ALL Statistical Tests
1/40
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
41 Terms
ONE CATEGORICAL VARIABLE
What does the Pearson chi-square goodness-of-fit test evaluate? (chisq.test() or goodnessOfFitTest())
It tests whether observed frequencies match a specified probability distribution.
H₀: observed data are consistent with the specified distribution
H₁: observed data are not consistent with the distribution
Typical use-case: are all levels of a categorical variable equally likely?
What are the null and alternative hypotheses?
H₀: all class probabilities are equal
H₁: not all class probabilities are equal
What is the chi-square test statistic, and how are degrees of freedom calculated?
O = observed frequency
E = expected frequency
Larger X² values correspond to a lower probability of H₀ being true
Degrees of freedom: k − 1
where k is the number of categories (levels of the categorical variable)
*exact rejection region depends on degrees of freedom
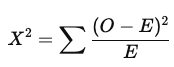
What is Cohen’s W, and how is it interpreted (3)? - Effect size
Cohen’s W measures the size of the deviation from the expected distribution:
0.10 = small effect
0.30 = medium effect
0.50 = large effect
*Larger values correspond to a larger deviation from the specified probability distribution under H0
What are the 2 assumptions of the chi-square goodness-of-fit test?
Expected frequencies are all at least 5 in each category
in case of violation: use the exact goodness of fit test
Observations are independent
TWO CATEGORICAL VARIABLES
What does the chi-square test of independence/association evaluate? (chisq.test() or associationTest())
It tests whether there is a relationship (association) between two categorical variables.
H₀: the variables are independent (no association)
there is no relationship between the categorical variables
H₁: the variables are not independent (association exists)
there is a relationship between the categorical variables
What is the test statistic for a chi-square test of independence and how are degrees of freedom calculated?
O = observed frequency
E = expected frequency
Larger X² values correspond to lower probability of H₀ being true
Degrees of freedom: df = (r−1)(c−1)
r = number of rows, c = number of columns in contingency table
the number of levels in both categorical variables
*exact rejection region depends on degrees of freedom
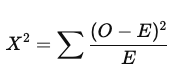
What is Cramer’s V and how is it interpreted? (cramersV())
Cramer’s V measures the strength of association between two categorical variables
Values range from 0 (no association) to 1 (perfect association).
Interpretation scale:
0–0.15: very weak
0.15–0.20: weak
0.20–0.25: moderate
0.25–0.30: moderately strong
0.30–0.35: strong
0.35–0.40: very strong
*larger values correspond to a larger deviation from the specified probability distribution under H0
What are the assumptions of the chi-square test of independence?
Expected frequencies are all at least 5
if violated: use Fisher’s exact test (fisher.test())
Observations are independent
if violated: use McNemar’s test (mcnemar.test())
ONE CONTINUOUS VARIABLE
What does a one-sample t-test evaluate? (t.test() or oneSampleTTest())
Formal name: Student’s t-test
It tests whether the mean of a sample differs significantly from a known or hypothesized population mean.
H₀: population mean equals a specific value
H₁: population mean does not equal a specific value
*If the population standard deviation is known: t-test becomes z-test
What is the t-test statistic, and how are degrees of freedom calculated?
xˉ = sample mean
μ0 = hypothesized population mean
s = sample standard deviation
n = sample size
Degrees of freedom: df = N − 1
N is the number of observations in the dataset
*exact rejection regions depend on degrees of freedom
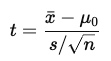
What is Cohen’s d and how is it interpreted? (cohensD()) - Effect size
Cohen’s d measures the magnitude of difference between the sample mean and the population mean:
0.20 = small effect
0.50 = medium effect
0.80 = large effect
*larger values correspond to a greater difference from the value under H0
What are the 2 assumptions of a one-sample t-test?
The continuous variable is normally distributed (check with Shapiro-Wilk test, histogram, Q-Q plot)
if normality is violated: use the Wilcoxon signed-rank test (wilcox.test())
Observations are independent
What are key properties of the t-distribution used in t-tests?
The t-distribution has thicker tails than the normal distribution (to account for extra uncertainty in small samples)
As sample size increases, the t-distribution approaches the normal distribution
Larger absolute t-values indicate more extreme results, corresponding to a lower probability of H₀ being true
TWO NON-PAIRED CONTINUOUS VARIABLES
What does an independent samples t-test evaluate? (t.test() or independentSamplesTTest())
Formal name: Student’s independent samples t-test
It compares the means of two independent groups to see if they are significantly different
H₀: population means of both groups (samples) are equal
H₁: population means of both groups (samples) are not equal
What is the test statistic for an independent samples t-test and how are degrees of freedom calculated?
Values further away from zero (i.e., higher absolute values) correspond to a lower probability of H0 being true
Degrees of freedom: df = N−2
N is the total number of observations across both groups (in the data set)
*exact rejection regions depend on degrees of freedom
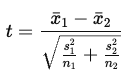
What is Cohen’s d and how is it interpreted for independent samples t-tests? (cohensD()) - Effect size
Cohen’s d measures the magnitude of difference between group means:
0.20 = small effect
0.50 = medium effect
0.80 = large effect
What are the 3 assumptions of the independent samples t-test?
The continuous variable is normally distributed in both groups
Check with Shapiro-Wilk test, histogram, Q-Q plot
If violated: use Wilcoxon rank sum test (wilcox.test())
Homoskedasticity: variances are equal between groups (the variance is the same in both groups)
Check with Levene’s test (leveneTest())
If violated: use Welch’s t-test
Observations are independent
TWO PAIRED CONTINUOUS VARIABLES
What does a paired samples t-test evaluate? (t.test() or pairedSamplesTTest())
Formal name: Student’s paired samples t-test
It compares the means of two related groups (e.g., before vs. after measurements on the same subjects).
H₀: the mean difference between the paired groups is zero
the difference between the population means for both samples is zero
H₁: the mean difference is not zero
the difference between the population means for both samples is not zero
What is the test statistic for a paired samples t-test and how are degrees of freedom calculated?
dˉ = mean of the differences
sd = standard deviation of the differences
n = number of paired observations
*Values further away from zero (i.e., higher absolute values) correspond to a lower probability of H0 being true
Degrees of freedom: df = N−1
N is the number of observations in the data set
*exact rejection regions depend on degrees of freedom
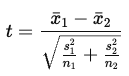
What is Cohen’s d and how is it interpreted for paired samples t-tests? - Effect size
Cohen’s d measures the size of the mean difference:
0.20 = small effect
0.50 = medium effect
0.80 = large effect
*larger values correspond to a greater difference difference in means
What assumptions does the paired samples t-test have?
The differences between paired observations are normally distributed
Check with: Shapiro-Wilk test, histogram, Q-Q plot
If normality is violated: use the Wilcoxon signed-rank test (wilcox.test())
(???? chat said:)
Observations within pairs are related; observations across pairs are independent
CONTINUOUS VARIABLES FOR TWO OR MORE GROUPS
What does a one-way ANOVA test evaluate? (aov(), then summary())
Full name: analysis of variance
ANOVA tests whether three or more group means are significantly different.
H₀: all population means are equal
the population means are the same for all groups
H₁: at least one population mean is different
the population means are not the same for all groups
What is the ANOVA test statistic and how are degrees of freedom calculated?
Test statistic: F
Higher F values = lower probability of H₀ being true
2 degrees of freedom:
Between groups: G − 1
G = number of groups
Within groups: N − G
N = total number of observations
*exact rejection regions depend on degrees of freedom
What do you do after a significant ANOVA?
Use post-hoc tests to determine which groups are significantly different from each other:
Pairwise comparisons: TurkeyHSD() or posthocPairwiseT()
Planned comparisons: for contrasts of a prior interest
Specify comparisons of interest
*Adjust p-values for multiple comparisons using Bonferroni correction: p′ = p*m
m = total number of comparisons
What is eta squared (η²) and how is it interpreted? - Effect size
Eta squared measures the proportion of variance explained by the group differences:
0.01: small
0.06: medium
0.14: large
*larger values correspond to more unequal means
(Alternate scale: 0.02 / 0.13 / 0.26 from lecture slides) (?? sooo which scale)
What assumptions does ANOVA make?
Normality of residuals: the residuals are normally distributed
Check with: Shapiro-Wilk test (shapiro.test()), histogram, QQ plot
If violated: use Kruskal-Wallis sum test (kruskal.test())
Homogeneity of variance: the variance is the same in both groups
Check with: leveneTest())
If violated: use Welch’s one-way test (oneway.test())
Independence of residuals: The residuals are independent
CONTINUOUS VARIABLES FOR MULTIPLE CATEGORICAL VARIABLES WITH TWO OR MORE GROUPS
What does a factorial ANOVA test? (aov(), then summary())
Full name: analysis of variance
A factorial ANOVA tests the effects of two or more categorical independent variables on a continuous dependent variable. It evaluates:
Main effects of each factor
Interaction effects between factors
What are the null and alternative hypotheses for a factorial ANOVA?
Multiple sets of null and alternative hypotheses:
Main effect of predictor A:
H₀: all group means of A are equal
the population means are the same for all groups of predictor A
H₁: at least one group mean differs
the population means are not the same for all groups predictor A
Main effect of predictor B:
H₀: all group means of B are equal
the population means are the same for all groups of predictor B
H₁: at least one group mean differs
the population means are not the same for all groups predictor B
Interaction between predictor A and predictor B: (A*B????)
H₀: the effect of A is the same at all levels of B
the population means for predictor A are the same for all groups of predictor B
H₁: the effect of A differs depending on the level of B
the population means for predictor A are not the same for all groups predictor B
**Make sure you know what main effects and interaction effects look like in a graph (see Section 16.2 of the book)
What do you do after a significant main or interaction effect in ANOVA?
Use post-hoc tests to determine which groups are significantly different from each other:
Pairwise comparisons (TukeyHSD() or posthocPairwiseT())
Planned comparisons for contrasts of a prior interest:
*Adjust p-values for multiple comparisons using a Bonferroni correction: 𝑝′ = 𝑝*𝑚
𝑚 = the total number of comparisons
What is the test statistic and how are degrees of freedom calculated in factorial ANOVA?
Test statistic: F
higher values correspond to a lower probability of H0 being true for a model term
Degrees of freedom:
Factor A: R−1
R is the number of groups for predictor A
Factor B: C−1
where C is the number of groups for predictor B
Interaction A and B: (R−1)(C−1)
Residuals: N − (R*C)
N is the total number of observations
*exact rejection regions depend on degrees of freedom
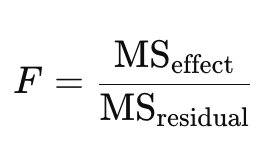
What is the relationship between sums of squares (SS), means of squares (MS), and the F-statistic in ANOVA?
Sums of Squares (SS) measure the total variability:
SSbetween: variability between group means
SSwithin (residual): variability within groups
Mean Squares (MS) are averages of sums of squares: MS = SS / df
F-statistic is the ratio of these mean squares: F = MSbetween / MSwithin
A higher F-value suggests that between-group differences are large relative to within-group variation, which may indicate a significant effect.
What is partial eta squared, and how is it interpreted? (etaSquared()) - Effect size
Partial η² measures the proportion of variance explained by one factor or interaction while controlling for others (0-1): Apply to both main effects and interaction effects.
0.01 = small
0.06 = medium
0.14 = large
*larger values correspond to more unequal means
Partial η² measures the effect size of individual model terms (main effects or interactions), controlling for other terms in the model
What are the assumptions of factorial ANOVA?
Residuals are normally distributed
Check with: Shapiro-Wilk test (shapiro.test()), histogram, QQ plot
Homogeneity of variance: The variance is the same in both groups
Check with: leveneTest()
Residuals are independent