BCH210 (2nd Half)
1/127
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
128 Terms
cause of membrane formation
a decrease in ionic interactions with water
FRAP
Fluorescence Recovery After Photobleaching
A way to see how fast lipids and proteins diffuse laterally in a membrane. Damage fluoro phores with bleach and see how long it takes for regular fluorophores to naturally diffuse and replace white/ bleached ones.
lipid raft
Cytoskeleton part of the membrane that lipid /protein interacts with
flip-flop diffusion
Heads moving to the other side of a bilayer
Flippase
FIIppase (I for Inside) where head facing outside of cell is flipped inside
Floppase
FlOppase ( O for Outside) where head facing inside of the cell is flipped outside
Scramblase
Head moves down concentration gradient in passive transport
Flippase conformational change
begins in ATP bound closed form open slightly to the outside of the cell, then to "flip" it turns into open form with a wider opening to the inside of the cell
Lipids can be either
hydrophobic or amphipathic
Lipids are diverse
liposome
head on inside and outside forming a water filled circle
Mirelle- Inside out
heads on the inside, tails sticking out (hydrophobic env) circular
Micelle - Normal
heads on outside tails on inside (polar solvent)
Triacylglyceride function
Store carbon for energy
fatty acid charge
amphipathic
signal transduction
cascade, amplification event throughout cell
hormone, primary messenger
reception of message, usually integral protein
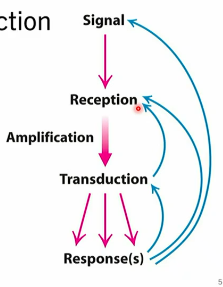
GPCRs
g proteins coupled receptors
7 TM segments
common signal receiving proteins
conformational changes release G proteins
binds many ligands
GPCR ligands
natural
serotonin, epinephrine, prostaglandins, dopamine, psilocin/psilocybin
synthetic
morphine, histamine, LSD
ncv int.s between side chains
beta adrenergic receptor
has inactive and active states w diff structures
ligand binding changes tm5
major conf change in tm6 release GalphaGTP →activates adenylyl cyclase→secondary messenger cAMP→cAMP activates other enzymes (eg.PKA)
Ras proteins
type of g alpha proteins
structural switches (change when gtp→gdp)
signals for cell proliferation and apoptosis→issue can lead to cancer
PTMs
can turn off/on an enzyme
enzyme linked receptors
integral membrane proteins
hormone attached
conf change enzymes now work inside of cell
phospholipid mediated signalling
phospholipases hydrolyse phospholipids to produce other 2nd messengers like diacykgkycerol (dag) or ip3 → release of Ca
competing hormones example
insulin and epinephrine
act on similar metabolic pathways- one turns on and the other turns off
phosphorylation of insulin receptor substrate 1→phosphorylation of beta adrenergic receptor by PKB
degraded no epi signalling insulin is stronger and will win
GLUT transporter
facilitated diffusion
glucose binding → conf change
transport conc. dependent and saturable (at high conc.)
beta barrel proteins
facilitated diffusion
integral membrane proteins single polypeptide chains forming barrel shape
hphilic inside, hphobic outside

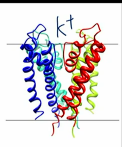
potassium ion channel
tetramer
v structure
selectivity filter (TVGYG) cocntributing to K+ binding
Which glucose transporter is responsive to insulin?
GLUT 4
What is one of the molecules that inhibits glycolysis?
Alanine
Lecture 16: Carbohhydrate Structure
Carbohydrate Numbering
carbon 1 is carbonyl (CHO) carbon, proceeds from there
carbohydrate configuration
multiple carbon chiral centres, multiple isomers
L or D assigned to chiral centre furthest from carbonyl
# of structures = 2^n
n= number of chiral centres
carbohydrate cyclization
hemiacetal aldehyde derivative
hemiketal ketone derivative
(aldehydes and ketons are very reactive, undergo nucleophilic attack)
alpha or beta based on whether hydroxl is up or down(?)
anomeric carbons are…
chiral
anomer = isomers hat differ at a new aymmetric carbon atom formed at a ring closure
haworth projections
let you see cyclic sugars in 3d
carbohydrate modification
sugar can be phosphorylated, methylated, or N-group added
hydrozyls or carbonyls may be removed
isomer
same formula differnt structure/organization
constitutional isomer
different order of functional group bonding
stereoisomers
same formula and order but can be enantionmer, diastereomer, (epimer, anomer)
enantiomer
non-superimposable mirror images
diastereomer
not mirror images.
epimer
anomer
epimer
differ at one asymmetric carbon
anomer
differ at a newly formed, asymmetric C in the ring
structure
why are the different positions impoertant
very specific binding in enzyme active site, may weaken interactions and fail to bind ligand
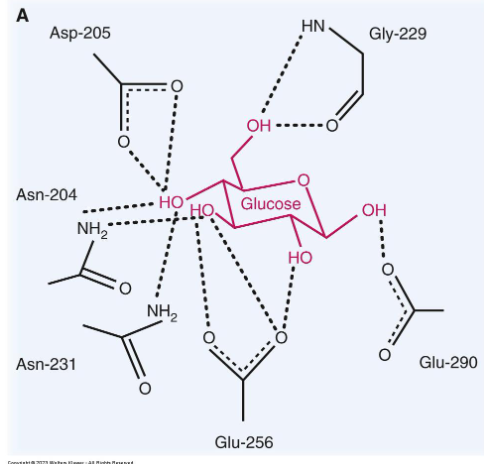
reducing sugars
sugar molcule with a (very reactive) free aldehyde or ketone group act as a reducing agent by giving e- to another molecule
to identify: 1) identify anomeric carbon (to right of ring O) 2) does it have a free OH group?
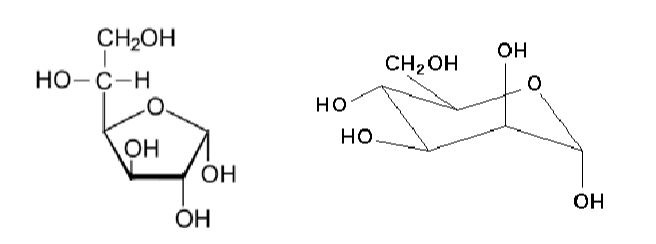
glycation
non ezymatic reaction covalent attachment of a sugar to a protein, lipid or nucleic acid molecule.
(basically glycosylation w/o an enzyme)
hemoglobin and glycation
> 6%
diagnostic tool to see if glycation “frosting” indicates diabetes
simple monosaccharides
simplest carbohydrate (CH20)n
aldoses or ketoses
D form sugars are biologically relevant
cyclization leads to 2 additional structures
reducing sugar can reform reactive linear structure
glycosidic bonds
forms as condensation / dehydration synthesis reaction
cleaving these bonds is hydrolysis reaction
monosaccharides are joined by glycosidic linkages to form disaccharides and etc
nomenclature (1→2) carbon 1 linked to carbon 2
can also be alpha/beta
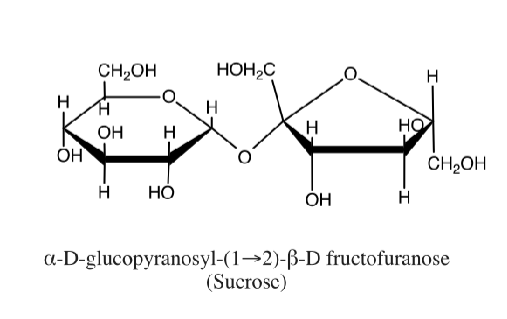
a or b glycodisic bond
attack from bottom→alpha conformation (point down)
attack from top → beta conformation (point up)
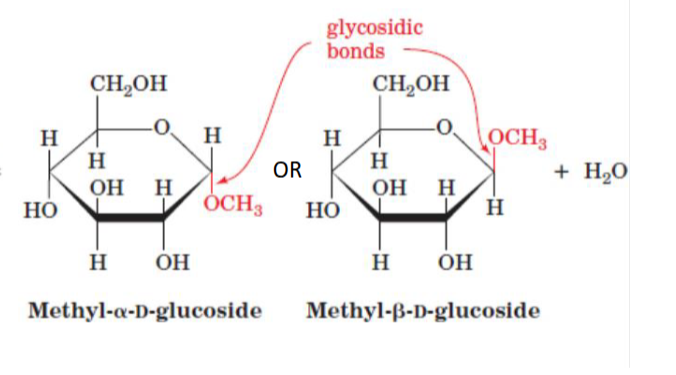
N or O linked glycosidic bond
intermolecular glycosisidic bonds formed b/w amine or hydroxyl and a reactive anomeric carbon
eg DNA, RNA bases, glycoproteins may be O-linked (ser/thr) or N-linked (asn)
complex carbs
mono, di, oligo, poly sacchs
oligosacch
3-20 sugars
poly sacchs
up to 1000s of sugars
linear or branched links
energy storage, cell strcutrue, recognition
aka glycans
homopolymer
sme monosacchs
heteropolymer
diff monosacchs
eg lactose glucose + galactose (B- 1,4)
starch
amylose and amylopectin
amylose
unbranched glucose units
α(1→4)-linkages
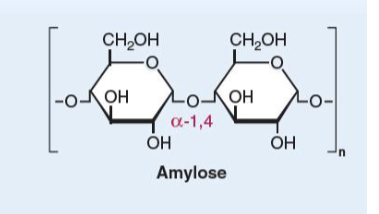
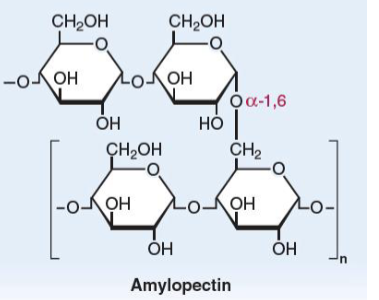
Amylopectin
linear glucose chains joined by α(1→4)-linkages.
α(1→6)-linkages at branch points once every 30
glucose units
α-amylase
enzyme secreted by the salivary glands and pancreas to degrade starch
Cleaves at random locations along the chains to give maltose and maltotriose
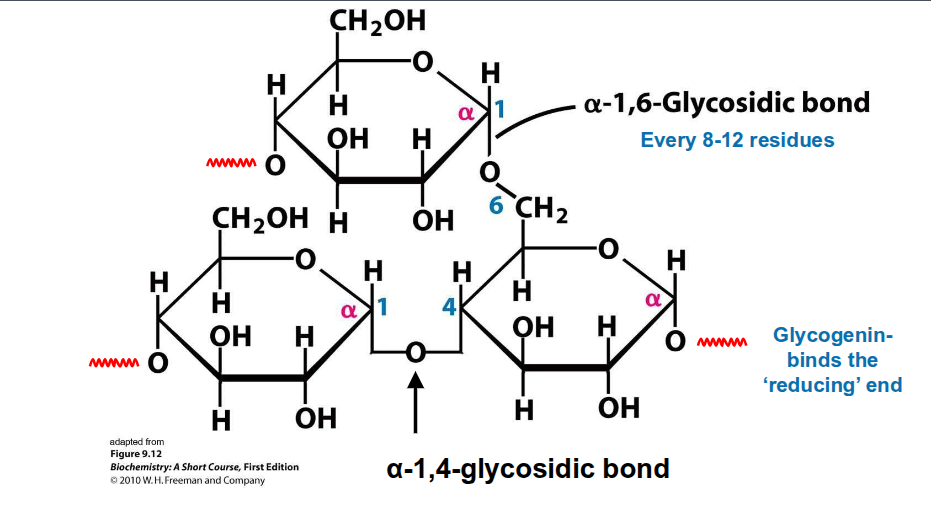
glycogen
A storage form of long, branched
chains of glucoselinear glucose chains joined by α(1→4)-
linkages.α(1→6)-linkages at branch points once
every 8-12 glucose units.
Contains a dimer of glycogenin at the
centre.Glucose units are added and removed
from the non-reducing endsFound in the liver and muscle
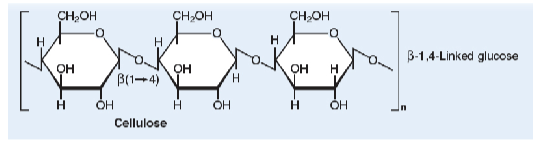
cellulose
most abundant organic
Unbranched chains of glucose units are
joined by β(1→4) linkages with many
hydrogen bonds
cellobiose
disaccharide of glucose linked by β(1→4).
cleavage to monosacchs
enzymes like lactase, maltase, sucrase
cells can only transport and use monosacchs for fuel
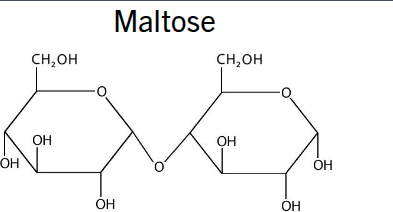
maltose
2 glucose alpha 1→2
lactose
glucose + galactose (B 1→4)
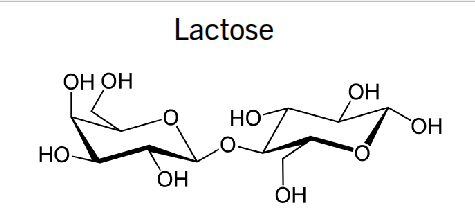
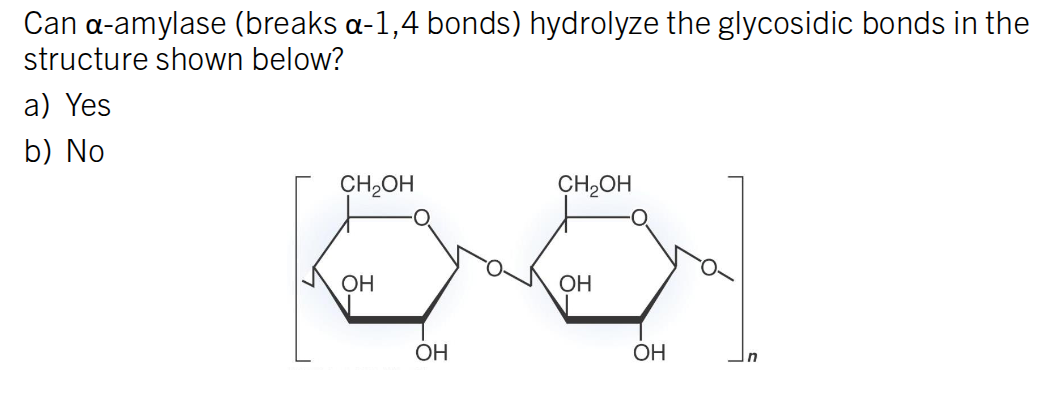
No. This is a B 1→4 bond. This is cellulose.
(Hydroxyl at C1 is pointing up)
challenge of sugar sequencing
many isomers
mass spec/visualization is hard to distinguish b/w sugars
multiple possible sugars
DNA 4 possible nt, Protein 20 possible monomers, carb 100s of possible at each position
glucose transport
glucose is polar and requires facilitated transport either down or against conc. gradient
types of glucose transporters
sodium glucose co transporters
2ndary active transport
allow sodium to go along gradient; glucose against gradient
glucose transporters
facilitated diffusion
fructose transpoorter
facilitated diffusion
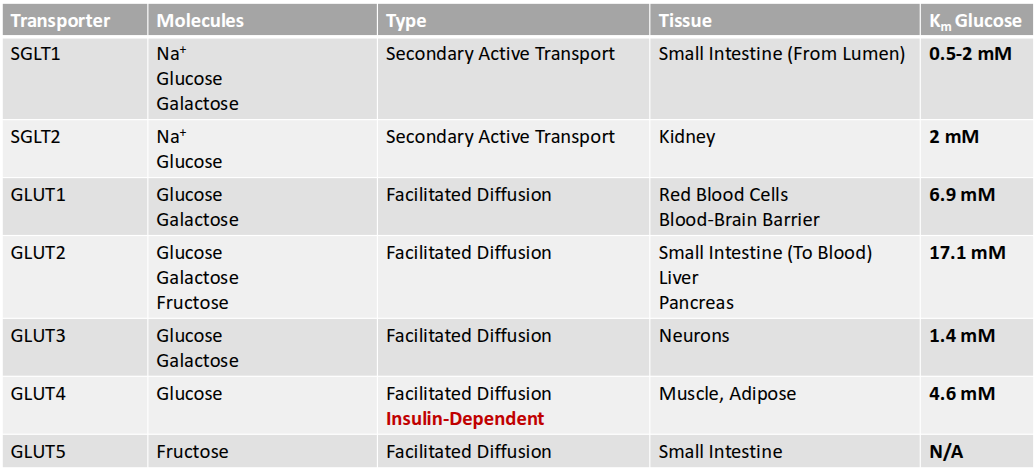
GLUT4
is expressed in adipose and muscle tissues.
insulin dependent
In response to insulin, this transporter is translocated to the surface of the cell, allowing glucose to enter. ‘opening a door”
GLUT1, 2, and 3
are expressed on cells that always need glucose
glut 1 & 3 seen brain; glucose moves through glut 1 to exit neuron and glut 3 to enter another
GLUT5
is a transporter that is specific for fructose.
FASTING state blood glucose
glucose moves most effectively in neurons and blood cells
liver, pancreas not releasing insulin, less flow
Km low → fast, free flowing
no insulin for receptor in muscles→ little glucose in muscles
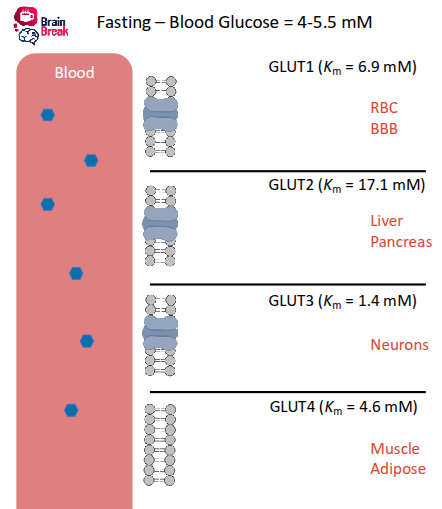
FED state blood glucose
move fast in blood
pancreas release insulin into bloodstream
glut 3 in neurons get lots of glucose in brain
insulin receptors GLUT 4 muslce cells use glucose and store as fats
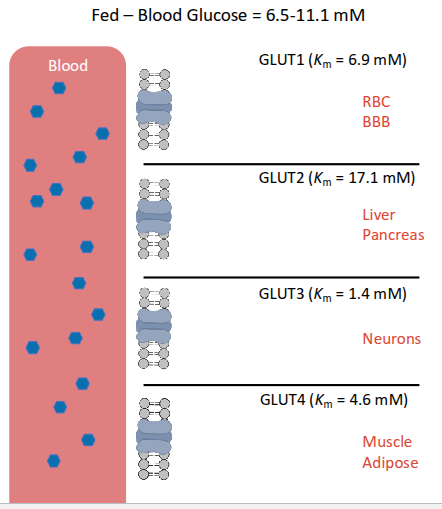
hypoglycemia
insulin sensitivity
low blood glucose level
decreased neural activity not enough glucose in the brain , fainting not enough neurons firing
lecture 17: carb metabolism
glycogen synhesis, glycogenolysis, glycolysis, anaerobic metabolism
anabolic state
insulin signaling increases glucose utilization
Increases glucose transport into cells
Increases the expression and activity of enzymes that use glucose as a
substrate
catabolic state
low amnt of glucose, energy
breaking down energy, eg stored as glycogen or fats
protein synthesis/netbreakdown might increase
hormones epinephrine and glucagon activate this state
D-glucose strcutre"*need to know
pyruvate strcutre"*need to know
glycilysis total substrates and products of the pathway
branching and why glycolysis pathway
glycolysis regulatory enzymes and how they are regulated
glycolysis
10 step catacbolic pathway found in cytoplasm using glucose and other simple monosacchs
glycolysis starts with…
glucose (6-carbons)
stage 1 glycolysis -
preparing
energy inputted
2 phosphorylations
Aldose
2^n stereoisomers
N=
Lecture 15: Intro to Metabolism
catabolism and anabolism are…
interrelated !
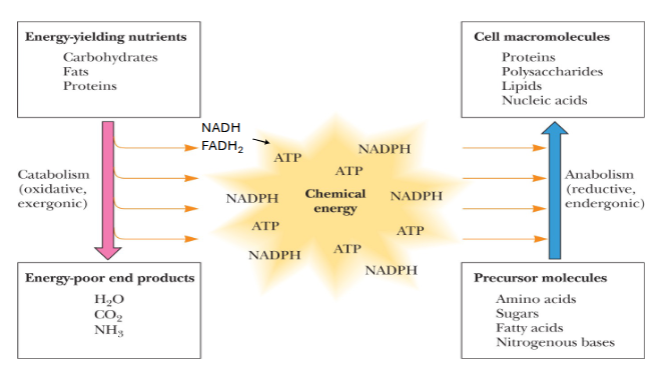
catabolism
energy yielding nutrients → energy poor products
milk the energy out of those damn nutrients
oxidative
exergonic (releases energy)
anabolism
precursor molecules → cell macromolecules
reductive
endergonic (uses energy)
catabolism is oxidative or reductive?
oxidative.
chemical energy in metabolism
includes ATP, NADPH, NADH(?), FADH2(?)
metabolism is highly …
complex !
metabolic “map” can be broken down into linear, cyclic, or branched athways
catabolism of compounds (does?)
involved in metabolism
releases free energy which can be stored in ATP or other high energy intermediates
anabolism of compounds (does?)
synthesize larger macromolecules using simple building blocks and energy
what influences metabolic flux in pathways?
Gibbs free energy changes and enzymes
influence the conversion of metabolites through the pathways
determining flux through a metabolic pathway (4)
The presence of enzymes
Metabolite concentration
ATP availability
Changes in Gibbs free energy
*in order
Gibbs Free Energy Eqn
ΔG = ΔG°′+ RT ln([P]/[S])
gibbs free energy change = standard free energy change +(8.314 J/mol*K)(temperature)(ln conc. at equillibrium)
![<p><span>ΔG = Δ</span><span style="color: rgb(255, 255, 255);"><span>G°′</span></span><span>+ RT ln([P]/[S])</span></p><p><span style="color: rgb(255, 248, 248);"><span>gibbs free energy change = standard free energy change +(8.314 J/mol*K)(temperature)(ln conc. at equillibrium)</span></span></p>](https://knowt-user-attachments.s3.amazonaws.com/2d396ece-bc98-43b9-8179-08b4da606c27.png)