3,4: Biopolymers, The Central Dogma of Molecular
1/12
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
13 Terms
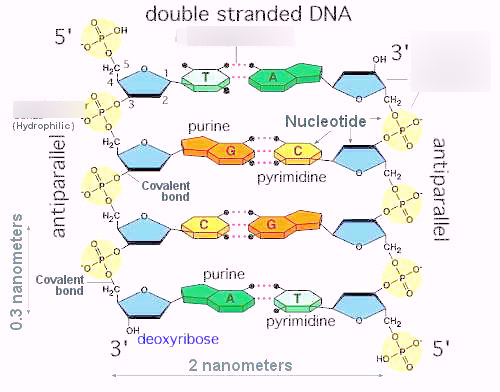
What are the three main components of a DNA nucleotide, and how do DNA and RNA differ in their sugars and bases?
Each DNA nucleotide has three parts:
Phosphate group – gives the DNA backbone a negative charge.
Sugar – in DNA it’s deoxyribose (a 5-carbon sugar missing an oxygen at the 2′ carbon).
Nitrogenous base – the “letters”: Adenine (A), Thymine (T), Cytosine (C), Guanine (G).
👉 RNA difference
Sugar = ribose (with the oxygen still present).
Base = uses Uracil (U) instead of Thymine (T).
What types of bonds hold nucleotides together in nucleic acids, and what type of bond attaches bases to sugars?
Base to sugar → β-N-glycosidic bond
Sugar to phosphate to sugar → phosphodiester bond.
Links nucleotides into a chain.
Always connects the 5′ phosphate of one sugar to the 3′ hydroxyl (–OH) of the next sugar.
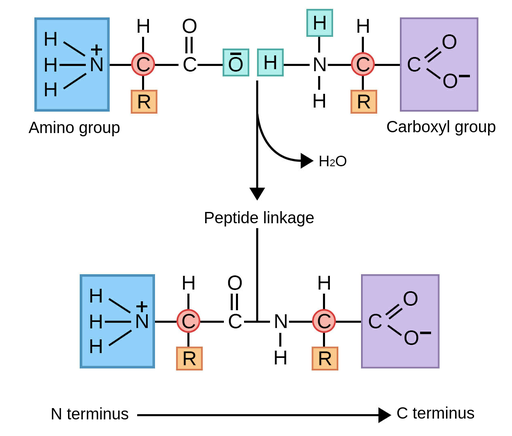
What are the key structural groups of an amino acid, and how are amino acids linked together into proteins?
Amino group (–NH₃⁺)
Carboxyl group (–COO⁻)
R group (side chain) → gives each amino acid its special properties (polar, non-polar, acidic, basic).
Bond: amino acids join by peptide bonds (formed by dehydration synthesis, meaning water is removed).
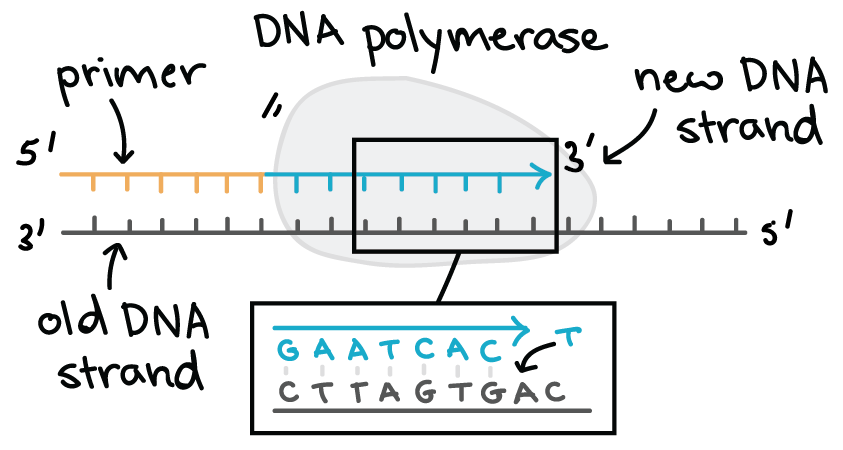
Describe nucleic acid’s directionality (5′→3′ vs N→C)?
Each nucleic acid strand (DNA or RNA) has two distinct ends based on the carbon numbering in the sugar:
5′ end → has a phosphate group attached to the 5′ carbon.
3′ end → has a hydroxyl (–OH) group on the 3′ carbon.
Nucleotides are linked by phosphodiester bonds between the 3′–OH of one sugar and the 5′ phosphate of the next.
During replication or transcription:
The template strand is read 3′ → 5′,
The new strand is synthesized 5′ → 3′.
DNA strands run antiparallel: one strand 5′ → 3′, the other 3′ → 5′.
How is DNA structured into a double helix, and how does this differ from RNA structure?
DNA consists of two antiparallel strands forming a right-handed double helix, held together by hydrogen bonds between complementary bases (A–T, G–C). Its deoxyribose sugar makes it stable for long-term genetic storage, while RNA is single-stranded, contains ribose and uracil, and is more reactive and short-lived.
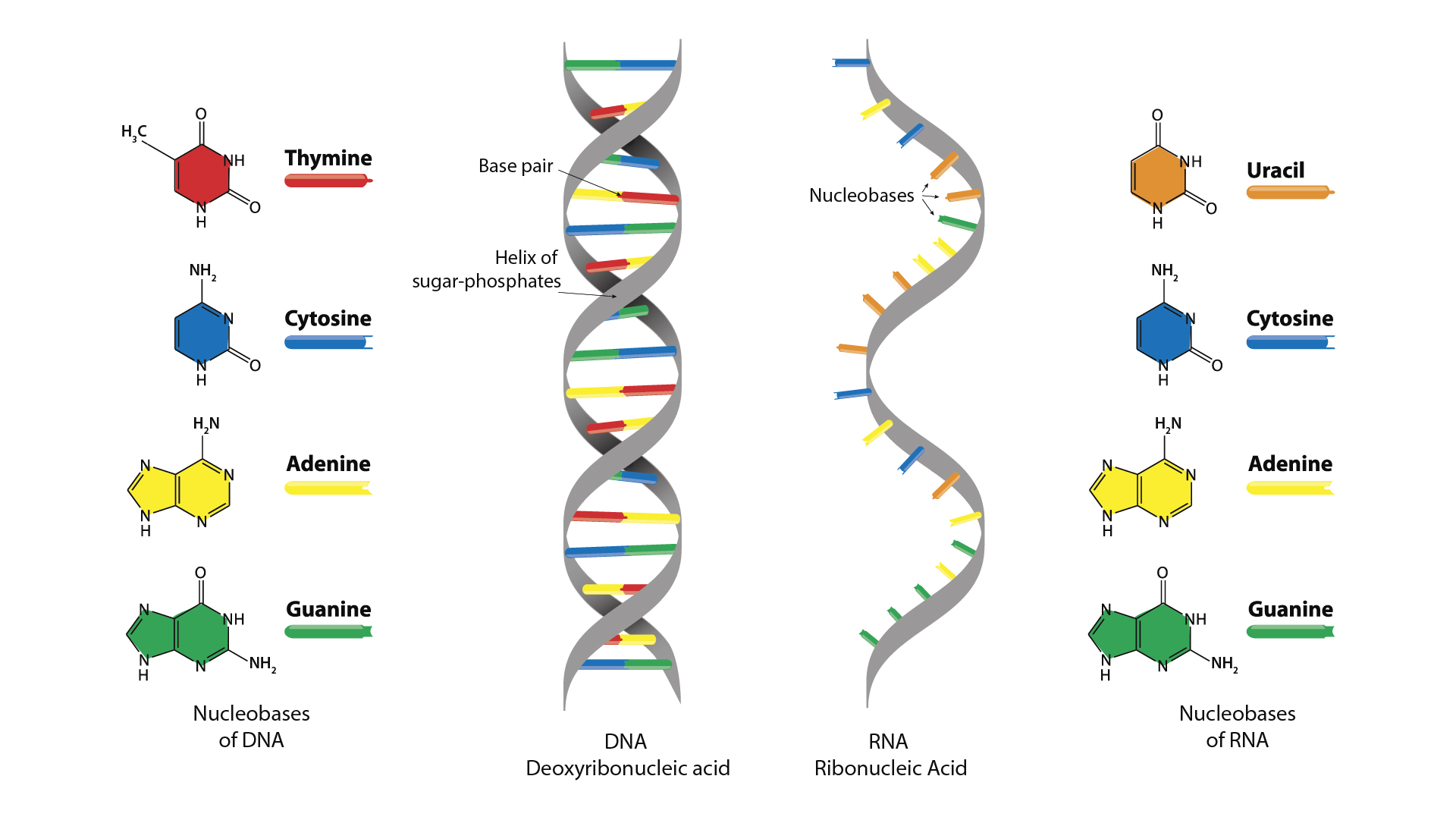
Why is DNA more stable than RNA, and what roles do RNA hairpins and loops play in protein synthesis
Sugar: DNA has deoxyribose (no 2′–OH), making it less reactive and less likely to break; RNA’s 2′–OH makes it unstable.
Base: DNA uses thymine (T) instead of uracil (U), which is chemically more stable and easier to repair.
Strands: DNA is double-stranded, so bases are protected inside the helix; RNA is single-stranded and more exposed.
How can the charge of DNA be exploited in laboratory techniques like electrophoresis, precipitation, and spectrophotometry?
Electrophoresis
DNA has a negatively charged backbone.
In a gel with an electric field, DNA runs toward the positive (+) electrode.
Precipitation with alcohol
Add salt (Na⁺) → shields negative backbone.
Add ethanol/isopropanol → DNA becomes insoluble and clumps, so you can see/collect it.
Spectrophotometry (UV absorbance)
Shine UV light → DNA, RNA, and proteins absorb differently.
At 260 nm: DNA & RNA absorb strongly (bases absorb UV).
At 280 nm: proteins absorb (aromatic amino acids).
What is the central dogma of molecular biology, and how does genetic flow between DNA, RNA and proteins
The central dogma of molecular biology explains how genetic information flows inside a cell.
Information is stored in DNA, copied into RNA through transcription, and then translated into proteins.
So the direction of flow is:
DNA → RNA → Protein
(Replication → Transcription → Translation)
What are the genome, transcriptome, and proteone, and how do they differ between various cell types
The genome is all the DNA in a cell
The transcriptome is all the RNA molecules being made from that DNA — it shows which genes are active.
The proteome is all the proteins produced in that cell.
The genome stays the same in all cells, but the transcriptome and proteome differ depending on the cell type and its function.
What are the key features of the universal genetic code, and what is meant by it being triplet, non-overlapping, and common?
The genetic code translates mRNA codons (three-base sequences) into amino acids.
It is triplet because each amino acid is coded by three nucleotides (a codon), non-overlapping because codons are read one after another with no shared bases, and common (universal) because nearly all organisms use the same codons to specify the same amino acids.
This shared system allows genes to be read across different species.
Compare bacterial vs eukaryotic genomes.
Feature | Bacteria (Prokaryotes) | Eukaryotes |
|---|---|---|
DNA shape | Usually single, circular molecule | Multiple, linear chromosomes |
Location | In nucleoid (no nuclear membrane) | In a nucleus (membrane-bound) |
Histones | Absent (may have histone-like proteins) | Present — DNA wound around histones → chromatin |
Genome size | Small, few million base pairs | Large, billions of base pairs |
Non-coding DNA | Very little; genes packed closely | Large amounts of non-coding and regulatory DNA |
Extra DNA | Often have plasmids (circular mini-DNA) | May have mitochondrial DNA (and chloroplast DNA in plants) |
Replication origin | Single origin of replication | Multiple origins per chromosome |
Cell division | Binary fission | Mitosis and meiosis |
Describe the effects of DNA/RNA mutations on protein mutations
Mutations = permanent changes in DNA/RNA sequence.
Types:
Point mutation (SNP) = single base change.
Insertion = extra base added.
Deletion = base removed.
Duplication = segment copied twice.
Effects on protein:
Silent: codon changes, but amino acid is the same.
Missense: codon changes → different amino acid.
Nonsense: codon becomes stop → protein truncated.
Frameshift: insertion/deletion changes reading frame → all amino acids downstream change.
How can you use the genetic code table to translate an mRNA (or DNA) sequence into its corresponding amino acid (peptide) sequence?
To translate, first find the start codon (AUG) on the mRNA
Then, divide the sequence into groups of three bases (codons) and use the genetic code table to match each codon to its amino acid.
Stop translating when you reach a stop codon (UAA, UAG, or UGA).
If given DNA, first convert it to mRNA by replacing T → U, then follow the same process.
Shortcut:
1⃣ Find AUG → start
2⃣ Split into triplets
3⃣ Use table to find amino acid for each
4⃣ Stop at stop codon