IMED1002 - Chromosome Changes - Structure (L29)
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
27 Terms
Chromosome Variants
- deletion is not just of a single gene and can be a deletion of a section of the chromosome that held thousands of genes
Chromosome Structure Changes (in terms of telomere and centromere)
- following meiosis, only get survival of cell if chromosomes have one centromere and two telomeres (VERY IMPORTANT TO KNOW)
- lack of centromere: acentric, nowhere for spindle to attach, so DNA lost
- Two centromeres: dicentric, typically not fully incorporated into progeny cell
- Loss of telomeres impacts DNA stability and replication
- telomeres are GC rich repeats at the ends of chromosomes (caps) that protect the ends of chromosomes.
- if you don't have telomeres, these ends of chromosomes are called naked and naked DNA is really recombinogenic (lots of recombination going to happen, hence not going to produce a viable cell).
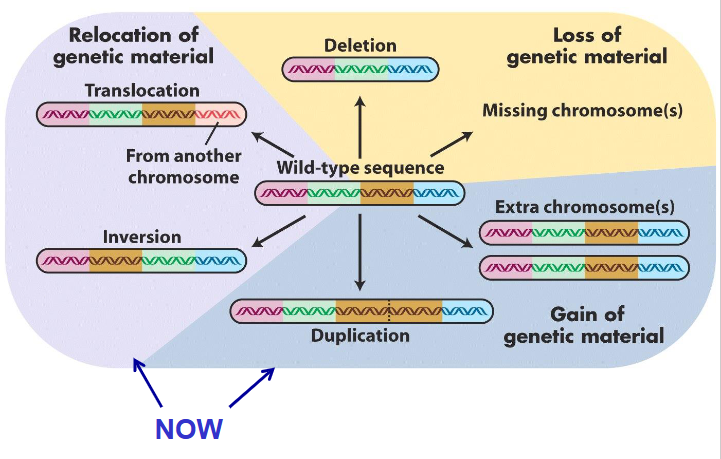
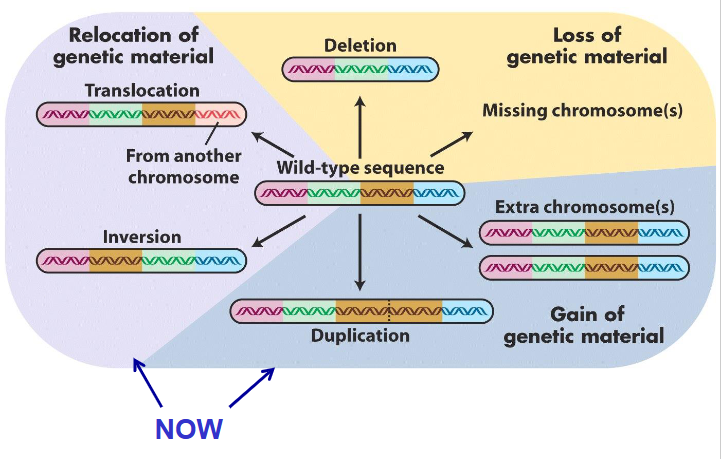
Variants that change the structure of an individuals chromosomes
Rearrangement 4 types:
- Deletion: loss of chr segment
- Duplication: doubling of chr segment
- Inversion: orientation within chr reversed
- Translocation: segment moved to different site
- deletions and duplications can affect gene balance
- Chromosome (chr) abbreviation (AB.CDEFG)
- A, B are regions of the chr and the dot (.) is the centromere
- note that ABCD are not single genes they are regions of the chromosomes, segments that contain hundreds of thousands of genes
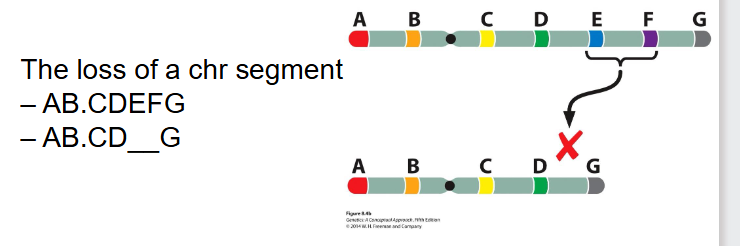
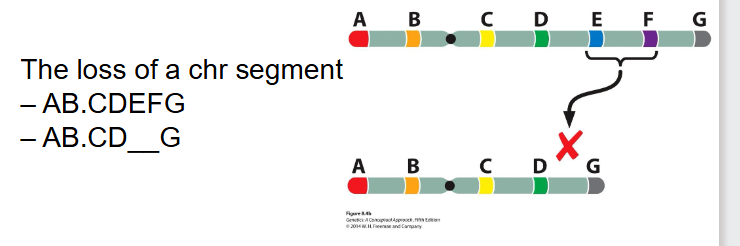
Deletions
- the loss of a chr segment
AB.CDEFG turns into AB.CD__G
- multigenic deletions: several or many genes lost
- if have multigenic on both homologues - not viable
- Often multigenic deletion on one chromosome also not viable
- Gene dosage effect, and/or expression of deleterious recessive allele
Effects of Deletions
- Phenotypic consequences depend on which genes located in deleted region
- Deletion of centromere = acentric chr will not segregate and will be lost
- Homozygous deletions: many are lethal
- Heterozygotes may have multiple defects due to imbalance: Pseudodominance (recessive allele expressed when dominant allele is deleted)
- Haploinsufficient gene (some genes must be present in two copies for normal function)
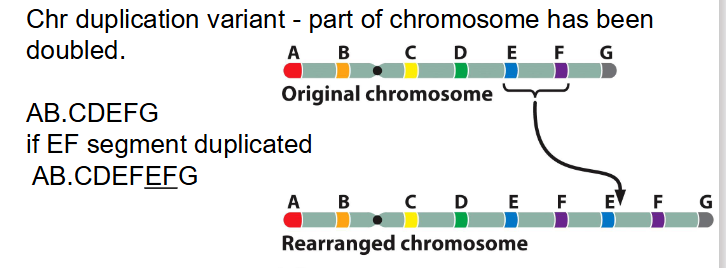
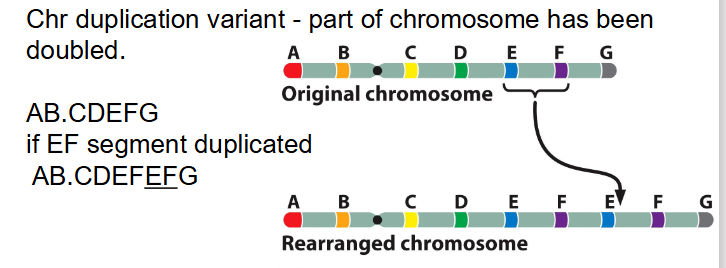
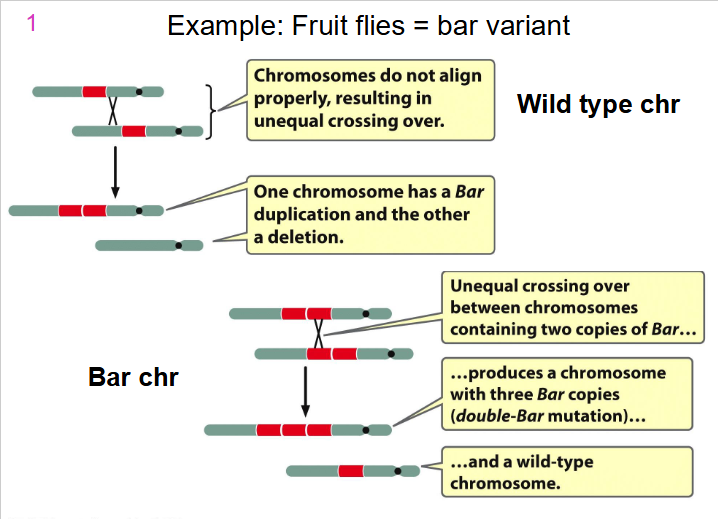
Duplications
- Chr duplication variant - part of chromosome has been doubled
AB.CDEFG becomes AB.CDEFEFG if the EF segment is duplicated
- if duplication is immediately adjacent = tandem duplication
- if duplication is some distance = displaced duplication (e.g AB.CDEFG(EF)
- duplication may be in the same orientation or inverted
e.g AB.CDEF(FE)G

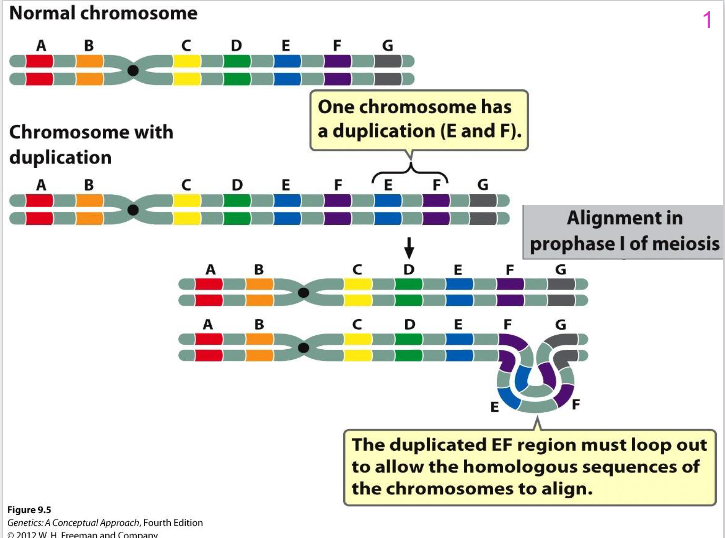
Effects of Duplication
- individual homozygous for duplication carries duplication on both homologous chr
- Heterozygous = 1 normal chr and 1 chr with duplication problems arise in chr pairing in meiosis
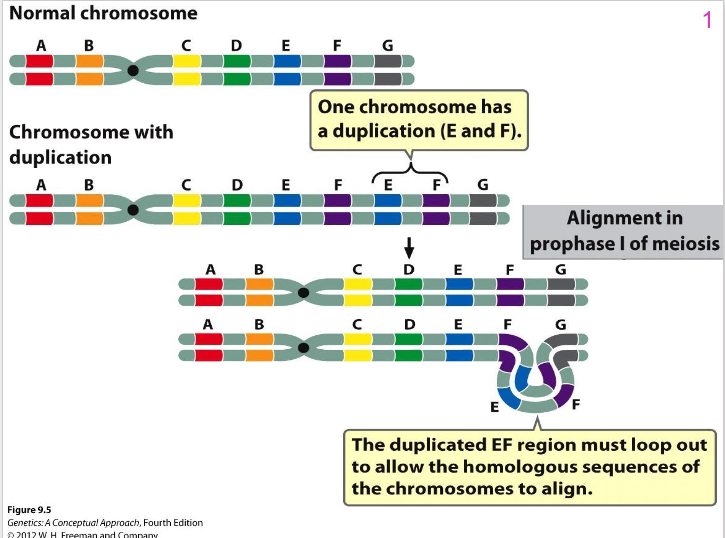
- Chr loop and twist - regions line up (see diagram)
- Characteristic loop structure is one way can detect duplications

Chromosome Loop Diagram
DIAGRAM ON SLIDE 10
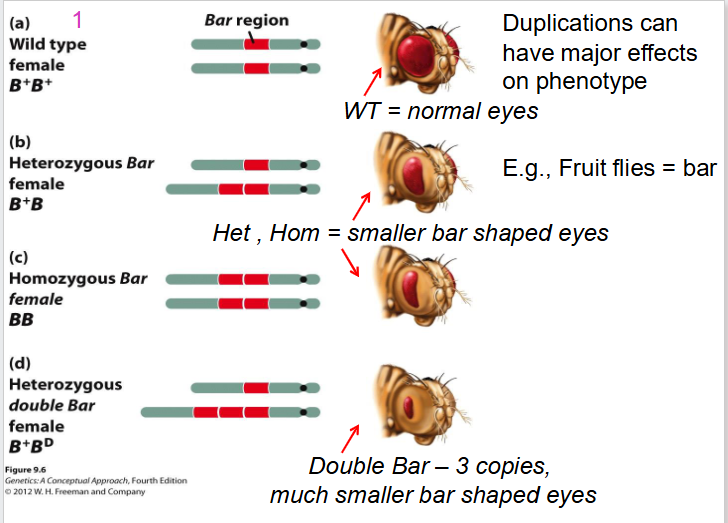
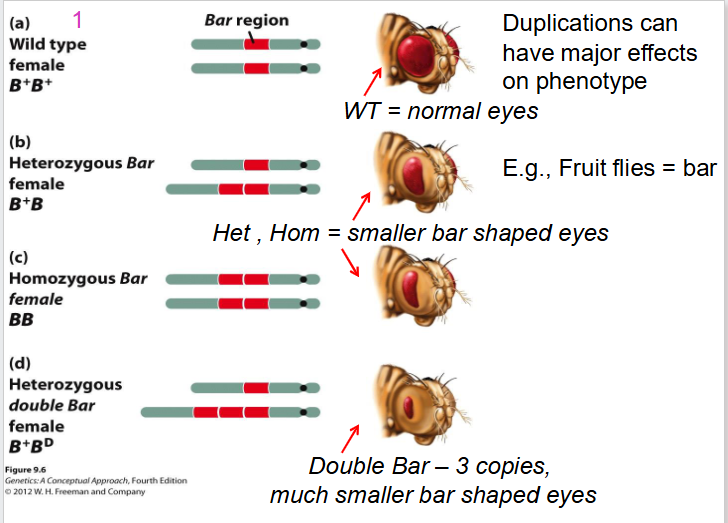
Effect of Duplications on Fruit Flies
DIAGRAM ON SLIDE 11
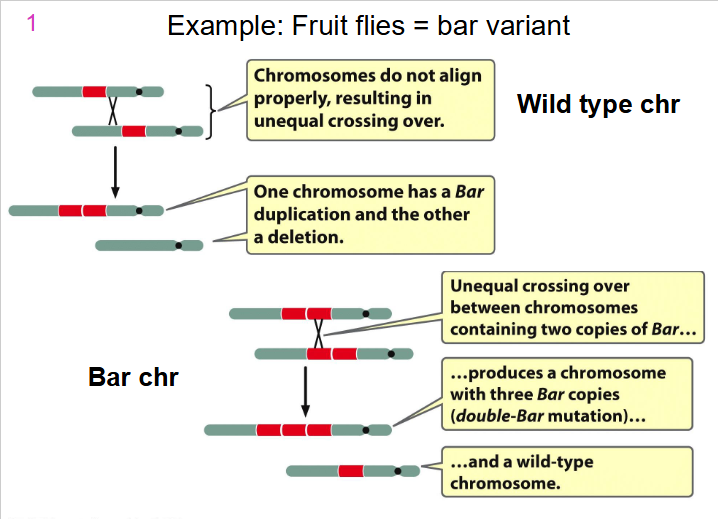
Example: Fruit Flies = bar variant
DIAGRAM ON SLIDE 12
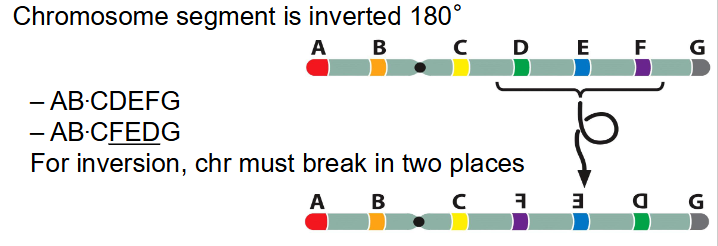
Inversions
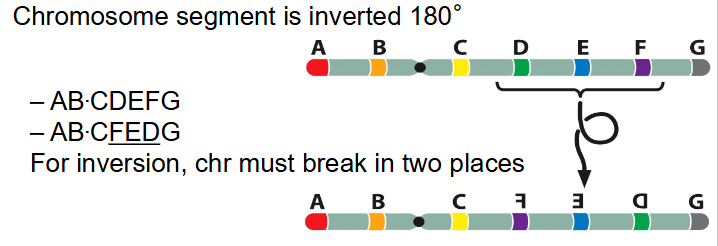
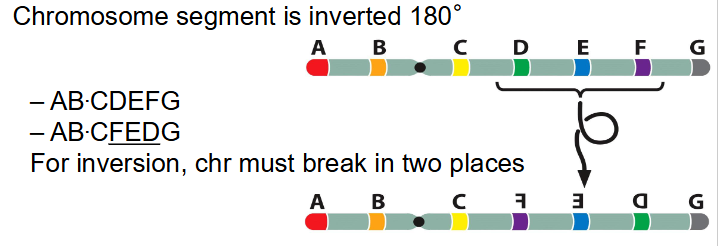
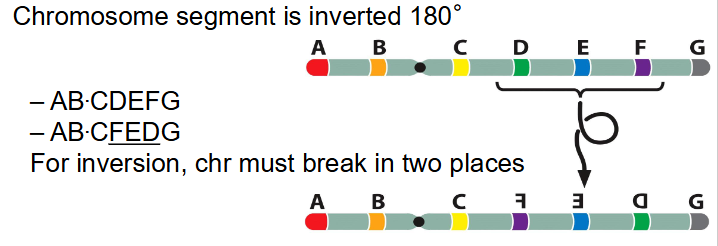
- chromosome segment is inverted 180 degrees
- AB.CDEFG becomes AB.C(FED)G
- for inversion, chr must break in two places
Effect of Inversion
- haven't lost any genetic material, still often have pronounced effects
- May break a gene into two parts
- If between genes, inverted gene order
- Regulation of gene expression is position-dependent
- Mis-expression (position effect)
Types of Inversions
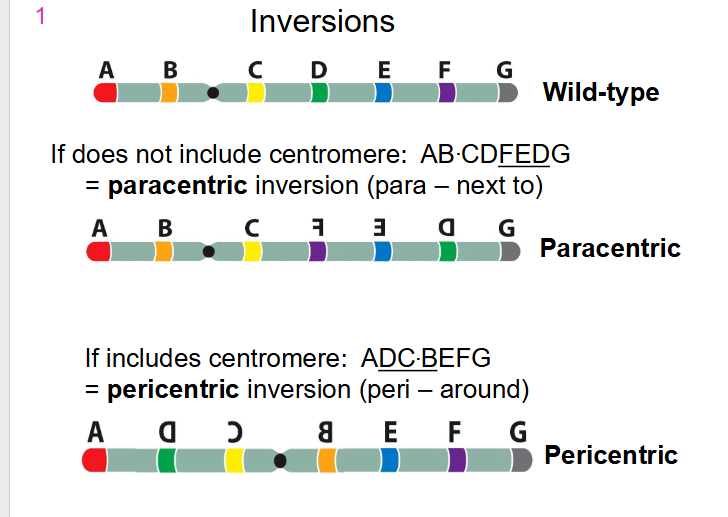
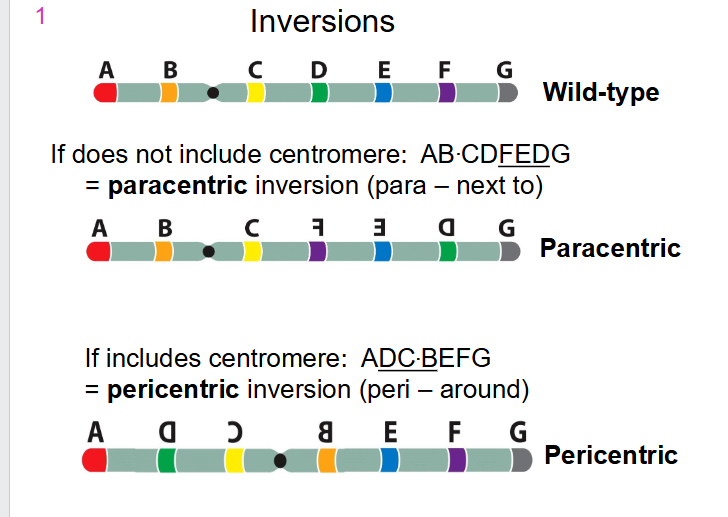
- if does not include centromere: AB.CD(FED)G we call it a paracentric inversion (para means "next to")
- If includes centromere: A(DC.BEF)G we call it a pericentric inversion (peri means "around)
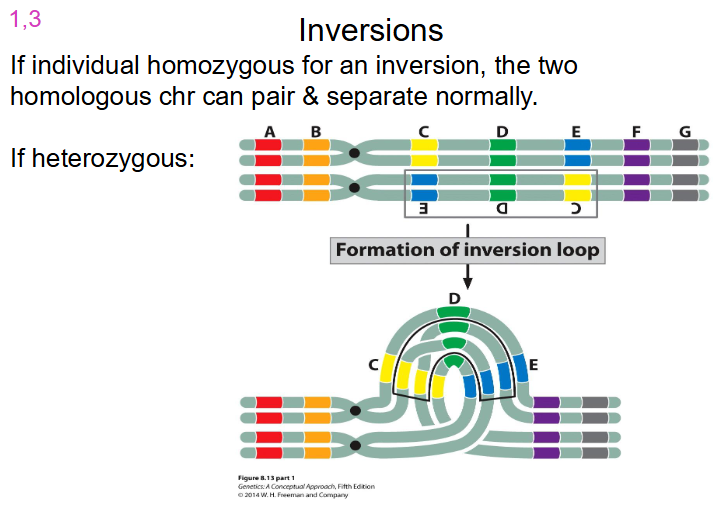
Inversions that are Homozygous and Heterozygous
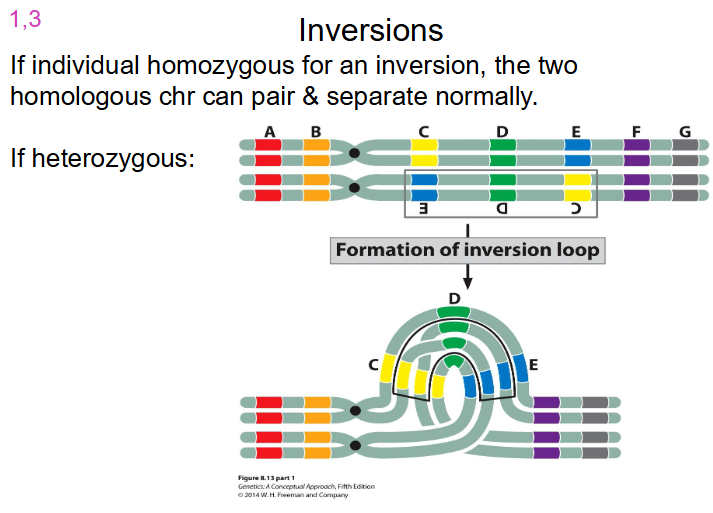
- if individual homozygous for an inversion, the two homologous chr can pair and separate normally
- if its heterozygous, look at diagram
- problems occur when we have crossing over of non-cystic chromatids
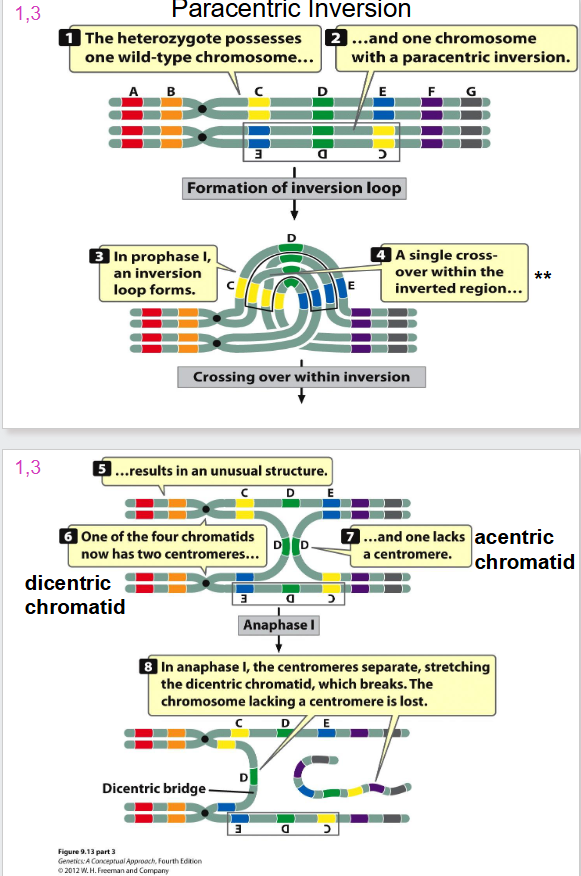
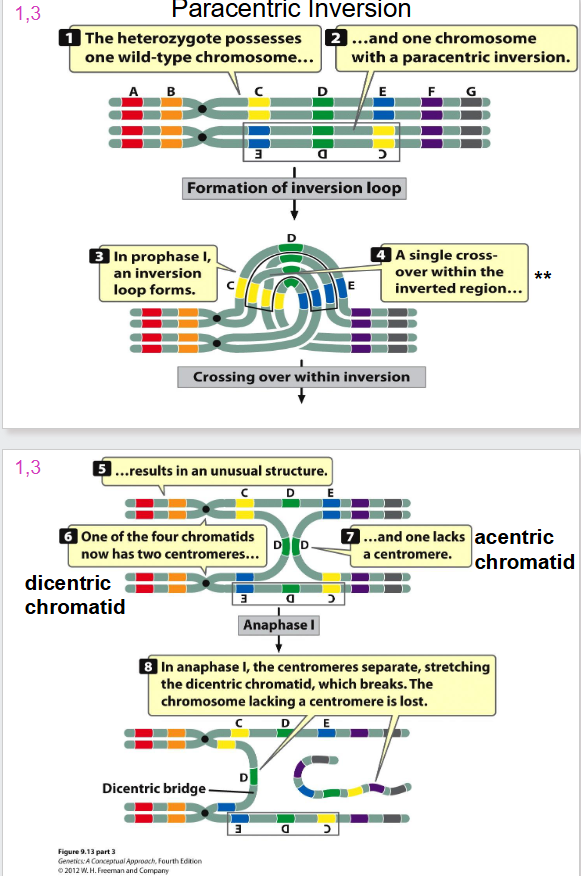
Paracentric Inversion
- formation of inversion loop and a crossing over event occurs
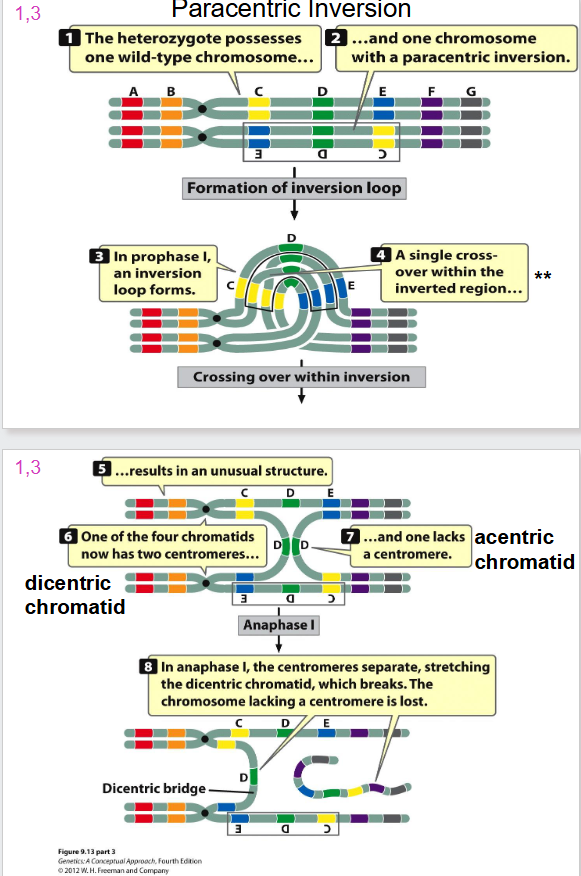
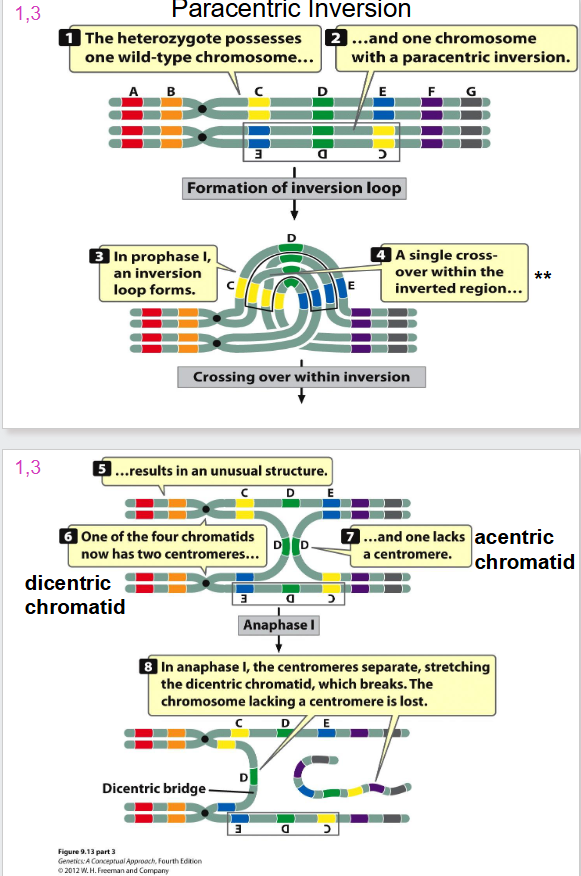
Problem with Inversion and Crossing Over
- when one sister chromatid is wild type, and other has dicentric inversion we have crossing over
- a dicentric bridge is formed
- when you have two centromeres, the spindles form and rip chromatid apart
- the acentric chromatid will not attach to any spindles and hence is lost
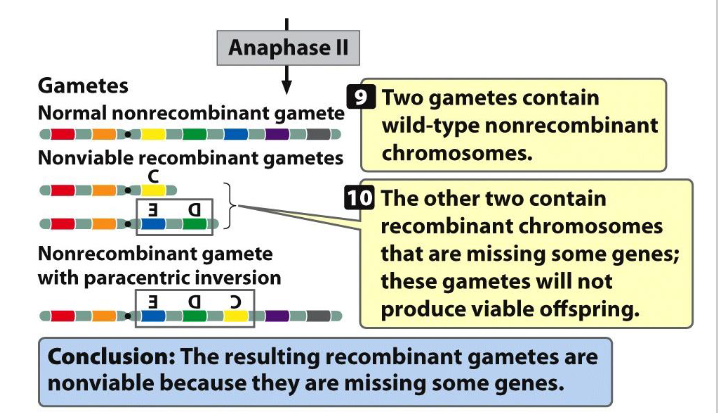
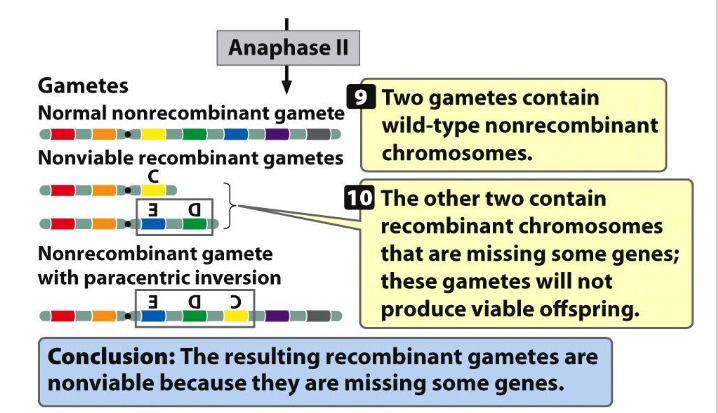
Paracentric Inversion SUMMARY
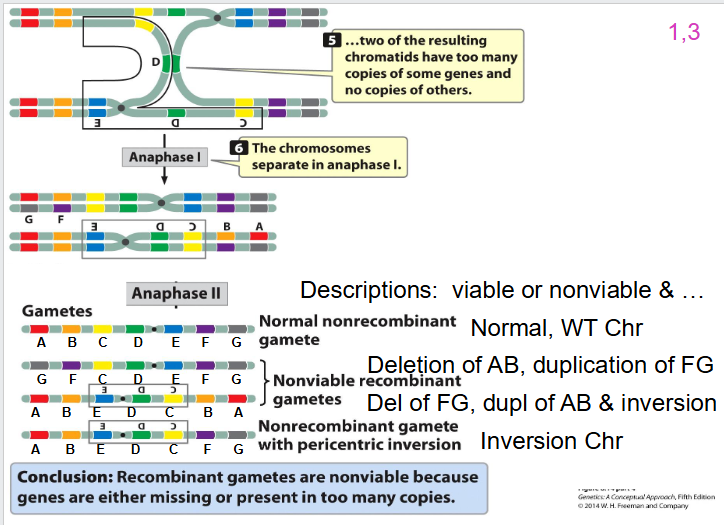
- need to be able to tell the 4 different gametes that can be formed if you're a heterozygous with a paracentric inversion
- 50% of gametes will be non-viable: you'll have that acentric chromosome that's lost and that dicentric chromosome thats broken in two, it does not have a telomere on one of the ends. they will not produce a viable gamete, a viable cell
- you'll have one gamete that has a normal wild type non-recombinant chromosome
- and you'll have one that is non-recombinant that has the paracentric inversion
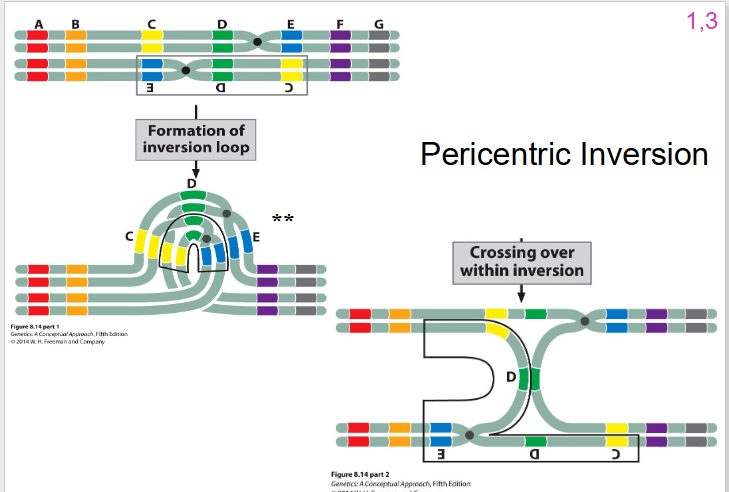
Pericentric Inversion DIAGRAM
- still formation of inversion loop
- still one crossing over event that can occur, but the outcomes are going to be different to the paracentric inversion
- we're going to have one that is wild type non-recombinant
- we're going to have one that is inverted non-recombinant
- what happens to the other two is different
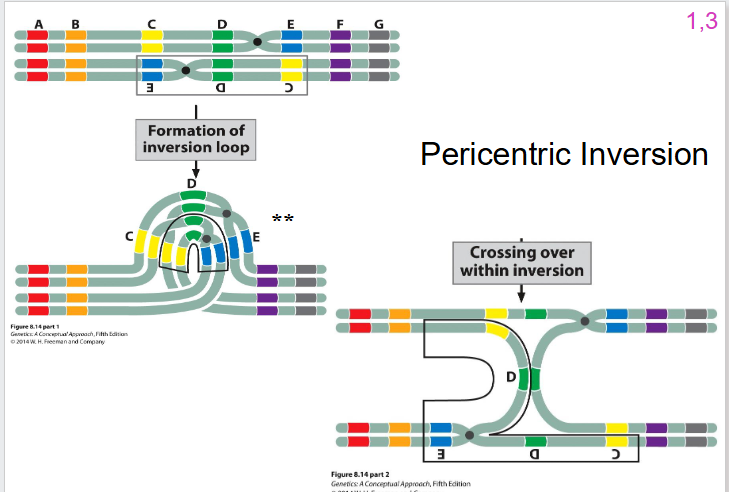
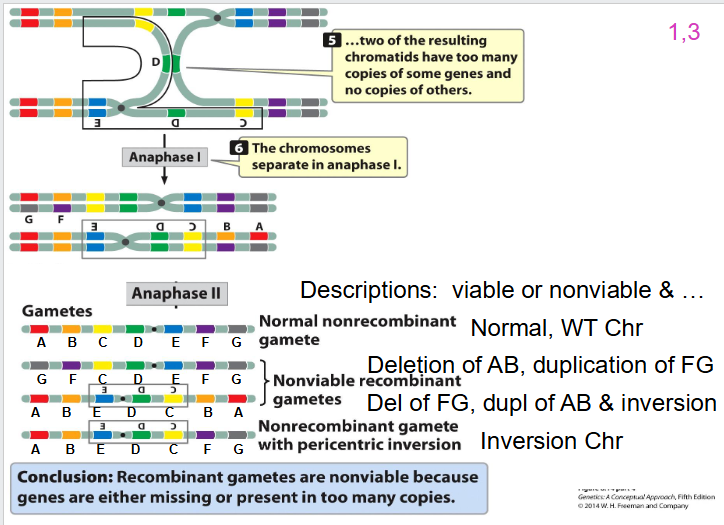
Pericentric Inversion Other two
- still going to have 50% of gametes that are viable, 50% not viable
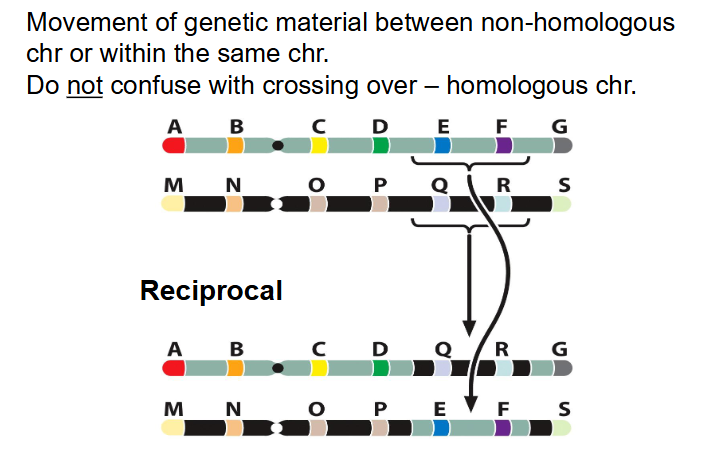
Translocations
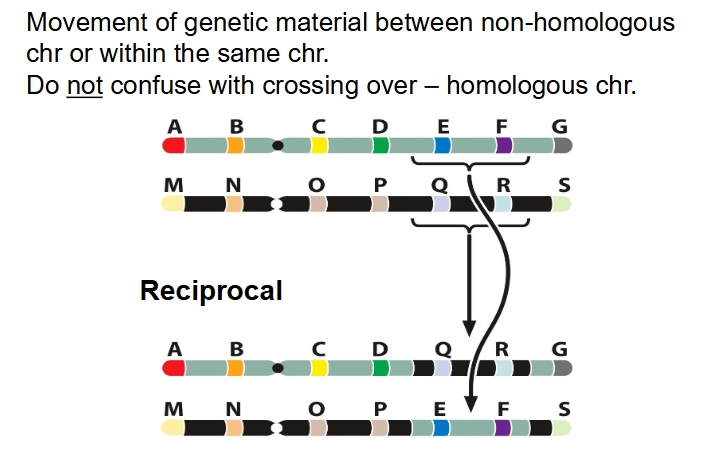
- Movement of genetic material between non-homologous chr or within the same chr
- do not confuse with crossing over - homologous chr

Translocation Types
- Nonreciprical: material moves from one chr to another without reciprocal exchange.
AB.CDEFG and MN.OPQRS (EF MOVES):
becomes AB.CDG and MN.OPEFQRS
- reciprocal: two way exchange (more common)
AB.CDEFG and MN.OPQRS
AB.CDQRG and MN.OPEFS
- usually, theres no change in gene dosage effects. we aren't changing the number of genes, we're just changing their position

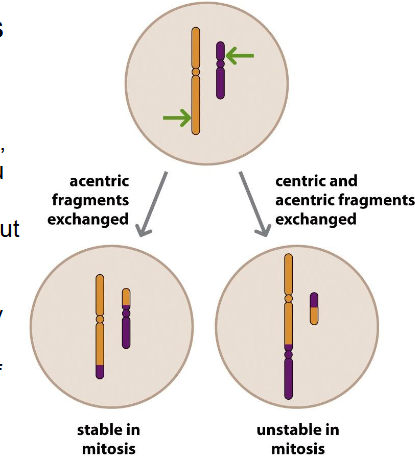
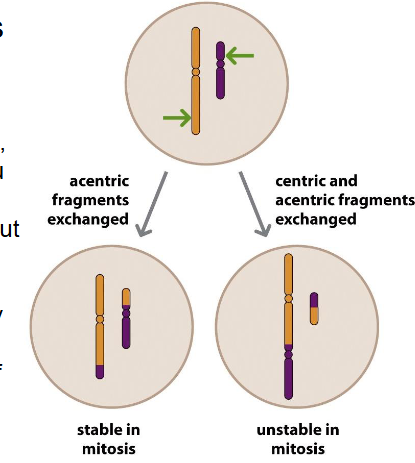
Translocation is common in
- usually between non-homologous chromosomes
- more common then expected (1 in 500)
- most carriers are healthy, but increased risk of offspring having unbalanced chr translocations and of infertility or miscarriage
- there are also examples of cancers and other disorders caused by translocations
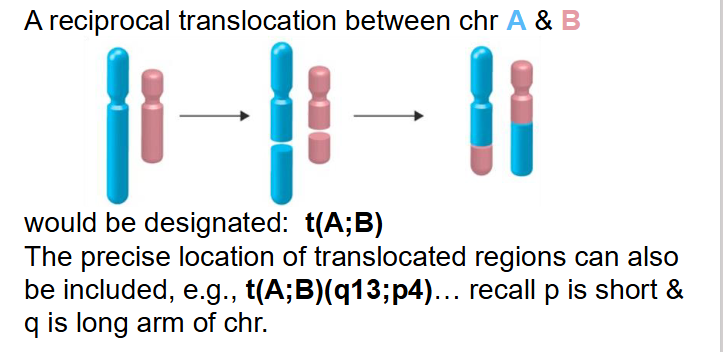
ISCN Designation for Translocations
- reciprocal translocation between chr A and B
- would be designated t(A;B)
- The precise location of translocated regions can also be included, e.g t(A;B)(q13;p4)... recall p is short and q is long arm of chr
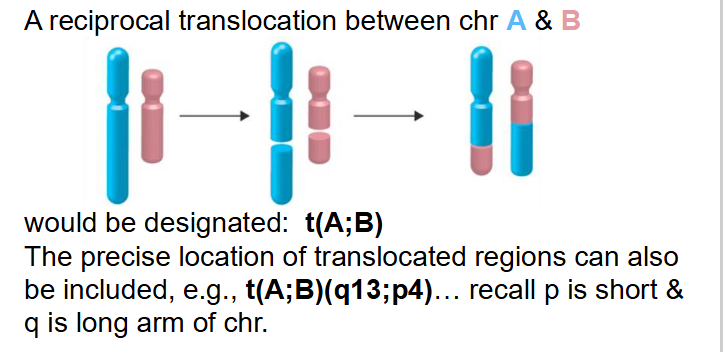
EXAMPLE OF NOMENCLATURE BEING USED
DIAGRAM ON SLIDE 25
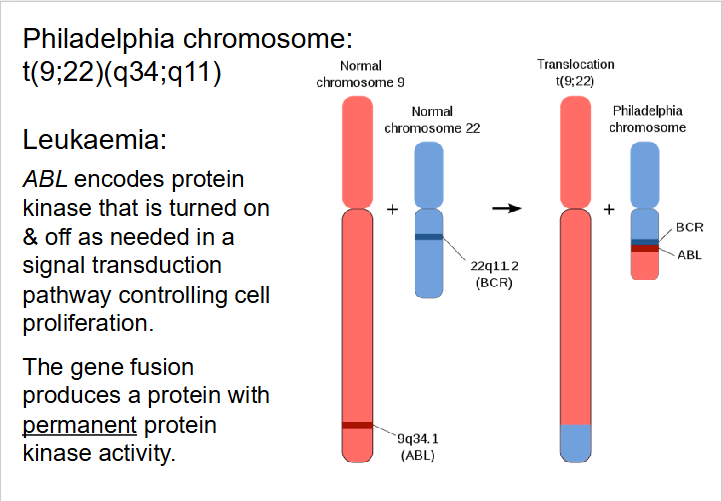
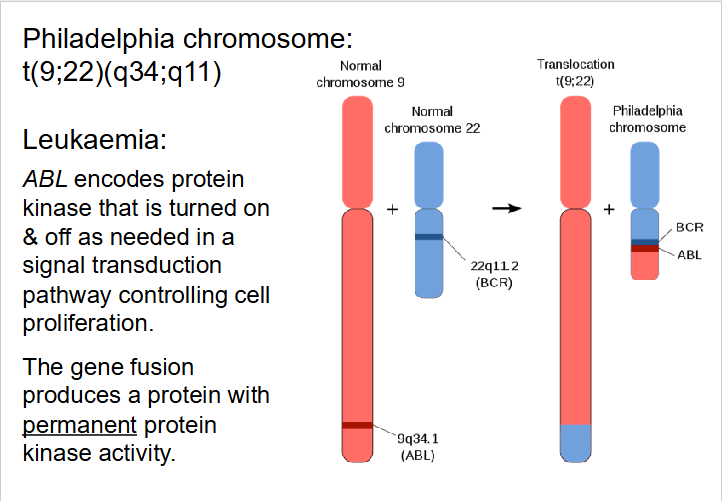
Effects of Translocation
- can impact phenotype in several ways
- Can physically link genes that were formally on different chromosomes
- Change gene expression as genes may be under control of different regulatory sequences
- Chromosomal breaks may take place within a gene
- Deletions frequently accompany translocations
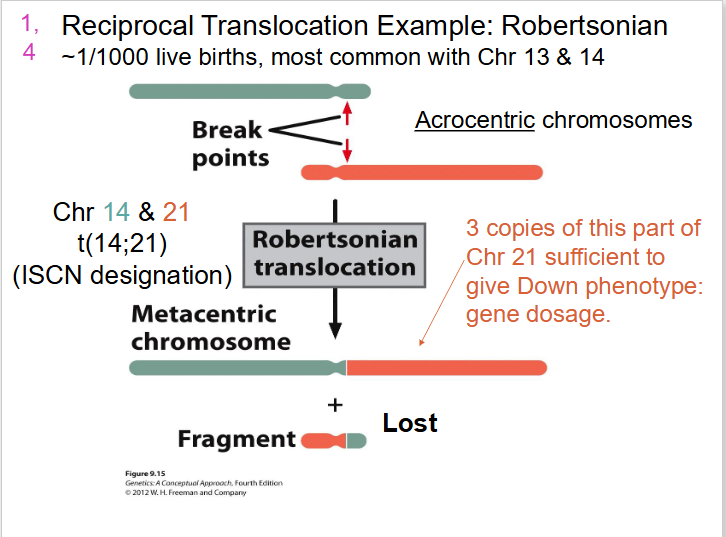
- Robertsonian translocation (break close to centromere): e.g rare form of Down Syndrome phenotype results from translocation events - may be transmitted, parent to child
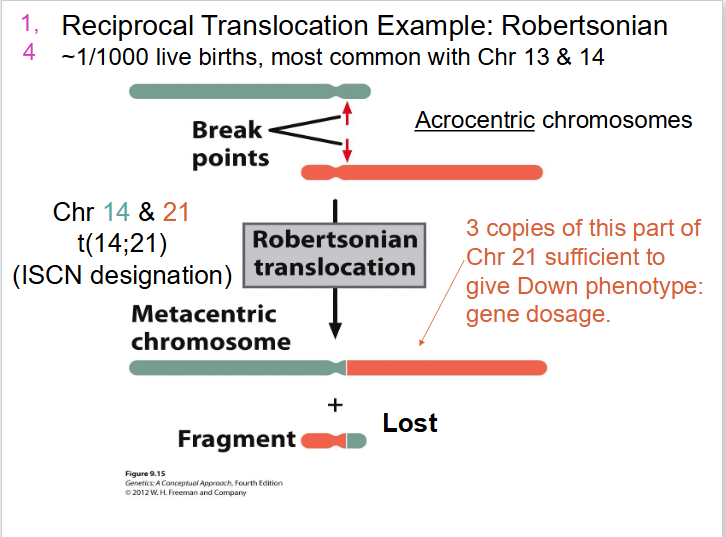
Reciprocal Translocation Example: Robertsonian
- Acrocentric chromosomes
- 3 copies of this part of Chr 21 sufficient to give down phenotype: gene dosage
- we end up with one really long one and one really short one, so short that the cell won't detect it and it will be lost. the long one contains most of genes from 21 and 14
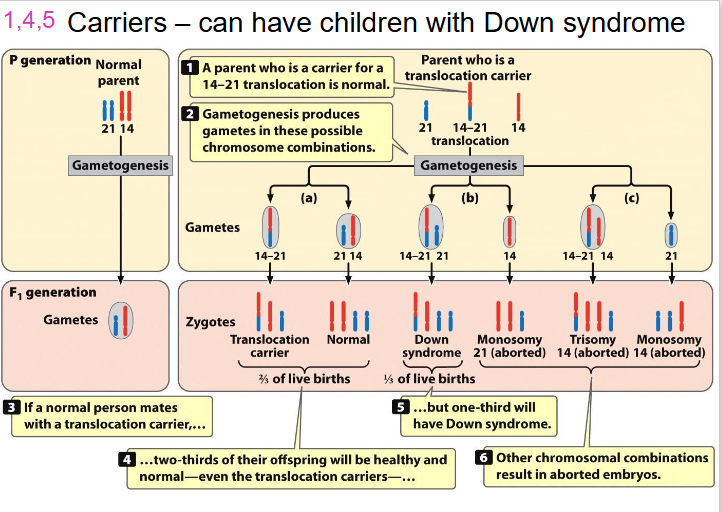
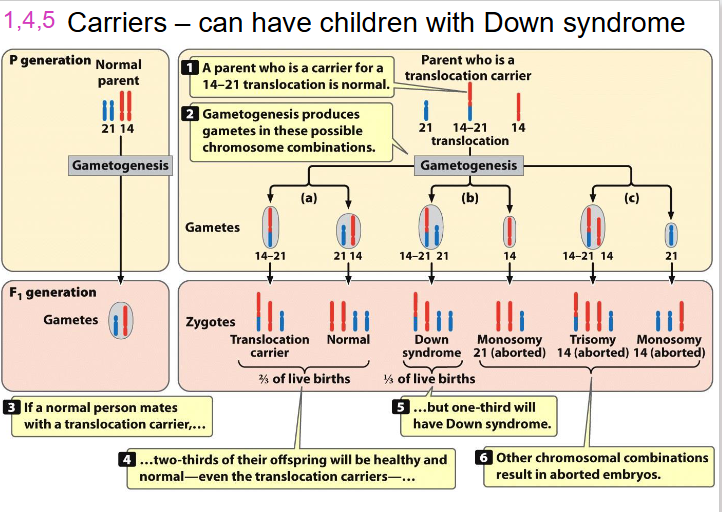
Carriers - can have children with Down Syndrome
LEARN THIS AGAIN, since its part of learning outcomes, there is a video on youtube she put on LMS
- trisomy 14 will be aborted, its not viable
- trisomy 21 wil be viable
- basically translocation can make it act like trisomy