Bio 2960-- Eukaryotic Transcription + Transcription Regulation
0.0(0)
Card Sorting
1/23
There's no tags or description
Looks like no tags are added yet.
Last updated 6:15 PM on 4/30/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
24 Terms
1
New cards
ways in which eukaryotic and prokaryotic transcription differ
* In eukaryotes, transcription and translation do not occur at the same time nor the same place. In prokaryotes, transcription and translation occur simultaneously in the cytoplasm. In eukaryotes, transcription occurs in the nucleus while translation occurs in the cytoplasm.
* Eukaryotes have more complex transcription factors and regulatory factors that are called general transcription factors. They also have more varied cis acting factors.
* Eukaryotes have extensive mRNA processing before translation occurs. This occurs in the cytoplasm
* Eukaryotes have larger genomes, so their DNA is packaged, which makes the DNA more difficult for RNA polymerase to reach, and thus requires all of these transcription factors to get transcription going
* Eukaryotes have more complex transcription factors and regulatory factors that are called general transcription factors. They also have more varied cis acting factors.
* Eukaryotes have extensive mRNA processing before translation occurs. This occurs in the cytoplasm
* Eukaryotes have larger genomes, so their DNA is packaged, which makes the DNA more difficult for RNA polymerase to reach, and thus requires all of these transcription factors to get transcription going
2
New cards
chromatin
the eukaryotic genome is vast, but must still fit into the nucleus. So, there is a lot of packaging of eukaryotic DNA.
Chromatin refers to the whole genome, specifically the DNA that is wrapped around the histone proteins. So, the word chromatin includes all the of the DNA and proteins that go into containing that DNA and wrapping it, and that makes up chromosomes.
Chromatin refers to the whole genome, specifically the DNA that is wrapped around the histone proteins. So, the word chromatin includes all the of the DNA and proteins that go into containing that DNA and wrapping it, and that makes up chromosomes.
3
New cards
nucleosomes
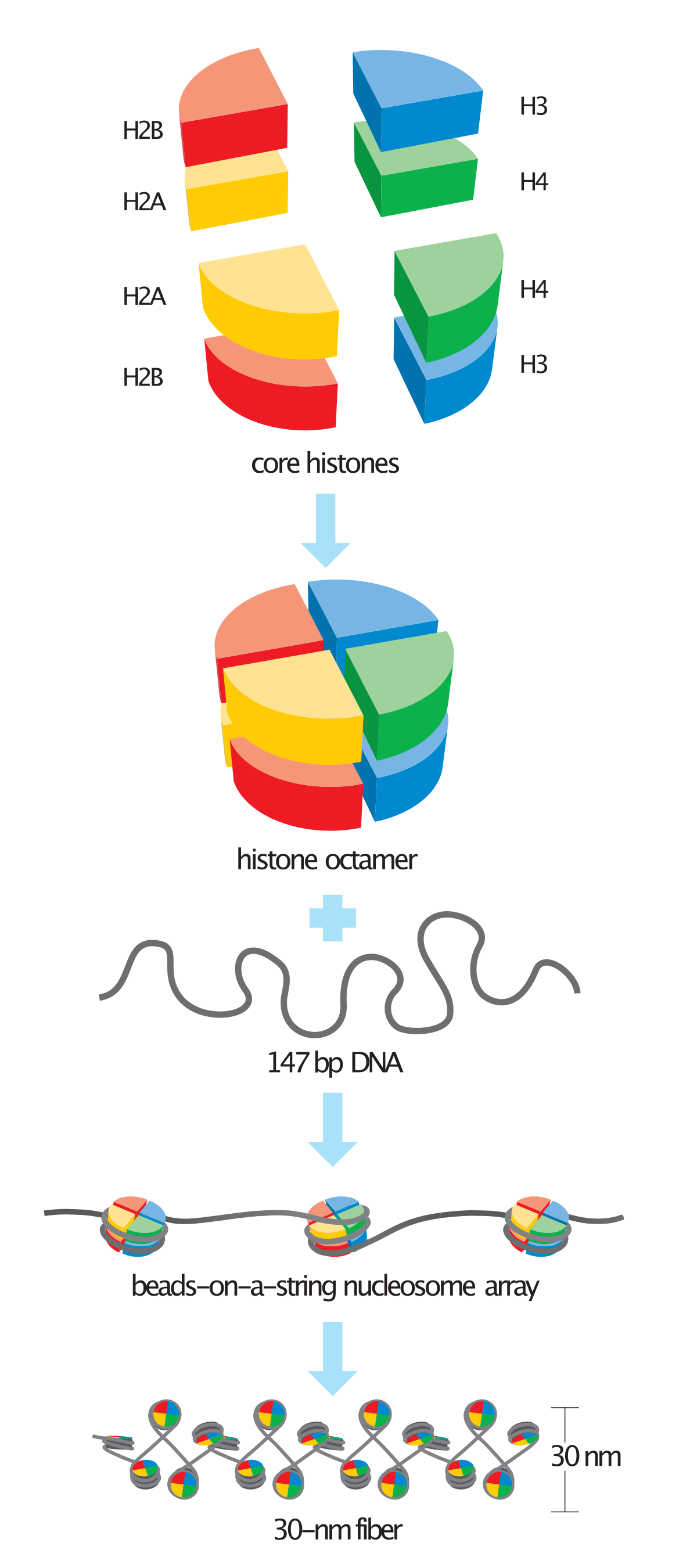
Chromatin is packaged into nucleosomes, which is DNA wrapped around one histone protein.
There are 4 types of histone proteins, and 2 of each, so they form a **histone octomer** that the DNA wraps around. These 8 histones come together to form one core histone protein.
Most of the DNA wraps around the histone, but the little part that doesn’t is called **linker DNA**, and it connects the nucleosomes together.
Histones interact with DNA in the **minor groove**. Interactions are specific to sequences that are A-T rich. They interact with the DNA in the minor groove via these alpha helices.
There are 4 types of histone proteins, and 2 of each, so they form a **histone octomer** that the DNA wraps around. These 8 histones come together to form one core histone protein.
Most of the DNA wraps around the histone, but the little part that doesn’t is called **linker DNA**, and it connects the nucleosomes together.
Histones interact with DNA in the **minor groove**. Interactions are specific to sequences that are A-T rich. They interact with the DNA in the minor groove via these alpha helices.
4
New cards
C terminal end vs N terminal end of nucleosomes
At the **C terminal end** of histones, there are alpha helices that interact with the C terminal ends of other histones. This is how the 8 histone subunits interact with each other in the core of the nucleosome.
At the **N terminal end**, there are tails that are conserved but are subject to modifications like acetylation and methylation. These modifications regulate chromatin structure and availability.
At the **N terminal end**, there are tails that are conserved but are subject to modifications like acetylation and methylation. These modifications regulate chromatin structure and availability.
5
New cards
amino acids that mainly make up histones
**Lysine** and **Arginine** are the amino acids most commonly found in histones. These two amino acids are positively charged, so they give histones an overall positive charge.
They are subject to modifications, **specifically lysine in the N terminal tails of the histones**. Lysine is subject to modifications like acetylation, methylation, and phosphorylation. These modifications regulate chromatin structure and packaging.
They are subject to modifications, **specifically lysine in the N terminal tails of the histones**. Lysine is subject to modifications like acetylation, methylation, and phosphorylation. These modifications regulate chromatin structure and packaging.
6
New cards
3 types of modifiers
1. **Writers**-- the proteins that actually covalently add on the acetyl and methyl groups to amino acids. Examples include histone acetyl transferases and methyl transferases.
2. **Erasers**-- the proteins that remove methyl and acetyl groups from the amino acids. Examples include demethylases and deactylases.
3. **Readers**-- the proteins that recognize acetylation and methylation on amino acids. Examples include methyl readers and acetyl readers.
7
New cards
bacterial vs eukaryotic transcription regulation
in prokaryotes, **the ground state of transcription is on**. This is because the promoter is always available and ready for RNA Polymerase to bind, without any outside help.
In eukaryotes however, **the ground state is off**. This is because eukaryotic DNA is packaged so much that the promoter is not just out and RNA polymerase cannot easily access this promoter region. There must be some outside help that unwinds the DNA, but without it, transcription cannot occur. In eukaryotes, promoters are protein-DNA complexes that are hidden in the chromatin structure.
In eukaryotes however, **the ground state is off**. This is because eukaryotic DNA is packaged so much that the promoter is not just out and RNA polymerase cannot easily access this promoter region. There must be some outside help that unwinds the DNA, but without it, transcription cannot occur. In eukaryotes, promoters are protein-DNA complexes that are hidden in the chromatin structure.
8
New cards
structure of eukaryotic genes
transcribed regions, translated vs untranslated regions, introns and exons, and promoter, but the promoter is broken up into two different regions
9
New cards
structure of promoter
Recall the promoter is the region of DNA at which RNA Polymerase II binds. In general, in eukaryotes, this is the TATA region.
1. **Core promoter**-- is a sequence of TATA that is about 25 base pairs upstream of the transcription start site. These are the minimum sequences required to recruit RNA polymerase and begin transcription. In test tubes, this is the only part of the promoter that is present, and the only thing required for transcription to begin.
2. **Nearby elements**-- these are parts of the promoter that are farther away, either upstream or downstream of the core promoter. These nearby elements are required for transcription inside the cells. This includes enhancers and other regulatory elements.
1. **Core promoter**-- is a sequence of TATA that is about 25 base pairs upstream of the transcription start site. These are the minimum sequences required to recruit RNA polymerase and begin transcription. In test tubes, this is the only part of the promoter that is present, and the only thing required for transcription to begin.
2. **Nearby elements**-- these are parts of the promoter that are farther away, either upstream or downstream of the core promoter. These nearby elements are required for transcription inside the cells. This includes enhancers and other regulatory elements.
10
New cards
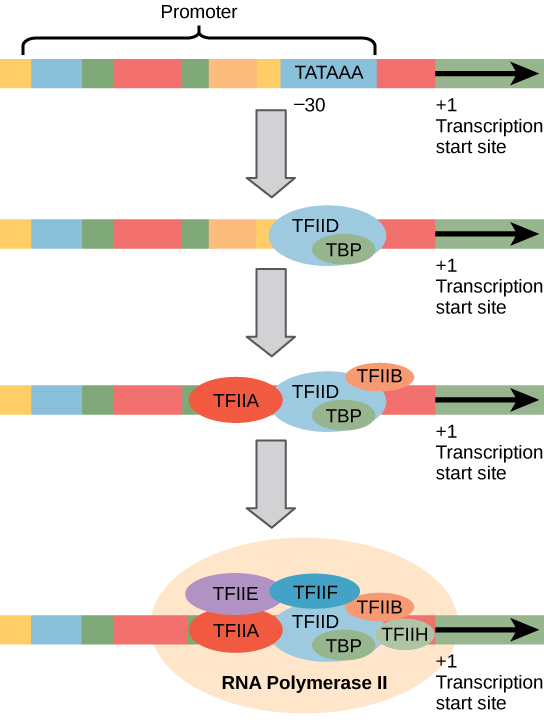
Initiation of transcription in test tubes
Recall that only the core promoter is present in test tubes. Nonetheless, a complex of general transcription factors and RPII must bind to the promoter region for transcription to begin.
1. The **TATA binding protein** (which is apart of TFIID) recognizes the -25 region that contains the TATA sequence and binds to it.
2. **Transcription Factor IIB** binds to the TATA binding protein already there.
3. **RPII** binds to the developing protein complex (TFIIB). **It does not actually bind to the DNA like in prokaryotes!!**
4. **Transcription Factor IIH** binds to the complex. TFIIH acts as a helicase and unwinds the DNA, and acts as a kinase and phosphorylates the C terminal domain of RNA polymerase. This allows RPII to escape the promoter tether that is preventing RPII from beginning transcription before all the GTF are present.
1. The **TATA binding protein** (which is apart of TFIID) recognizes the -25 region that contains the TATA sequence and binds to it.
2. **Transcription Factor IIB** binds to the TATA binding protein already there.
3. **RPII** binds to the developing protein complex (TFIIB). **It does not actually bind to the DNA like in prokaryotes!!**
4. **Transcription Factor IIH** binds to the complex. TFIIH acts as a helicase and unwinds the DNA, and acts as a kinase and phosphorylates the C terminal domain of RNA polymerase. This allows RPII to escape the promoter tether that is preventing RPII from beginning transcription before all the GTF are present.
11
New cards
phosphorylation of C-terminal domain of RPII
Recall that TFIIH phosphorylates the C terminal domain of RPII with its kinase activity. Before phosphorylation
12
New cards
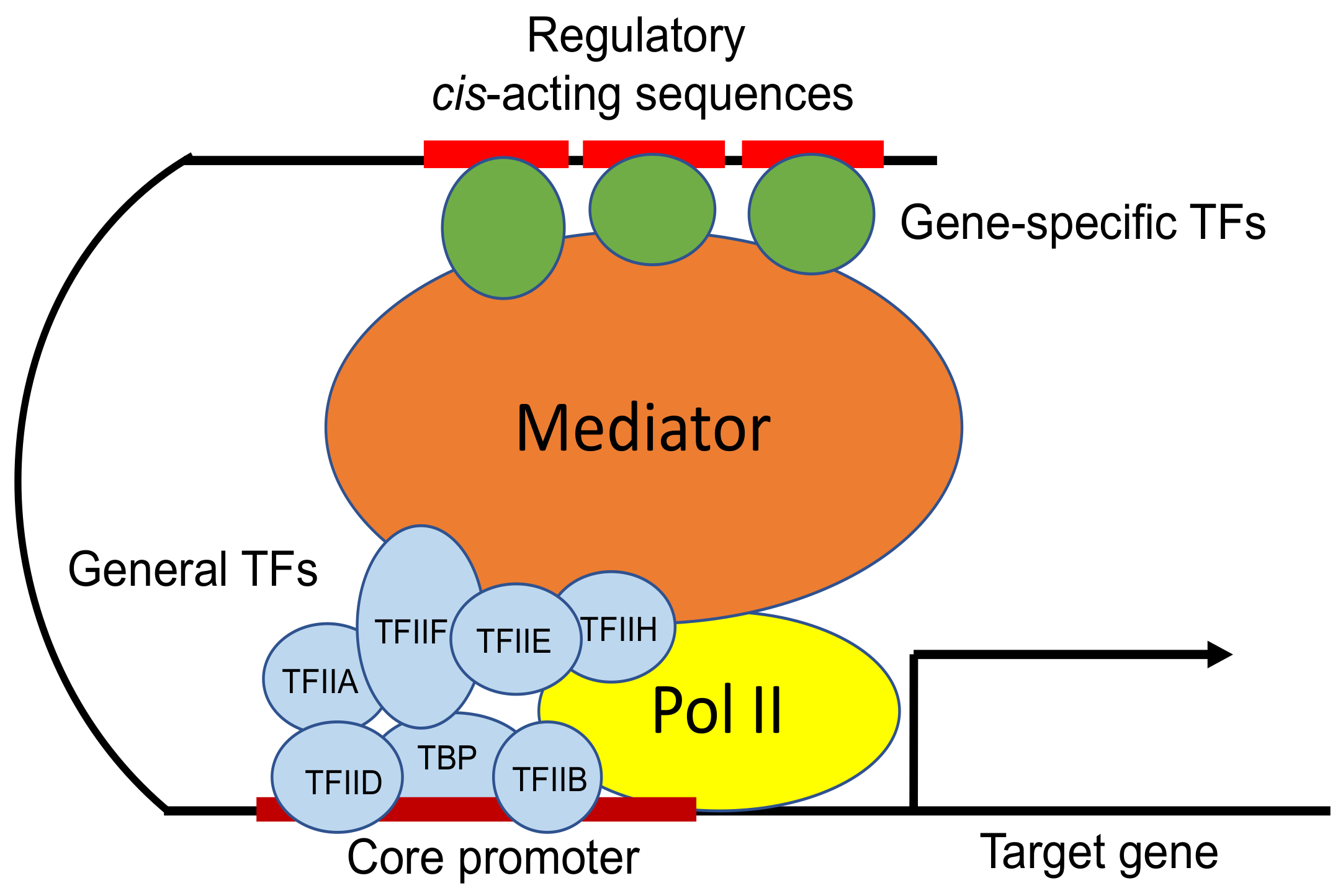
Initiation of transcription in cells
Before all of the transcription factors can bind, histone modifications must happen first since the promoter region is not easily accessible to RPII.
1. **Histone acetyltransferases** (HAT) add acetyl groups to the lysines on the N terminal tails of the histone proteins. This loosens the chromatin structure.
2. **Chromatin remodeling complexes** rearrange histones/nucleosomes to allow the promoter region to be outside of histone wrapping and available for proteins to bind to it. These remodeling complexes use energy from ATP hydrolysis to restructure the nucleosomes.
3. Now that the promoter region is exposed, general transcription factors and RPII bind in the order described in the steps of transcription in test tubes.
4. **Mediators** are proteins required for transcription in cells. They interact with and bind to RPII, other general transcription factors, and activator proteins bound to enhancers.
1. **Histone acetyltransferases** (HAT) add acetyl groups to the lysines on the N terminal tails of the histone proteins. This loosens the chromatin structure.
2. **Chromatin remodeling complexes** rearrange histones/nucleosomes to allow the promoter region to be outside of histone wrapping and available for proteins to bind to it. These remodeling complexes use energy from ATP hydrolysis to restructure the nucleosomes.
3. Now that the promoter region is exposed, general transcription factors and RPII bind in the order described in the steps of transcription in test tubes.
4. **Mediators** are proteins required for transcription in cells. They interact with and bind to RPII, other general transcription factors, and activator proteins bound to enhancers.
13
New cards
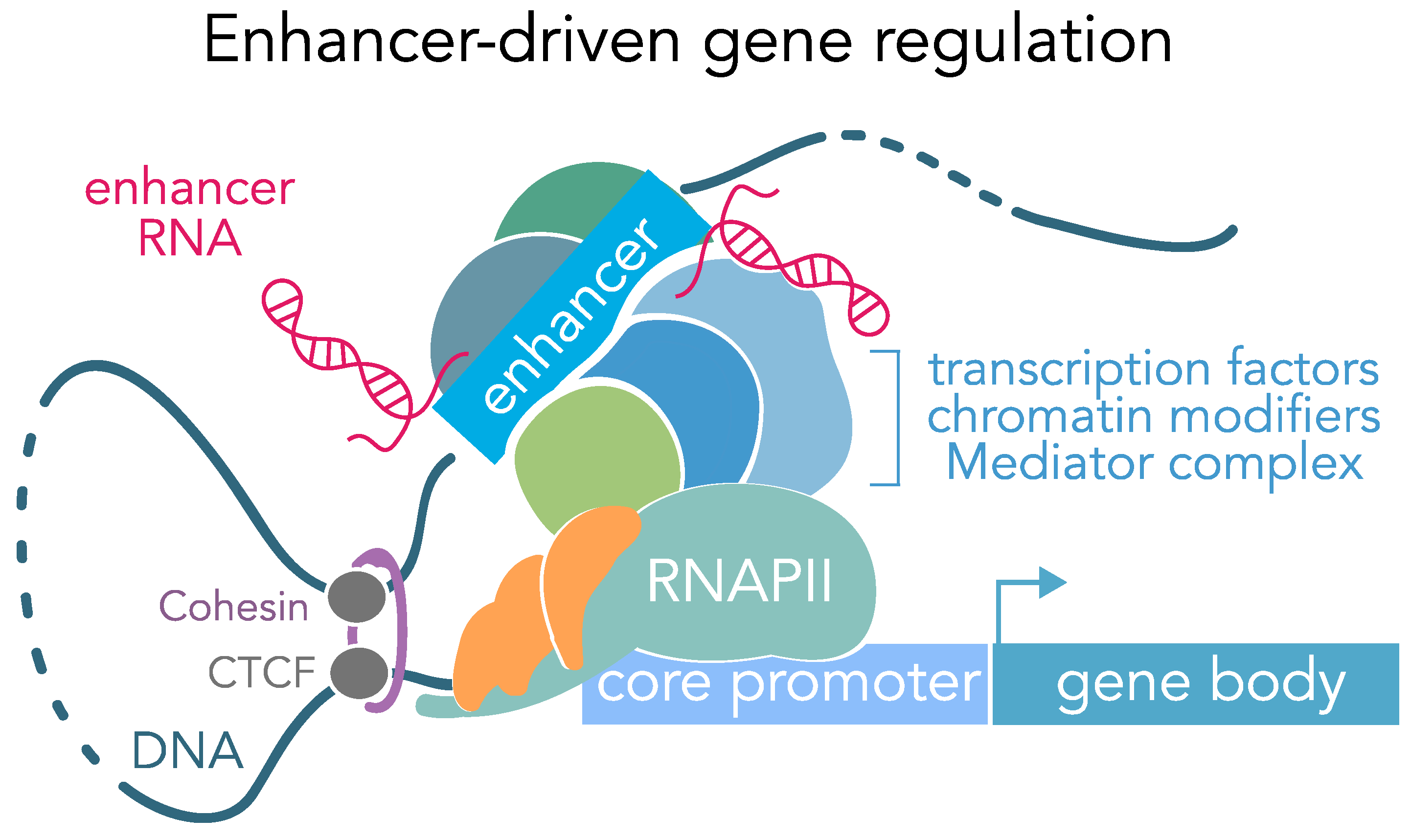
enhancer
cis-acting DNA sequences located far from the core promoter but are still apart of the entire promoter region. They work from far away to increase transcription and activators bind to them. Although they are far away, the DNA loops around so that they may be in contact with the core promoter region. Although they may be far from the main promoter, they are specific to genes and only activate specific genes.
Activator proteins bind to enhancer sequences.
Activator proteins bind to enhancer sequences.
14
New cards
activator proteins
bind to enhancer regions and help attract RPII to the promoter region to begin transcription. But keep in mind, they are **not required** for transcription to begin.
Are made up of 2 separable domains-- the DNA-binding domain and activation domain.
The **DNA-binding domain** has specificity and binds to specific regions of DNA (AKA enhancers). So, activators only differ by the enhancers that they bind.
The **activation domain** is the same on all activator proteins and is where co-activator proteins bind.
Co-activators are proteins like general transcription factors, histone code writers (HATS), and chromatin remodeling complexes.
Although the enhancers that these activator proteins bind can be thousands of nucleotides upstream or downstream of the core promoter region, they still end up coming into contact with the protein complex bound to the TATA box. This is because the **DNA loops around**, which brings the enhancer region and associated proteins right next to the TATA box and co-activators bound to the TATA box.
Are made up of 2 separable domains-- the DNA-binding domain and activation domain.
The **DNA-binding domain** has specificity and binds to specific regions of DNA (AKA enhancers). So, activators only differ by the enhancers that they bind.
The **activation domain** is the same on all activator proteins and is where co-activator proteins bind.
Co-activators are proteins like general transcription factors, histone code writers (HATS), and chromatin remodeling complexes.
Although the enhancers that these activator proteins bind can be thousands of nucleotides upstream or downstream of the core promoter region, they still end up coming into contact with the protein complex bound to the TATA box. This is because the **DNA loops around**, which brings the enhancer region and associated proteins right next to the TATA box and co-activators bound to the TATA box.
15
New cards
how activator proteins work
activator proteins restructure chromatin in a way that promotes transcription, and recruit general transcription factors. These general transcription factors then recruit RPII, and allow for transcription to begin.
16
New cards
transcription regulation in yeast
Yeast use galactose as a carbon source, so when galactose is present, in needs to be broken down. However, if galactose is not around, the cells do not want to waste their energy and transcribe the genes for the enzyme anyways.
The genes that code for these enzymes are regulated by an activator called **Gal4**, which is **bound to an enhancer upstream** of the core promoter. **When galactose is not present**, a protein called **Gal80** binds to the activation domain of Gal4. This prevents any general transcription factors from binding to Gal4 and transcribing the genes to break galactose. When galactose is present, it binds to Gal80, and this prevents Gal80 from entering the nucleus (where Gal4 is) and thus from binding to Gal4. This allows transcription factors to bind to Gal4 and begin transcription for the genes that encode the enzymes that break down galactose.
The genes that code for these enzymes are regulated by an activator called **Gal4**, which is **bound to an enhancer upstream** of the core promoter. **When galactose is not present**, a protein called **Gal80** binds to the activation domain of Gal4. This prevents any general transcription factors from binding to Gal4 and transcribing the genes to break galactose. When galactose is present, it binds to Gal80, and this prevents Gal80 from entering the nucleus (where Gal4 is) and thus from binding to Gal4. This allows transcription factors to bind to Gal4 and begin transcription for the genes that encode the enzymes that break down galactose.
17
New cards
acetylation and methylation
**acetylation**-- covalently adding an acetyl group to the N terminal tails of histones. This leads to **increased** transcriptional activity as it “loosens” the chromatin structure
**methylation**-- covalently adding a methyl group to the N terminal tails of histones. Leads to **decreased** transcriptional activity by recruiting proteins involved in gene repression or preventing the binding of transcription factors.
**methylation**-- covalently adding a methyl group to the N terminal tails of histones. Leads to **decreased** transcriptional activity by recruiting proteins involved in gene repression or preventing the binding of transcription factors.
18
New cards
regulation during elongation
after RPII’s tail gets phosphorylated, its promoter tether is removed, and transcription can begin. However, many proteins bind to this phosphorylated tail and move along with RPII as it transcribes. Specifically, histone acetyltransferase (HAT) and histone methyltransferase (HMT) travel with RPII.
**HAT** adds acetyl groups to activate the chromatin so that RPII can transcribe the DNA.
**HMT** methylates the histones, which recruits histone deactylase to remove acetyl groups once RPII is done transcribing to deactivate the chromatin to its normal state.
**HAT** adds acetyl groups to activate the chromatin so that RPII can transcribe the DNA.
**HMT** methylates the histones, which recruits histone deactylase to remove acetyl groups once RPII is done transcribing to deactivate the chromatin to its normal state.
19
New cards
transcription repression-- Yeast Example
Yeast prefer to use glucose as a carbon source over galactose. So, if glucose is present, they would use all of that first. When glucose is around, it induces the binding of the **Mig1** repressor. Mig1 binds to the Mig1 site, and then a **histone deactylate co-repressor** binds to Mig1, and this keeps chromatin deactylated. Acetyl groups are either removed or prevented from being added. This prevents the encoding of genes that encode for enzymes that break down galactose.
20
New cards
transcription repression-- Growth Factor Example
Growth factor is signal for cell division. However, if no growth factor is present, the cell should not divide. Thus, the presence of growth factor regulates cell division. E2F is a transcription factor that controls the genes responsible for cell division.
**In the absence of growth factor**, E2F does not phosphorylate RB, so the genes that are behind cell division are not turned on. When RB is not phosphorylated, it associates with a histone deacetylase, which further turns off the genes responsible for cell division.
**In the presence of growth factor**, a kinase phosphorylates RB, making RB incapable of interacting with E2F. Instead, RB interacts with a histone acetylase, which turns on the genes required for cell division, and thus allows cell division to proceed.
**In the absence of growth factor**, E2F does not phosphorylate RB, so the genes that are behind cell division are not turned on. When RB is not phosphorylated, it associates with a histone deacetylase, which further turns off the genes responsible for cell division.
**In the presence of growth factor**, a kinase phosphorylates RB, making RB incapable of interacting with E2F. Instead, RB interacts with a histone acetylase, which turns on the genes required for cell division, and thus allows cell division to proceed.
21
New cards
transcription repression-- permanent repression
there are some genes that are just never expressed. These long ranges of repressed chromatin are called **heterochromatin**. These genes are never transcribed, and this is caused by non-histone chromosomal proteins that interact with the DNA and silence it.
22
New cards
X inactivation
when the original cells of a eukaryote are made, one of the X chromosomes is silenced, and its associated genes are silenced. Then, as the cells divide and create more cells, the new cells will have the same genes on and off as the cell that they originated from. Once cells differentiate and are localized to specific regions of the body, there will be genes that are only expressed in certain regions of the body because that is where those cells are. Thus occurs in females only since we have two X chromosomes. This occurs because each X chromosomes codes for something different.
Most commonly observed in cats and fur colors.
Most commonly observed in cats and fur colors.
23
New cards
epigenetics
the study of how lifestyle can impact gene expression
In epigenetic effects, DNA base sequences are not altered like they are in mutations. Rather, the base sequences remain the same, and transcription does not occur.
Conversely, in mutations, DNA base sequences are changed and transcription still occurs.
In epigenetic effects, DNA base sequences are not altered like they are in mutations. Rather, the base sequences remain the same, and transcription does not occur.
Conversely, in mutations, DNA base sequences are changed and transcription still occurs.
24
New cards
methods of repressing chromatin in epigentics
1. **Histone tail methylation**. Histone methyltransferase (HMT)can add methyl groups to the Lysines on the histone tails. This silences the gene by preventing it from opening up and allowing RPII to bind.
2. **Methylation of 5’ end of cytosine if this cytosine is followed by guanine**. If a C is followed by a G, DNA methyltransferase (DMT) comes and adds a methyl group to that cytosine base itself. Specific proteins then detect this methylation, and recruit histone deactylases to the area to remove any histone groups to further repress expression of the gene.