotation by the γ subunit is critical because it brings about three significant conformational changes: (1) βHC to βDP (ADP bound), (2) βDP to βTP (ATP bound), and (3) βTP to βE. Change from βDP to βTP is accompanied by the formation of ATP; change from βTP to βE allows for release of ATP from ATP synthase.
Proton motive force to make ATP
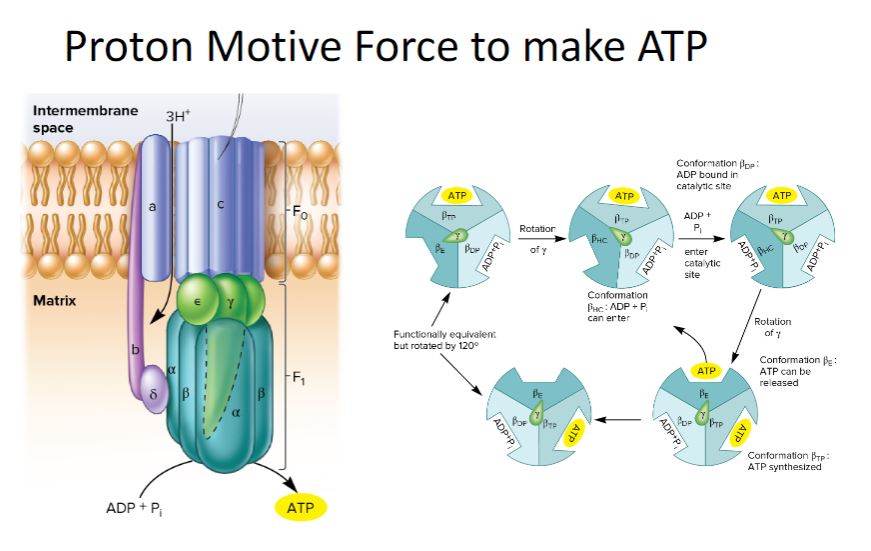
93
New cards
-It is possible to estimate the number of ATP molecules synthesized per pair of electrons donated by an NADH or FADH2 to the ETC and ultimately used to reduce an atom of oxygen to H2O. For historical reasons, this is called the phosphorus to oxygen (P/O) ratio. Prior to the acceptance of the chemiosmotic hypothesis, the concept of the P/O ratio was used as a measure of the number of ATP molecules (phosphorus) generated per oxygen (O) reduced as NADH and FADH2 were oxidized. With the acceptance of the chemiosmotic hypothesis, it was recognized that the important measurement was actually the number of protons transported across the membrane per NADH oxidation and the number of protons needed to drive the synthesis of a single ATP. Nonetheless, the amount of ATP synthesized as the result of the oxidation of one NADH or FADH2 is still often referred to as a P/O ratio. The best estimates of the number of protons moved across the membrane due to NADH oxidation is 10; six electrons are thought to be transferred when FADH2 oxidation initiates electron flow to oxygen. It is currently thought that four protons are consumed during ATP synthesis; three are used by ATP synthase and one is used for transport of ATP, ADP, and Pi. Thus the P/O ratio is estimated to be 2.5 for NADH and 1.5 for FADH2. Given this information, the maximum ATP yield of aerobic respiration in a eukaryote can be calculated (figure 7.17). Substrate-level phosphorylation yields at most two ATP molecules per glucose converted to pyruvate (figure 7.5). Two GTP (ATP equivalent) are generated by substrate-level phosphorylation during the two turns of the TCA cycle needed to oxidize two acetyl-CoA molecules (figure 7.8). Thus during aerobic respiration of glucose, at most four ATP molecules are made by substrate-level phosphorylation. Most of the ATP made during aerobic respiration is generated by oxidative phosphorylation. Up to 10 NADH (2 from glycolysis, 2 from pyruvate conversion to acetyl-CoA, and 6 from the TCA cycle) and 2 FADH2 (from the TCA cycle) are generated when glucose is oxidized completely to 6 CO2. Assuming a P/O ratio of 2.5 for NADH oxidation and 1.5 for FADH2 oxidation, the 10 NADH could theoretically drive the synthesis of 25 ATP, while oxidation of the 2 FADH2 molecules would add another 3 ATP for a maximum of 28 ATP generated via oxidative phosphorylation. Thus oxidative phosphorylation accounts for seven times more ATP than does substrate-level phosphorylation.
-The maximum total yield of ATP during aerobic respiration by eukaryotes is 32 ATP. Bacterial ETCs are often shorter and therefore transport fewer protons across the plasma membrane. Thus their ETCs have lower P/O ratios, and their ATP yields are smaller. For example, E. coli, with its truncated ETC, has a P/O ratio around 1.3 when using the cytochrome bo path at high oxygen levels and a ratio of only about 0.67 when employing the cytochrome bd branch (figure 7.13) at low oxygen concentrations. Page 164 Two other factors prevent the yield of ATP from reaching its theoretical maximum when glucose is catabolized by aerobic respiration. One is that the PMF generated by electron transport is used for functions other than ATP synthesis (e.g., transport and bacterial flagella rotation). The second is related to the amphibolic nature of the pathways used to catabolize glucose. Recall that an important function of these pathways is the generation of precursor metabolites for anabolism. For each molecule of glucose degraded, numerous precursor metabolites are made, and as each is made, a microbe must “decide” if that metabolite is needed for anabolism or if it can continue the catabolic process. If the precursor metabolite is used for biosynthesis, fewer NADH and FADH2 molecules are made and fewer ATP molecules are generated. Cells must carefully monitor and regulate the flow of the carbons from glucose such that ATP production is appropriately balanced with biosynthesis. The mechanisms for this regulation are described in chapters 6 and 11.
-The maximum total yield of ATP during aerobic respiration by eukaryotes is 32 ATP. Bacterial ETCs are often shorter and therefore transport fewer protons across the plasma membrane. Thus their ETCs have lower P/O ratios, and their ATP yields are smaller. For example, E. coli, with its truncated ETC, has a P/O ratio around 1.3 when using the cytochrome bo path at high oxygen levels and a ratio of only about 0.67 when employing the cytochrome bd branch (figure 7.13) at low oxygen concentrations. Page 164 Two other factors prevent the yield of ATP from reaching its theoretical maximum when glucose is catabolized by aerobic respiration. One is that the PMF generated by electron transport is used for functions other than ATP synthesis (e.g., transport and bacterial flagella rotation). The second is related to the amphibolic nature of the pathways used to catabolize glucose. Recall that an important function of these pathways is the generation of precursor metabolites for anabolism. For each molecule of glucose degraded, numerous precursor metabolites are made, and as each is made, a microbe must “decide” if that metabolite is needed for anabolism or if it can continue the catabolic process. If the precursor metabolite is used for biosynthesis, fewer NADH and FADH2 molecules are made and fewer ATP molecules are generated. Cells must carefully monitor and regulate the flow of the carbons from glucose such that ATP production is appropriately balanced with biosynthesis. The mechanisms for this regulation are described in chapters 6 and 11.
Summary
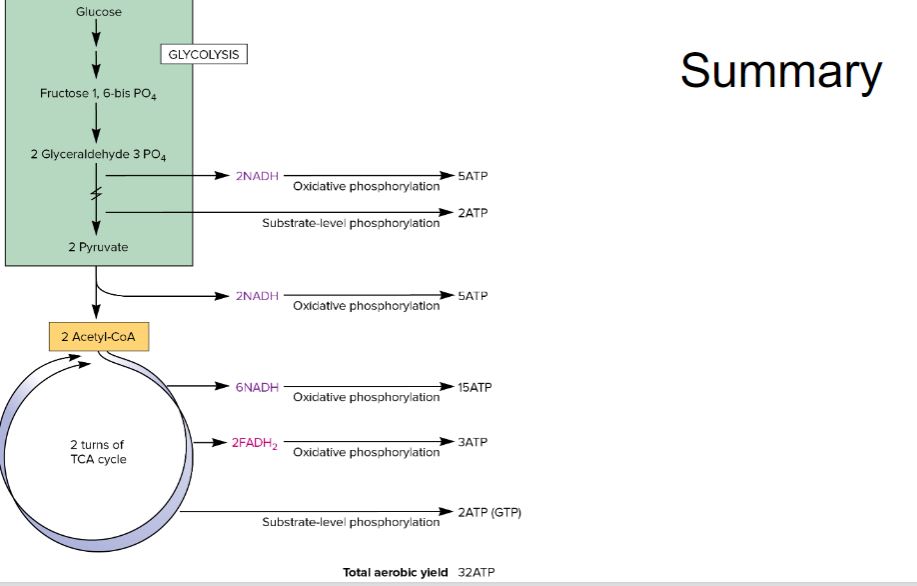
94
New cards
-Anaerobic respiration is the chemoorganotrophic process whereby a terminal electron acceptor other than O2 is used in the electron transport chain. It is carried out by many bacteria and archaea, and some eukaryotic microbes. The most common terminal electron acceptors used during anaerobic respiration are nitrate, sulfate, and CO2, but metals and a few organic molecules can also be reduced
Anaerobic respiration
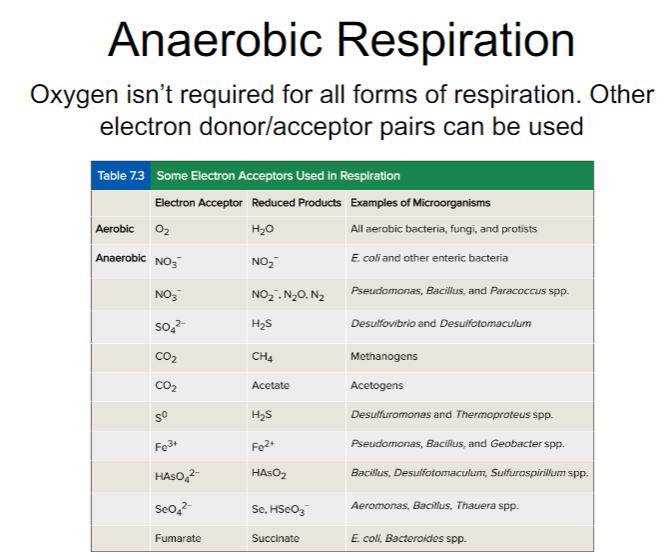
95
New cards
-In both aerobic and anaerobic respiration processes, the oxidation of organic substrates generates NADH and FADH2, which are used as the electron donors to the ETC. However, for each NADH or FADH2 oxidized, anaerobic respiration yields less ATP. The lower ATP yield is due to the fact that all terminal electron acceptors used during anaerobic respiration have less positive reduction potentials than O2 (figure 7.10). Thus the difference in standard reduction potentials between NADH and these electron acceptors is smaller than the difference between NADH and O2. This results in a shorter ETC with fewer protons transported to the periplasm per oxidation of NADH or FADH2 as compared to aerobic respiration. The smaller PMF and ATP yield generated by anaerobic respiration explains why facultative anaerobes capable of both aerobic and anaerobic respiration will use aerobic respiration whenever possible. An example of a facultative anaerobe that can carry out both anaerobic respiration and aerobic respiration is Paracoccus denitrificans. Figure 7.12 shows the ETC used by this microbe during aerobic respiration. Under anoxic conditions, P. denitrificans uses NO3− as its electron acceptor, reducing it to gaseous dinitrogen (N2). As shown in figure 7.18, the anaerobic ETC is more complex than the aerobic chain. The anaerobic ETC is branched and uses different electron carriers, some of which are located in the periplasmic space. Not as many protons are pumped across the membrane during anaerobic growth, but nonetheless, a PMF is established.
-The anaerobic reduction of NO3− makes this nitrogen unavailable for assimilation into the cell. That is, it can’t be taken up by the cell to construct N-containing molecules such as amino acids and nucleotides. Therefore this process is called dissimilatory nitrate reduction. When dissimilatory nitrate reduction leads to the production of gaseous compounds such as N2 that are released into the atmosphere, the process is also called denitrification. As can be seen in figure 7.18, denitrification carried out by P. denitrificans is a multistep process with four enzymes participating. These enzymes function sequentially to reduce NO3− to nitrite (NO2−), then nitrite to nitric oxide (NO), followed by NO to nitrous oxide (N2O), and finally N2O to N2.
-image below-
Paracoccus denitrificans Electron Transport Chain Used During Anaerobic Respiration. This branched ETC is made of both membrane and periplasmic proteins. Nitrate is reduced to gaseous nitrogen (N2) by the collective action of four different reductases that receive electrons from CoQ and cytochrome c. Locations of proton movement are indicated. Four protons are pumped into the periplasm by complex I, two by nitrate reductase (Nar), and two by complex III. However, two protons are used by nitric oxide reductase (Nor) to reduce nitric oxide to nitrous oxide. Thus six protons, net, are used to create a PMF. FP, Flavoprotein; Nir, nitrite reductase; Nos, nitrous oxide reductase, which has an unusual copper (Cu)–sulfur center.
-The anaerobic reduction of NO3− makes this nitrogen unavailable for assimilation into the cell. That is, it can’t be taken up by the cell to construct N-containing molecules such as amino acids and nucleotides. Therefore this process is called dissimilatory nitrate reduction. When dissimilatory nitrate reduction leads to the production of gaseous compounds such as N2 that are released into the atmosphere, the process is also called denitrification. As can be seen in figure 7.18, denitrification carried out by P. denitrificans is a multistep process with four enzymes participating. These enzymes function sequentially to reduce NO3− to nitrite (NO2−), then nitrite to nitric oxide (NO), followed by NO to nitrous oxide (N2O), and finally N2O to N2.
-image below-
Paracoccus denitrificans Electron Transport Chain Used During Anaerobic Respiration. This branched ETC is made of both membrane and periplasmic proteins. Nitrate is reduced to gaseous nitrogen (N2) by the collective action of four different reductases that receive electrons from CoQ and cytochrome c. Locations of proton movement are indicated. Four protons are pumped into the periplasm by complex I, two by nitrate reductase (Nar), and two by complex III. However, two protons are used by nitric oxide reductase (Nor) to reduce nitric oxide to nitrous oxide. Thus six protons, net, are used to create a PMF. FP, Flavoprotein; Nir, nitrite reductase; Nos, nitrous oxide reductase, which has an unusual copper (Cu)–sulfur center.
Anaerobic respiration 2
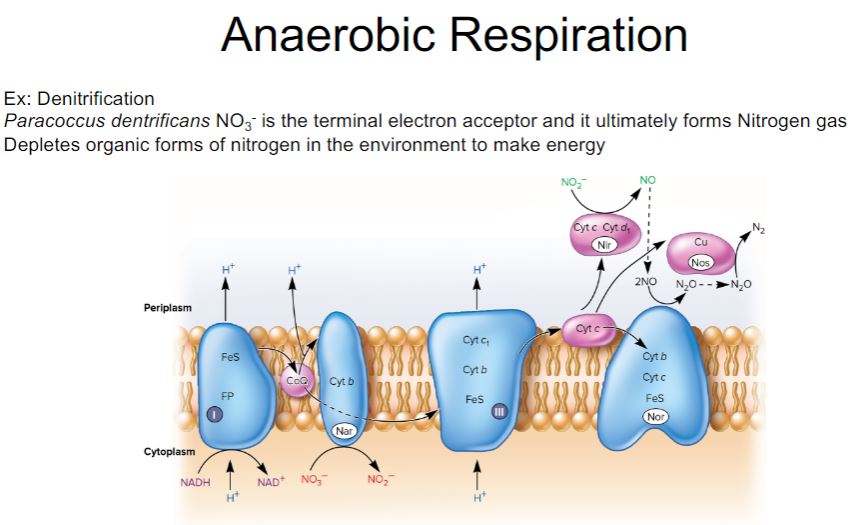
96
New cards
-Despite the tremendous ATP yield obtained by oxidative phosphorylation, some chemoorganotrophic microbes never respire (i.e., use an ETC to generate a PMF) and others may be unable to respire under certain conditions. Microbes completely incapable of respiration lack ETCs. Those that can carry out aerobic respiration may repress the synthesis of ETC components under anoxic conditions, making anaerobic respiration impossible. Finally, some facultative anaerobes can respire both aerobically and anaerobically but may find themselves in an environment that lacks both oxygen and the terminal electron acceptor they use for anaerobic respiration (e.g., nitrate). For each of these microbes, NADH produced by the Embden-Meyerhof pathway reactions during glycolysis (figure 7.5) must still be oxidized back to NAD+ even though there is no ETC to accept electrons from NADH. If NAD+ is not regenerated, the oxidation of glyceraldehyde 3-phosphate will cease and glycolysis will stop. Many microorganisms solve this problem by slowing or stopping pyruvate dehydrogenase, the enzyme complex that feeds pyruvate into the TCA cycle as acetyl-CoA. Instead, they use pyruvate or one of its derivatives as an electron acceptor for the reoxidation of NADH in a fermentation process (figure 7.19). There are many kinds of fermentations, and they often are characteristic of particular microbial groups (figure 7.20). A few of the more common fermentations are introduced here.
Fermentation
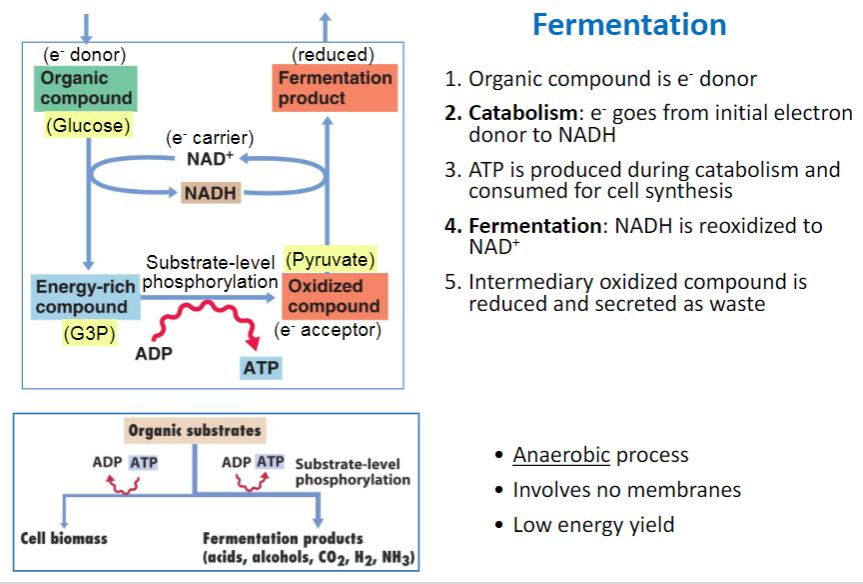
97
New cards
-Some fungi, protists, and bacteria ferment sugars to ethanol and CO2 in a process called alcoholic fermentation. Pyruvate is decarboxylated to acetaldehyde, which is then reduced to ethanol by alcohol dehydrogenase with NADH as the electron donor (figure 7.20, number 2). This fermentation produces the ethanol desired by brewers making beer and ale and by vintners making wine.
Ethanol fermentation pathway
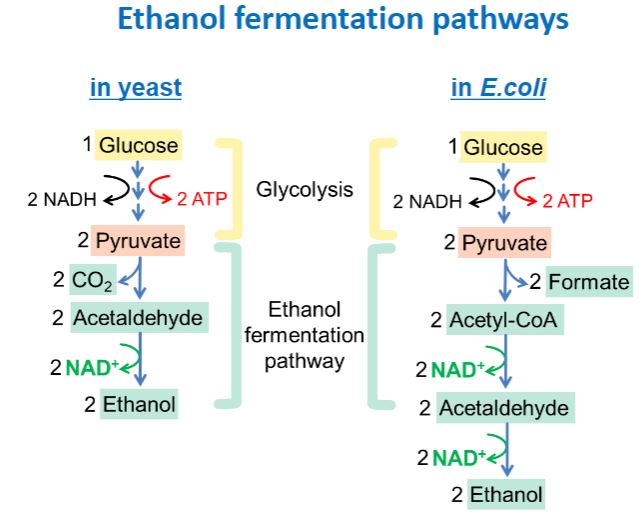
98
New cards
-fermentation pathways are named after the major acid or alcohol produced. The most common fermentation is lactic acid (lactate) fermentation, the reduction of pyruvate to lactate (figure 7.20, number 1). It occurs in some bacteria, protists, and in animal skeletal muscle. Lactic acid fermenters can be separated into two groups. Homolactic fermenters use the Embden-Meyerhof pathway and directly reduce almost all their pyruvate to lactate with the enzyme lactate dehydrogenase. Heterolactic fermenters use the pentose phosphate pathway to form products other than lactate; many also produce ethanol and CO2. Lactic acid bacteria are important to the food industry,
Ethanol fermentation in yeast and lactic acid fermentation
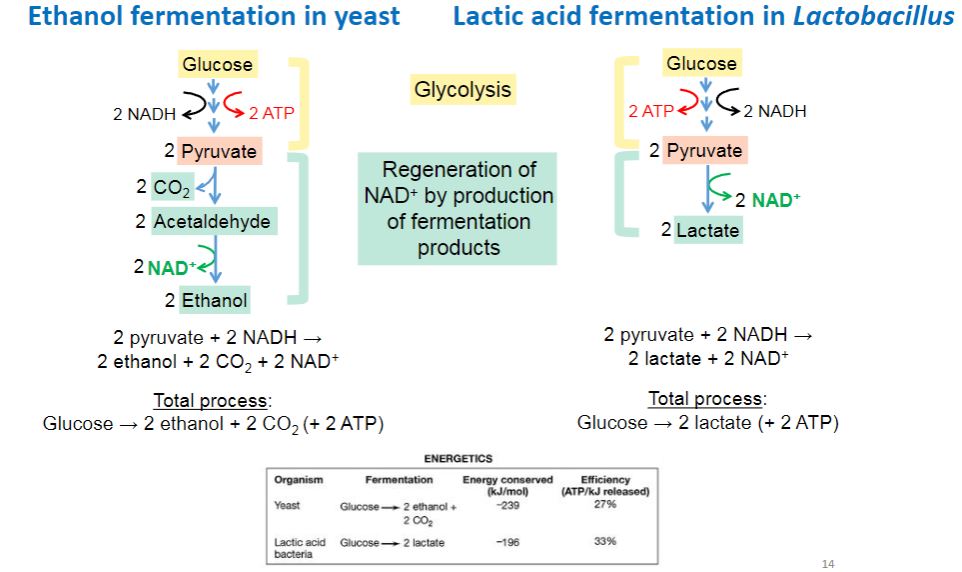
99
New cards
-Many bacteria, especially members of the family Enterobacteriaceae, metabolize pyruvate to numerous products using several pathways simultaneously. One such complex fermentation is the mixed acid fermentation, so called because it results in the excretion of a mixture of acids, particularly acetic, lactic, succinic, and formic acids as well as ethanol. Members of the genera Escherichia, Salmonella, and Proteus carry out mixed acid fermentation.
-mixed acid fermentation
A type of fermentation carried out by members of the family Enterobacteraceae in which ethanol and a complex mixture of organic acids are produced.
-mixed acid fermentation
A type of fermentation carried out by members of the family Enterobacteraceae in which ethanol and a complex mixture of organic acids are produced.
Mixed acid fermentation
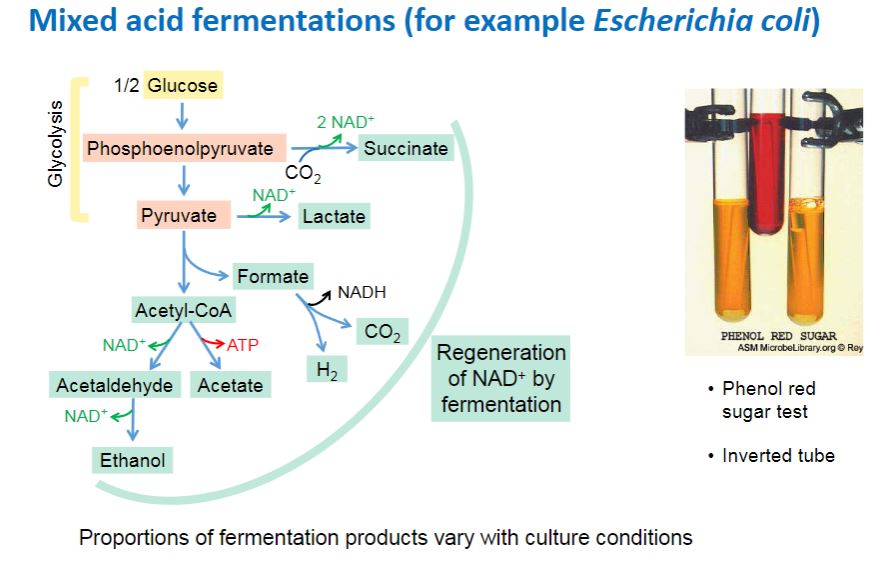
100
New cards
a
Secondary fermentation