Lecture 7- DNA replication in eukaryotes
1/35
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
36 Terms
{INITIATION}
Why does initiation only happen once per cycle?
So the right number of chromosomes is produced.
What are the 4 stages of the cell cycle?
G1, S, G2, M (mitosis begins),
Where do origins of replication arise from?
Sections of DNA called Autonomously Replicating Sequences (ARS).
In G1 phase, what binds to the replication origin region?
Origin Recognition Complex of proteins (ORC).
In G1 phase, what accumulate in the nucleus and what binds to ORC?
Accessory proteins (licensing factors) accumulate in the nucleus.
Cdc6 and Cdt1 bind to ORC
In G1 phase, what are 2 helicases loaded onto by Cdt1? What leaves? What does this produce?
They are loaded onto dsDNA.
Cdc6/Cdt1 leave.
Produces ‘licensed’ pre-replication complex.
During S phase, how is pre-replication complex activated?
Additional proteins are added turning this to an active initiation complex/ replisome progression complex.
Does G1 or S phase have helicase loading?
G1 phase.
{ELONGATION}
What are the 5 main eukaryotic DNA polymerases?
α, β, γ, δ, ε
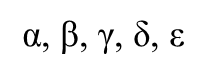
What is the function of polymerase a?
Has its own primase activity and polymerase so can make its own primers (it’s attached to a primase subunit)
No proofreading 3’-5’ exonuclease.
Not processive; doesn’t associate with eukaryotic sliding clmap protein PCNA (proliferating cell nuclear antigen).
What loads PCNA onto primer/dna ready for a different Pol to bind?
Replication factor C (RPC)
Role of polymerase β
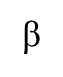
Involved in repair
Role of polymerase γ
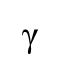
Replicates mitochondrial/mtDNA.
Role of polymerase sigma?
For lagging strand. Associates with PCDNA (clamp protein) and has proofreading.
Role of polymerase ε
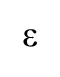
For leading strand. Associates with PCNA and has proofreading.
What can pol sigma do about the presence of RNA primers?
Can displace them but can’t release them as rNMPs.
When pol sigma displaces RNA primers, what is produced? How is this dealt with?
An RNA ‘flap’.
Flap endonucleoase-1 (FEN1) recognises 5’ flaps and cleaves off a short section, leaving a nick. DNA ligase removes the nick.
How are NB primers digested?
By RNAse H2, with 1 RNA nt left. Then FEN1 removes last RNA nt.
What happens if an origin isn’t activated (silent origins)?
Replication forks from 1 origin will pass through another origin (pasive replication).
At the end of replication for linear chromosomes, what does the lagging strand experience?
Last RNA primer may not be at the extreme 3’ end of the DNA→missed section.
Even if it’s at the end, it will be removed because it’s RNA → ssDNA section.
How many nucleotide are lost per replication?
10 nucleotide. At some point you will start deleting genes and the chromosome will die.
What’s the solution to the problem of chromosome shrinkage?
Telomeres.
Why do telomeres end this problem?
At the end of DNA, the sequence TTAGGG (meaningless DNA) creates a cap at the end so doesn’t matter if it’s eaten away because it doesn’t encode for anything.
What is the Hayflick limit?
Once the telomeric DNA is gone, gene loss, cells stop dividing, cell death.
How do embryonal, cancer, stem cells and other types divide beyond Haflick limit?
New telomeric DNA is synthesised by a ribonucleoprotein called telomerase, containing RNA strand with sequence CUAACCUAAC, acting as a template for synthesis of new telomeric repeats, growing the telomere.
4 steps in process of telomerase synthesising.
telomerase binds and extends ssDNA.
pol a (primase) binds and adds primer
pol sigma binds primer and extends it
dna ligase fills in nick.
Features of replication of mitcohondrial DNA?
2-10 copies of circular mtDNA per mitochondrion.
Unidirectional replication (1 fork).
In mtDNA what synthesises the leading strand?
Pol Y
What is the lagging strand in mtDNA composed of?
RNA Okazaki fragments.
Describe 3 stages of sigma replication (‘rolling circle’) in bacteriophages.
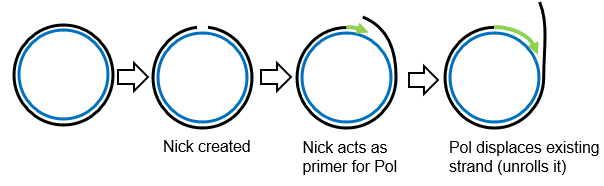
What is the general process of RNA genome replication in animal/plant viruses?
RNA-dependent RNA polymerase (RNA replicase) is used.
The plus strand RNA is copied directly to make the minus strand, which is used as a template for making more plus strands.
No primer or proof reading required.
How are retroviruses replicated?
reverse transcriptase creates a DNA strand using RNA as template and tRNA(Lys) as a primer.
Second DNA strand synthesized using first as template → incorporated into host genome.