Components of a Cell
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
85 Terms
DNA must be protected (it is fragile) and accessed in a controlled way. the cell isolates DNA in a dedicated compartment → —
the nucleus
Key consequences:
The nucleus is the main site of —-.
It is also where RNA is made (—-), since RNA is a working copy of DNA instructions.
DNA replication
transcription
Only small amounts of DNA exist outside the nucleus (—)
mitochondria
Once instructions exist, the cell needs a place to do the work. The working space → —
Cytoplasm and Cytosol
Everything outside the nucleus is the —
cytoplasm.
The fluid part of the cytoplasm is the —
cytosol.
Why the cytosol matters:
It is where:
Most proteins are —
Most proteins are degraded
Most intermediary metabolism happens (breaking down things into AA, nucleotides, etc)
synthesized
Instructions (RNA) are useless unless they are turned into —
functional molecules (proteins)
— read RNA and build proteins.
Ribosomes
Why the Endoplasmic Reticulum (ER) exists
Problem:
Some proteins must:
—-
Be secreted (AKA be in vesicles)
Solution:
Make them directly into a membrane system → the ER.
why does this solve the problem?
because we need to get the vesicles from somewhere and the ER buds off the vesicle to enclose the proteins that need to be secreted
Key first-principle insight:
Proteins enter the ER while they are being synthesized, not after.
what is meant by this?
when we talk about these secreted proteins, they first start being made in the cytoplasm like everybody else, but then they finish translation in the ER
why do we need some free ribosomes and some attached to the ER
some will be fully translated in the cytoplasm so we need free ribosomes
some will start in cytoplasm and then finish in ER, so we need ribosomes there too to finish the job
smooth er is mostly for
calcium storage
Making molecules isn’t enough, they must be modified and delivered correctly. Sorting and shipping → —
The Golgi Apparatus
The ER sends proteins to the —
Golgi apparatus
What the Golgi does:
Receives proteins from the ER
—- them
Sorts and dispatches them
Chemically modifies
t or f
Only proteins that enter the ER can ever go to the Golgi.
Proteins made in the cytosol never go to the Golgi.
t
the only way into the Golgi is:
👉 inside a vesicle
t or f
All proteins start on ribosomes in the cytosol.
t
At the beginning, every ribosome is:
Free in the cytosol
Making the first few amino acids of a new protein
if this protein needs to be secreted-HOW DOES IT KNOW IT NEEDS TO GO TO THE ER?
A protein contains address information in its amino acid sequence.
Think of this as a shipping label embedded in the protein itself.
Only proteins with an ER signal sequence go to the ER.
for a secreted protein:
As the protein is being synthesized, an— emerges
ER signal peptide
the ER signal peptide is recognized by the —
Signal Recognition Particle (SRP) (this is a whole other separate player just floating aroudn scanning for the signal, it is not part of the ribosome)
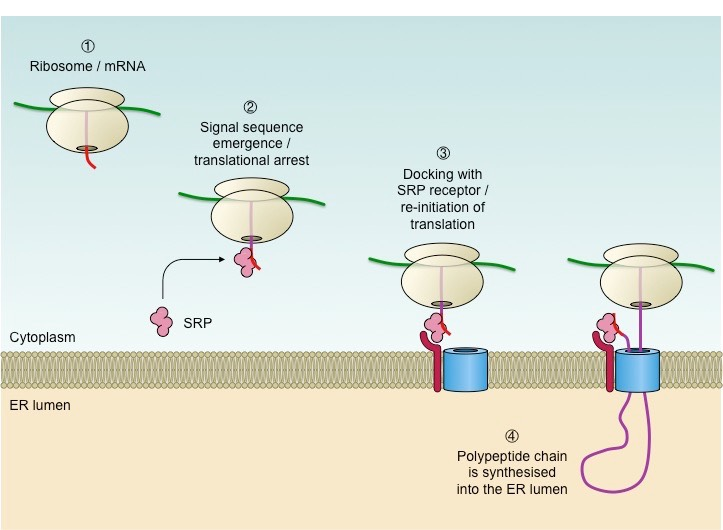
— pauses translation
Signal Recognition Particle
after SRP gets involved and pauses translation, what happens next?
The ribosome is moved to the ER membrane
This secreted translation pathway exists because:
Secreted proteins MUST be put in—
Vesicles bud only from the endomembrane system (ER → Golgi → membrane)
vesicles
By endomembrane system, we mean vesicles are capable from budding from those organelles, therefore, if we need to get it to the Golgi, it MUST have gotten a vesicle from the ER
what does “secreted protein” actually mean?
A secreted protein is a protein whose final functional location is outside the cell.
(blood, extracellular matrix, insulin, etc)
Why can’t the cell just make secreted proteins in the cytosol and let it drift out?
Problem #1: The plasma membrane is a barrier
Proteins are large and polar
They cannot cross lipid bilayers by diffusion
So if you make insulin in the cytosol:
It is trapped inside the cell
There is no hole it can just walk through
The only safe way to move a protein across a membrane is:
—-
Keep it enclosed by a membrane (vesicle) the entire time
a vesicle is
A small bubble of membrane
esicles cannot form out of thin air.
They must:
Bud off from—
an existing membrane
First-principle problem
Cells need a constant supply of ATP to drive function
Solution
Use a specialized compartment to:
Maintain proton gradients
Run redox reactions efficiently
this specialized compartment is —
the mitochondria organelle
First-principle problem
Cells must:
Break down worn-out organelles
Digest material taken up from outside
Do this without destroying themselves
Solution:
Create a sealed compartment full of digestive enzymes:
→ This is the Lysosome
Why lysosomes MUST be vesicular
—- are dangerous
If released into cytosol → cell death
Must be isolated behind membranes
Digestive enzymes
The organelle itself is not “built” like a machine — it is a space.
Nucleus = membrane + contents
Mitochondrion = membranes + contents
Lysosome = membrane + contents
its the contents that are proteins that perform functions, the membrane is just to create a space, a compartment
ok
so basically, the lysosome itself, it just a membrane
it contains lysosomal enzymes (digestive) which were synthesized in the
a) cytoplasm
b) ER
ER
they need to travel somewhere specific so they get “shipped” to the lysosome by a vesicle system and then put into another vesicle (the lysosome itself)
First-principle problem
Material taken into the cell (endocytosis) must be:
Sorted
Recycled
Or sent for destruction
Solution
—
Create intermediate compartments:
→ Endosomes
are endosomes basically vesicles?
yes, again it’s literally just a membrane bubble that encloses a space
What endosomes do
Receive vesicles from the —
Sort contents:
Some back to the membrane
Some onward to —-
plasma membrane
lysosomes (destruction)
You don’t want everything you ingest to be immediately destroyed, this is why — are helpful
endosomes
First-principle problem
Some reactions:
Produce harmful byproducts (e.g. hydrogen peroxide)
Should not occur in cytosol
Solution
Isolate these reactions in small compartments:
→ This is what Peroxisomes are
difference between lysosome and peroxisome
lysosome is more for breaking things down, garbage can, more general
peroxisomes have the specific function of detoxifying harmful substances, they use a different system to break things down and a lot of toxic stuff goes on in there
t or f'
If every cell had the same organelle proportions, most cells would be inefficient
t
Why the Golgi sits near the nucleus
The Golgi receives proteins from the ER
ER is continuous with the —
Shorter distances = faster trafficking
nuclear envelope
This minimizes transport time- the point of this is to say that the position of organelles inside the cell matters too, it’s not random
The precursors of the first eukaryotic cells are thought to have been relatively simple cells that —like most bacterial and archaeal cells—have a—-
plasma membrane but no internal membranes
That one membrane did everything:
Pumped ions
Made ATP
Secreted proteins
Synthesized lipids
eukaryotic cells are a lot bigger, but why couldn’t this one-membrane-system just scale up?
So a large cell with only one membrane has a crisis:
Too little membrane area (surface area) to support the volume
Too many membrane-based tasks
Evolution’s solution: -
internal membranes
the only way to:
Increase membrane area
Without increasing cell size
is to: fold the plasma membrane inward (invagination) just like brain gyri, cristae, it’s to increase surface area
Why are mitochondria so different from every other membrane-enclosed organelle?
Because they did not evolve from membrane invagination.
They evolved from engulfed bacteria
So far you’ve learned about organelles that came from:
Plasma membrane folding inward
(ER, Golgi, lysosomes, endosomes, etc.)
Now mitochondria belong to a completely different category.
okay
Imagine an ancient cell:
It had no internal membranes
It engulfed a bacterium
But instead of digesting it, the two formed a —
mutually beneficial relationship
because
The bacterium:
Was good at generating ATP
Over evolutionary time:
The bacterium became a permanent resident
It turned into what we now call a —
This is endosymbiosis.
mitochondrion
Why mitochondria have their own genome
If mitochondria came from bacteria, then originally they:
Had their own —
Had their own ribosomes
Controlled their own replication
DNA
During engulfment:
The bacterium already had its own plasma membrane
The host cell wrapped a membrane around it
So mitochondria ended up with:
Inner membrane → —
Outer membrane → —
Inner membrane → original bacterial plasma membrane
Outer membrane → host-derived engulfing membrane
The mitochondrial matrix = former —
bacterial cytoplasm
t or f
No vesicles bud to mitochondria
No vesicles bud from mitochondria
Their membranes are not part of the endomembrane system
t
This isolation is not a design choice — it’s historical inevitability.
problem:
DNA needs protection and regulation, but gene products must reach the cytosol efficiently.
Solution :
—
Enclose DNA in a nucleus
Add nuclear pore complexes instead of vesicles
No membrane fusion or vesicles involved
for things that don’t go through pores or don’t go in vesicles, what is the only other option?
The transported protein molecule usually must unfold to snake through the translocator
For a cell to stay alive and function, it must be able to:
Hold a shape (not collapse like a water balloon)
Withstand forces (pushing, pulling, squeezing)
Organize its inside (proteins, organelles can’t float randomly)
Change shape (during growth, division, movement)
Move things internally (vesicles, chromosomes, organelles)
Move itself (migration, tissue formation, immune response)
What I the solution?
the cytoskeleton: which is a flexible but rigid network of proteins
Why not one solid structure like bones?
Because cells need:
Strength and flexibility
Stability and rapid change
what is the name of the microfilament?
actin
what is actin used for?
cell shape and movement (crawling, contraction)
One important property of actin filaments?
Rapidly assembling/disassembling
what is the name of the “microtubules”?
tubulin
what are the three structural elements of cell (proper names)
actin (microfilament)
tubulin (microtubule)
Intermediate filaments
its hard to come up with one borad term for what microtubules are used for but they’re really Important in (2)
forming the mitotic spindle to pull apart chromosome sin mitosis and meiosis
kinesin and dyenin are motor proteins of microtubules
when you see intermeditate filament, think —
Provides Mechanical strength
(the thing in nails and hair)
Keratin is an intermediate filament
Structures actin builds:
Dynamic protrusions
—-: broad, sheet-like extensions for crawling
—: thin spikes for sensing the environment
Highly ordered bundles
— (inner ear): rigid actin bundles tilt as rods → convert sound vibrations into signals
— (intestine): actin-supported projections increase surface area for absorption
Important structure in mitosis and meiosis:
Lamellipodia
Filopodia
Stereocilia
Microvilli
CONTRACTILE RING!
also acts in muscle contraction
Structures microtubules build:
-
-
-
-Mitotic spindle
-cilia
-flagella
-axons
in microfilaments, the subunit is:
in microtubules, the subunits are:
actin
alpha and beta tubulin (dimer)→ together called tubulin
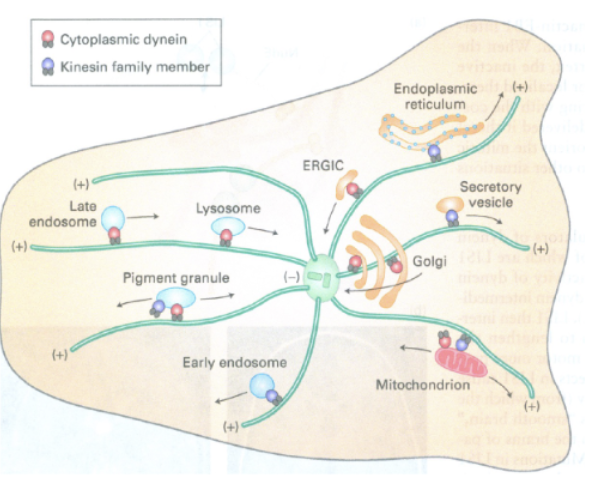
Cytoskeleton and Membrane Trafficking, why is this Important?
remember everything we just learnt about vesicle movement? Well, vesicles can’t just move on their own, they are carried on highways by microtubules and the cytoskeleton with motor proteins like kinesin, dyenin, myosin
not just vesicles, but also organelles like mitochondria, endosomes etc
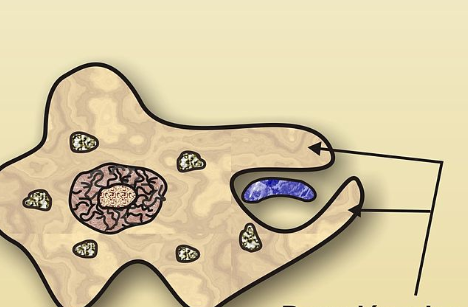
Cytoskeleton and Phagocytosis
Actin filaments rapidly polymerize at the site of the bacteria to push the membrane around it.
This forms pseudopods that wrap the particle. (ARMS)
principle behind dyes
Binds preferentially based on charge, hydrophobicity, or structure
hematoxylin binds:
negatively charged molecules (DNA)
what problem is in situ hybridization trying to solve?
In this tissue, which cells are expressing gene X, and how much?
it’s harder to get proteins so, the soltuion:
instead of detecting protein → detect mRNA directly
how is in situ hybriziation done?
fix cells: this preserves mRNA
section tissue
diffuse your fluorescent probe inside (probe is just a complimentary binding thing)
probe binds to mRNA (regardless that the cell is dead, it can base pair)
use a fluorescent microscope to visualize localization of expression (can quantify with softwares)
qPCR / RNA-seq → quantitative claims
FISH / ISH → spatial, cell-type, and — claims
localization
FISH is excellent for:
Which cells express gene X?
Where in the tissue is it expressed?
Is expression uniform or heterogeneous?
Is transcription active right now? (nuclear spots)
Does expression change in specific cell types?
okay
mRNA ≠ protein (always)
Western blots validate functional output
so basically, if they do something related to mRNA in the study, say that mRNA is just a proxy for protein so they should validate with protein study like —-
PROTEOMICS!
mass spectrometry is basically the RNA-seq equivalent
distinction between when you would use western blotting and immunohistochemistry
Western blot → how much protein (it’s extracted from cell, you get visual band output)
IHC → where protein is IN the cell (you preserve the netire cell, so you can see the relative amount as well as the spatial localization)
First principle: cells are dynamic, not static
Earlier techniques (fixation, staining, FISH, IHC):
Kill the cell
Give a snapshot
Issue?
But cells:
Move proteins
Assemble/disassemble structures
we want a ay to watch the dynamics in real time
solution?
genetically encoded fluorescence with GFP (no dyes, no antibodies needed)
Whenever the gene is transcribed → GFP is made
GFP fluorescence = gene expression (proxy for)
GFP as a reporter of gene expression -What question this answers
“When and where is this gene turned on?”
⚠ Important limitation:
GFP reports —, not native protein behavior
transcription only
FRET answers:
Are these two proteins physically interacting right now?
What FRET is used for
—
Signaling cascades
Protein complex assembly
Receptor–ligand interactions
FRAP measures:
Protein mobility and kinetics/turnover movement
FRAp stands for
Fluorescence Recovery After Photobleaching