DAT Biology
0.0(0)
Card Sorting
1/162
There's no tags or description
Looks like no tags are added yet.
Last updated 4:59 PM on 8/2/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
163 Terms
1
New cards
Ionic bonds
Transfer of e- from one atom to another where both atoms have different electronegativities
2
New cards
Electronegativity
Attraction an atom has for elections
3
New cards
Covalent bonds
* e- shared b/w atoms of similar electronegativities
\
* Can have single, double or triple bonds
\
* Can have single, double or triple bonds
4
New cards
Non-polar covalent bonds
Equal sharing of e- b/w two atoms of similar electronegativity
5
New cards
Polar covalent bonds
Unequal sharing of e- b/w two atoms of different electronegativities → forms a dipole
6
New cards
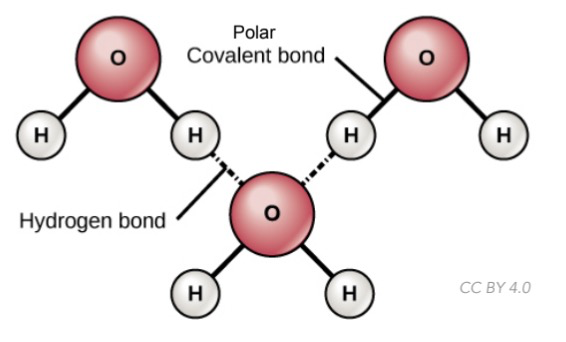
H-bonding
Weak bond b/w hydrogen attached to a highly electronegative atom and a negatively charged atom on another molecule (F, O or N)
7
New cards
5 Properties of H2O
* Excellent solvent
* High heat capacity
* Ice floats due to low density (water expands due to H-bonding that forms a lattice)
* Cohesion / surface tension: attracted to like substances
* Adhesion: attracted to unlike substances
* High heat capacity
* Ice floats due to low density (water expands due to H-bonding that forms a lattice)
* Cohesion / surface tension: attracted to like substances
* Adhesion: attracted to unlike substances
8
New cards
Macromolecules
Polymers formed from monomers (1 unit) e.g., polysaccharide
9
New cards
How are macromolecules formed?
==**Dehydration rxn:**== forms bonds; H2O by-product (monomers → polymers)
10
New cards
How are macromolecules broken down?
==**Hydrolysis:**== breaks bonds using H2O (polymers → monomers)
11
New cards
Examples of monosaccharides
* Glucose and fructose
* OH down = alpha
* OH up = beta
* OH down = alpha
* OH up = beta
12
New cards
Examples of covalent bonds
* Peptide bonds
* Glycosidic linkage
* Glycosidic linkage
13
New cards
How is the glycosidic linkage formed?
dehydration / condensation rxn
14
New cards
How many H2O molecules are lost for every glycosidic linkage formed?
1 H2O
15
New cards
Disaccharide
2 sugars joined by **glycosidic linkage**
16
New cards
What is **sucrose** made of?
glucose + fructose
17
New cards
What is **lactose** made of?
glucose + galactose
18
New cards
What is **maltose** made of?
glucose + glucose
19
New cards
Examples of 𝜶-glucose polysaccharides
* **Starch:** stores energy in plants
* **Glycogen:** stores energy in animals
* **Glycogen:** stores energy in animals
20
New cards
Examples of β-glucose polysaccharides
* **Cellulose:** walls of plant cells
* **Chitin:** β-glucose w/ nitrogen groups
* **Chitin:** β-glucose w/ nitrogen groups
21
New cards
Lipids
* ==**Functions:**== insulation, energy storage, form cholesterol and phospholipids in membranes, participate in endocrine signalling
\
* Covalent C-C bonds
\
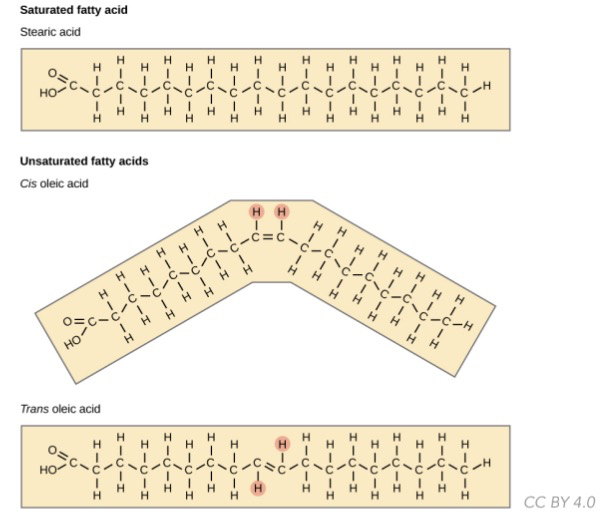
* ==**Triglycerides / triacylglycerols:**== 3 FA + glycerol backbone
* __Saturated:__ no double bonds; straight chains; unhealthy cuz chains stack densely(can form fat plaques)
* __Unsaturated:__ contains double bonds w/ kinks in chains; healthier cuz chains stack less densely; cis or trans
* ==**Phospholipids / diacylglycerols:**== 2 FA + phosphate group + glycerol backbone
* Amphipathic: both hydrophobic and hydrophilic
* ==**Steroids:**== three 6-membered rings + one 5-membered ring
* Hormones and cholesterol
\
* ==**Cell membrane fluidity:**== cell membranes change membrane FA composition to maintain steady degree of fluidity
* **Cold weather (rigid membranes):** __cholesterol & mono + polyunsaturated FA__ added into membrane to avoid cell membrane rigidity → increased fluidity
* **Warm weather (fluid & flexible membranes):** __cholesterol__ added into membrane to restrict movement/flexibility; __saturated FA__ tails become straight and pack tightly → decreased fluidity
\
* Covalent C-C bonds
\
* ==**Triglycerides / triacylglycerols:**== 3 FA + glycerol backbone
* __Saturated:__ no double bonds; straight chains; unhealthy cuz chains stack densely(can form fat plaques)
* __Unsaturated:__ contains double bonds w/ kinks in chains; healthier cuz chains stack less densely; cis or trans
* ==**Phospholipids / diacylglycerols:**== 2 FA + phosphate group + glycerol backbone
* Amphipathic: both hydrophobic and hydrophilic
* ==**Steroids:**== three 6-membered rings + one 5-membered ring
* Hormones and cholesterol
\
* ==**Cell membrane fluidity:**== cell membranes change membrane FA composition to maintain steady degree of fluidity
* **Cold weather (rigid membranes):** __cholesterol & mono + polyunsaturated FA__ added into membrane to avoid cell membrane rigidity → increased fluidity
* **Warm weather (fluid & flexible membranes):** __cholesterol__ added into membrane to restrict movement/flexibility; __saturated FA__ tails become straight and pack tightly → decreased fluidity
22
New cards
Lipid derivatives
* **Waxes:** esters of FA + alcohol for protective coating on skin (lanolin)
\
* **Carotenoids:** FA chains w/ conjugated double bonds; pigment producing colours in plants & animals
\
* **Adipocytes:** specialized fat cells
* __White fat cells:__ primarily contain triglycerides w/ thin layer of cytoplasm around it
* __Brown fat cells:__ have lots of cytoplasm, lipid droplets scattered throughout and lots of mitochondria
\
* **Glycolipids**: 2 FA + carbohydrate group + glycerol backbone
\
* **Lipoproteins:** lipid cores surrounded by phospholipids and apolipoproteins
\
* **Porphyrins / tetrapyrroles:** 4 joined pyrrole rings w/ a metal center atom
* Chlorophyll and hemoglobin
\
* **Carotenoids:** FA chains w/ conjugated double bonds; pigment producing colours in plants & animals
\
* **Adipocytes:** specialized fat cells
* __White fat cells:__ primarily contain triglycerides w/ thin layer of cytoplasm around it
* __Brown fat cells:__ have lots of cytoplasm, lipid droplets scattered throughout and lots of mitochondria
\
* **Glycolipids**: 2 FA + carbohydrate group + glycerol backbone
\
* **Lipoproteins:** lipid cores surrounded by phospholipids and apolipoproteins
\
* **Porphyrins / tetrapyrroles:** 4 joined pyrrole rings w/ a metal center atom
* Chlorophyll and hemoglobin
23
New cards
Proteins
* Monomer: amino acid
\
* Polymer: peptide
\
* Linkage: ==**peptide bonds (covalent bonds)**==
\
* Amino group + carboxyl groups + 𝜶 - Carbon bound to R chain (AA)
* Functions: storage, transport, defense (antibodies), enzymes **(exception: RNA e.g., ribosomes can act as an enzyme)**
\
* Polymer: peptide
\
* Linkage: ==**peptide bonds (covalent bonds)**==
\
* Amino group + carboxyl groups + 𝜶 - Carbon bound to R chain (AA)
* Functions: storage, transport, defense (antibodies), enzymes **(exception: RNA e.g., ribosomes can act as an enzyme)**
24
New cards
Factors affecting enzymatic activity
* Substrate and enzyme concentration
* Temperature
* pH
* Presence/absence of inhibitors
* Temperature
* pH
* Presence/absence of inhibitors
25
New cards
Cofactor
Non-protein molecule assisting enzymes → donate or accept electrons or fxnl groups
26
New cards
Holoenzyme
Union of cofactor + enzyme
27
New cards
Apoenzyme / apoprotein
When an enzyme is **not** combined w/ a cofactor
28
New cards
Inorganic cofactor
Metal ions (Fe2+ or Mg2+)
29
New cards
Coenzyme
Organic cofactor e.g., vitamins
30
New cards
Protein classification
* **Simple:** formed only of AA
* Albumins and Globulins: fxnl proteins
* Scleroprotein: structural protein
* **Conjugated:** simple protein + non-protein
* Lipoprotein: protein bound to lipid
* Albumins and Globulins: fxnl proteins
* Scleroprotein: structural protein
* **Conjugated:** simple protein + non-protein
* Lipoprotein: protein bound to lipid
31
New cards
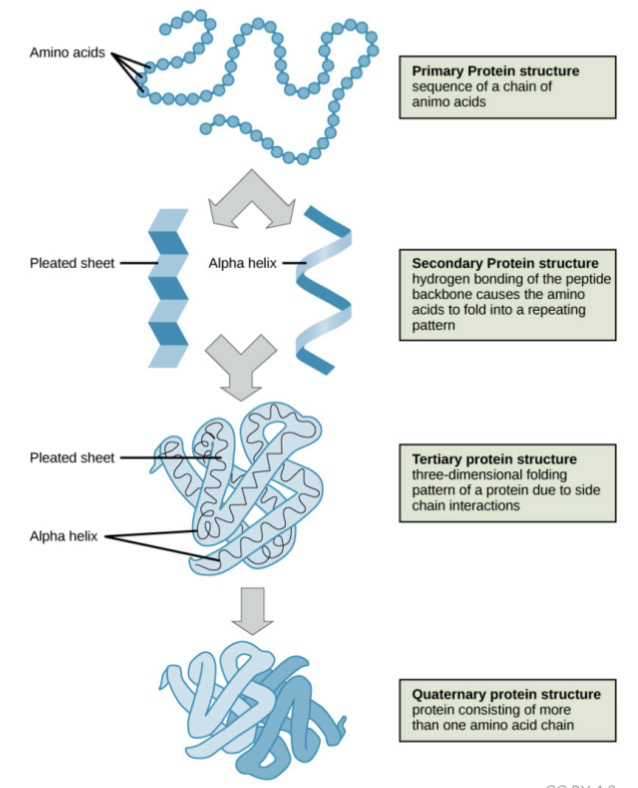
Protein Structure
* **Primary:** linear chain of AA connected by ==**peptide bonds**==
\
* **Secondary:** 3D shape due to **H-bonding** b/w amino and carboxyl groups of adjacent AA into ==𝜶-helices or β-sheets== / H-bonding of peptide backbone
\
* **Tertiary:** 3D structure due to ==**non-covalent interactions**== b/w AA side chains
* Also has **disulfide bonds:** covalent bond b/w cysteines
\
* **Quaternary:** 3D shape due to grouping of 2 or more separate peptide chains (e.g., 𝜶-helix + β-sheets)
\
* All proteins have a 1º structure, most have 2º & large proteins have 3º or 4º structures
\
* **Secondary:** 3D shape due to **H-bonding** b/w amino and carboxyl groups of adjacent AA into ==𝜶-helices or β-sheets== / H-bonding of peptide backbone
\
* **Tertiary:** 3D structure due to ==**non-covalent interactions**== b/w AA side chains
* Also has **disulfide bonds:** covalent bond b/w cysteines
\
* **Quaternary:** 3D shape due to grouping of 2 or more separate peptide chains (e.g., 𝜶-helix + β-sheets)
\
* All proteins have a 1º structure, most have 2º & large proteins have 3º or 4º structures
32
New cards
Non-covalent bonds
H-bonds, ionic bonds, hydrophobic interactions, Van Der Waals forces
33
New cards
Protein types
* **Globular**
\
* **Fibrous/structural proteins**
\
* **Membrane proteins:** membrane pumps, channels or receptors
\
* **Fibrous/structural proteins**
\
* **Membrane proteins:** membrane pumps, channels or receptors
34
New cards
Fibrous / Structural proteins
* Water insoluble
* Dominantly 2º structure
* Long polymers
* Maintain and add strength to cellular and matrix structure (collagen or keratin)
* Dominantly 2º structure
* Long polymers
* Maintain and add strength to cellular and matrix structure (collagen or keratin)
35
New cards
Globular proteins
* Somewhat water soluble
* Dominantly 3º structure
* Functions: enzymes, hormones, storage, antibodies, osmotic regulation
* Dominantly 3º structure
* Functions: enzymes, hormones, storage, antibodies, osmotic regulation
36
New cards
Protein denaturation
* Protein reversed back to its 1º structure
* Usually irreversible but can be reversed w/ removal of denaturing agent
* All folding information is encoded in primary structure
* Usually irreversible but can be reversed w/ removal of denaturing agent
* All folding information is encoded in primary structure
37
New cards
Protein digestion
Eliminates **all** protein structure, including 1º structure
38
New cards
Nucleic acids
* ==**Functions:**== encode, express and store genetic information
\
* **Monomer:** ==nucleotides== (nitrogen base + 5-carbon sugar + phosphate group)
* ==Nucleosides:== sugar + nitrogen base
\
* **Polymers:** ==nucleic acid== (DNA and RNA)
\
* ==**Linkage:**== phosphodiester bonds
\
### Nitrogenous bases
* DNA: adenine, thymine, guanine, cytosine
* A - T: 2 H-bonds
* G - C: 3 H-bonds
* RNA: adenine, uracil, guanine, cytosine
* A - U: 2 H-bonds
* G - C: 3 H-bonds
* ==**Chargaff’s Rule:**== A & T and G & C are always present in equal amounts
\
* **Monomer:** ==nucleotides== (nitrogen base + 5-carbon sugar + phosphate group)
* ==Nucleosides:== sugar + nitrogen base
\
* **Polymers:** ==nucleic acid== (DNA and RNA)
\
* ==**Linkage:**== phosphodiester bonds
\
### Nitrogenous bases
* DNA: adenine, thymine, guanine, cytosine
* A - T: 2 H-bonds
* G - C: 3 H-bonds
* RNA: adenine, uracil, guanine, cytosine
* A - U: 2 H-bonds
* G - C: 3 H-bonds
* ==**Chargaff’s Rule:**== A & T and G & C are always present in equal amounts
39
New cards
Purines
* 2 rings
* Adenine and Guanine (**Pur**e **A**s **G**old)\*\*
* Adenine and Guanine (**Pur**e **A**s **G**old)\*\*
40
New cards
Pyrimidines
* 1 ring
* Cytosine, Uracil, Thymine **(CUT**)**
* Cytosine, Uracil, Thymine **(CUT**)**
41
New cards
DNA
Deoxyribose sugar + 2 antiparallel strands of a double helix
42
New cards
RNA
* Ribose sugar
* Single stranded
* Single stranded
43
New cards
Cell Theory
* All living organisms are composed of one or more cells
* Cell is the basic unit of structure, fxn, and organization in all organisms
* All cells come from pre-existing, living cells
* Cells carry hereditary info
* Cell is the basic unit of structure, fxn, and organization in all organisms
* All cells come from pre-existing, living cells
* Cells carry hereditary info
44
New cards
RNA World Hypothesis
* Self-replicating RNA molecules were precursors to current life
* RNA stores genetic info like DNA and catalyzes rxns like an enzyme
* RNA played a major role in evol. of cellular life
* Evidence: RNA is unstable relative to DNA due to its extra OH group → more likely to participate in chemical rxns
* RNA stores genetic info like DNA and catalyzes rxns like an enzyme
* RNA played a major role in evol. of cellular life
* Evidence: RNA is unstable relative to DNA due to its extra OH group → more likely to participate in chemical rxns
45
New cards
Central Dogma of Genetics
* Biological info cannot be transferred backwards from protein to either protein or nucleic acid
* Info must travel from DNA → RNA → proteins
* Info must travel from DNA → RNA → proteins
46
New cards
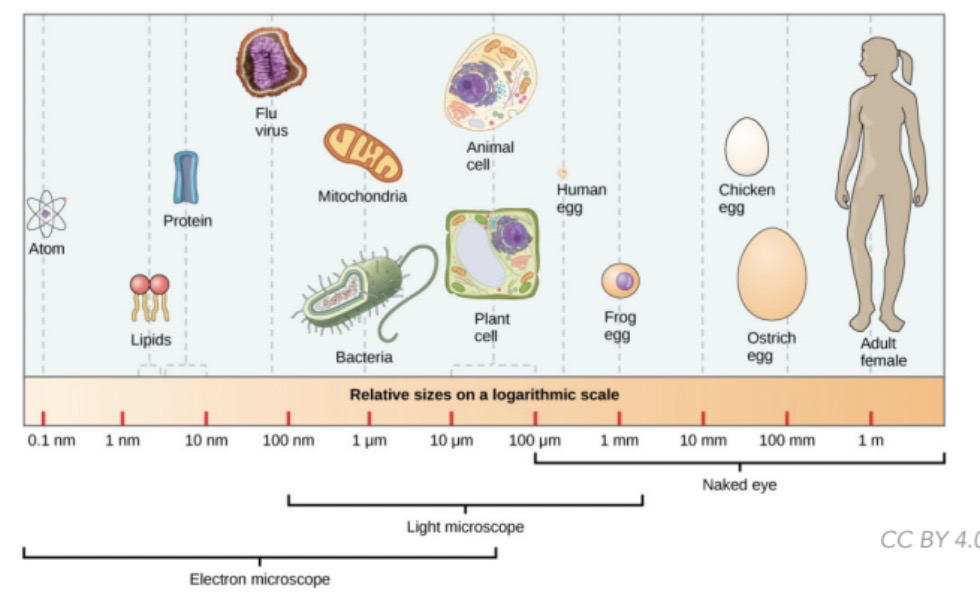
Elements that can be visualized w/ naked eye
==**100 µm - 1m**==
* Adult female
* Ostrich egg
* Chicken egg
* Frog egg
* Human egg
* Adult female
* Ostrich egg
* Chicken egg
* Frog egg
* Human egg
47
New cards
Elements that can be visualized w/ light microscope
==**100 nm - 1mm**==
* Mitochondria
* Bacteria
* Plant and animal cell
* Human egg
* Frog egg
* Mitochondria
* Bacteria
* Plant and animal cell
* Human egg
* Frog egg
48
New cards
Elements that can be visualized w/ electron microscope
==**0.1 nm - 10 µm**==
* Atom
* Lipids
* Protein
* Virus
* Bacteria
* Mitochondria
* Plant and animal cell
* Atom
* Lipids
* Protein
* Virus
* Bacteria
* Mitochondria
* Plant and animal cell
49
New cards
Microscopy techniques
* Stereomicroscope (light)
* Compound Microscope (light)
* Phase Contrast Microscope
* Confocal Laser Scanning Microscope and Fluorescence
* Scanning Electron Microscope (SEM)
* Cryo SEM
* Transmission Electron Microscope
* Electron Tomography
* Compound Microscope (light)
* Phase Contrast Microscope
* Confocal Laser Scanning Microscope and Fluorescence
* Scanning Electron Microscope (SEM)
* Cryo SEM
* Transmission Electron Microscope
* Electron Tomography
50
New cards
Stereomicroscope (light)
* Uses visible light to view surface of sample
\
* **Pro:** can view living samples
* **Con: l**ow resolution relative to a compound microscope
\
* **Pro:** can view living samples
* **Con: l**ow resolution relative to a compound microscope
51
New cards
Compound Microscope (light)
* Uses visible light to view thin section of sample
\
* Uses multiple lenses
\
* **Pro:** can view **some** living samples (single cell layer)
* **Con:** requires staining for good visibility
\
* Uses multiple lenses
\
* **Pro:** can view **some** living samples (single cell layer)
* **Con:** requires staining for good visibility
52
New cards
Phase Contrast Microscope
* Uses light phases and contrast for detailed observation of living organisms, including internal structures if thin
\
* **Pro:** good resolution and contrast
* **Con:** not ideal for thick samples and produces a ‘halo effect’ around perimeter of samples
\
* **Pro:** good resolution and contrast
* **Con:** not ideal for thick samples and produces a ‘halo effect’ around perimeter of samples
53
New cards
Confocal Laser Scanning Microscope / Fluorescence
* Used to observe thin slices while keeping a sample intact
* Common method for viewing chromosomes during mitosis
* Can be used w/o fluorescence → laser light is used to scan dyed specimen
\
* **Pro:** can observe specific parts of a cell using fluorescent tagged antibodies
* **Con:** can cause artifacts
* Common method for viewing chromosomes during mitosis
* Can be used w/o fluorescence → laser light is used to scan dyed specimen
\
* **Pro:** can observe specific parts of a cell using fluorescent tagged antibodies
* **Con:** can cause artifacts
54
New cards
SEM
* **Pro:** view surface of 3D objects w/ high resolution
* **Cons:** cannot use on living samples; preparation is extensive as sample needs to be dried and coated; costly
* **Cons:** cannot use on living samples; preparation is extensive as sample needs to be dried and coated; costly
55
New cards
Cryo SEM
* **Pro:** sample is not dehydrated co can observe in their ‘natural form’
* **Cons:** cannot use on living samples; samples must be frozen which can cause artifacts
* **Cons:** cannot use on living samples; samples must be frozen which can cause artifacts
56
New cards
TEM
* Used to view thin x-sections and internal structures within samples at very high magnification
\
* **Pro:** high resolution
* **Cons:** cannot use on living samples; preparation of sample is extensive; costly
\
* **Pro:** high resolution
* **Cons:** cannot use on living samples; preparation of sample is extensive; costly
57
New cards
Electron Tomography
* Not a microscope but a technique used to build a 3D model of sample using TEM data
\
* **Pro:** can view objects in 3D and see objects relative to one another
* **Cons:** cannot use on living samples; preparation of sample is extensive; costly
\
* **Pro:** can view objects in 3D and see objects relative to one another
* **Cons:** cannot use on living samples; preparation of sample is extensive; costly
58
New cards
Centrifugation
* Used to separate a liquified sample into its different components by spinning it rapidly
\
* Spins and separates liquified cell homogenates into layers based on density
\
* Cells separate from most dense to lease dense
\
* **Cell fractionation:** largest component of cells pellet first at the bottom and progressively spin faster: nuclei layer → mitochondria → larger macromolecules/viruses/ribosomes
\
* Spins and separates liquified cell homogenates into layers based on density
\
* Cells separate from most dense to lease dense
\
* **Cell fractionation:** largest component of cells pellet first at the bottom and progressively spin faster: nuclei layer → mitochondria → larger macromolecules/viruses/ribosomes
59
New cards
Differential centrifugation
* Relies on density, shape, speed at which macromolecule travels
\
* Forms continuous layers of sediments where insoluble proteins are found in the pellet while soluble proteins remain in the supernatant liquid above the pellet
\
* Forms continuous layers of sediments where insoluble proteins are found in the pellet while soluble proteins remain in the supernatant liquid above the pellet
60
New cards
Density centrifugation
Only relies on density
61
New cards
Rate of reaction in equilibrium
Rate of formation of reactants and products is equal
62
New cards
Enzymes
* Catalysts lowering the activation energy of a rxn → ↑ rate of rxn
* Substrate specific
* Remain unchanged during rxn
* Have an active site that binds substrates via induced fit
* Catalyze forward and reverse reactions
* Have varying fxn depending on pH and temperature
* Substrate specific
* Remain unchanged during rxn
* Have an active site that binds substrates via induced fit
* Catalyze forward and reverse reactions
* Have varying fxn depending on pH and temperature
63
New cards
Prosthetic group
Cofactors that bind tightly or covalent to an enzyme
64
New cards
ATP
* Potential energy stored as chemical energy
\
* Source of activation energy
\
* Formed by phosphorylation of ADP using energy from glucose → ==**endergonic**==
\
* Broken apart via hydrolysis → ==**exergonic**==
\
* ATP stores energy generated from the ==exergonic reactions== in the electron transport chain, and can then be used to fuel ==endergonic reactions==
\
* Source of activation energy
\
* Formed by phosphorylation of ADP using energy from glucose → ==**endergonic**==
\
* Broken apart via hydrolysis → ==**exergonic**==
\
* ATP stores energy generated from the ==exergonic reactions== in the electron transport chain, and can then be used to fuel ==endergonic reactions==
65
New cards
Enzyme regulation
* Km (Michaelis constant)
* Allosteric enzymes
* Competitive inhibition
* Noncompetitive inhibition
* Uncompetitive/anti-competitive inhibition
* Allosteric enzymes
* Competitive inhibition
* Noncompetitive inhibition
* Uncompetitive/anti-competitive inhibition
66
New cards
Km (Michaelis constant)
* Substrate \[ \] at which the rate of rxn = 1/2 max velocity of the enzyme (Vmax)
* Inversely represents binding affinity
* Small Km = less substrate needed to reach Vmax → higher binding affinity
* High Km = more substrate needed to reach Vmax → low binding affinity
* Inversely represents binding affinity
* Small Km = less substrate needed to reach Vmax → higher binding affinity
* High Km = more substrate needed to reach Vmax → low binding affinity
![* Substrate \[ \] at which the rate of rxn = 1/2 max velocity of the enzyme (Vmax)
* Inversely represents binding affinity
* $$Small Km = less substrate needed to reach Vmax$$ → higher binding affinity
* $$High Km = more substrate needed to reach Vmax$$ → low binding affinity](https://knowt-user-attachments.s3.amazonaws.com/8ee2b4871f4441cb9d8ee6c04ea9fad8.jpeg)
67
New cards
Allosteric enzymes
* Have both an ==active site== for ==substrate binding== and an ==allosteric site== for an ==allosteric effector== (can be an activator or inhibitor
\
* ==**Allosteric site:**== secondary location where an effector binds
\
* ==**Active site:**== area of an enzyme where the substrate binds
\
* ==**Allosteric site:**== secondary location where an effector binds
\
* ==**Active site:**== area of an enzyme where the substrate binds
68
New cards
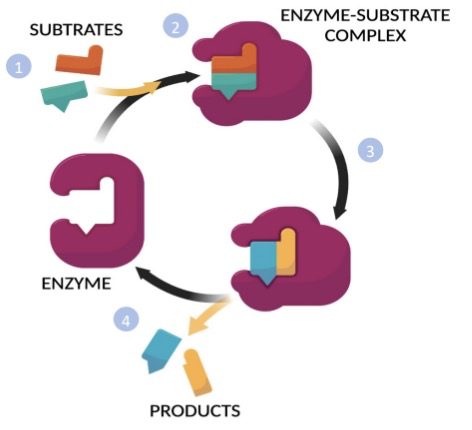
Mechanism of enzyme rxn
1. Substrates (aka reactants) enter the active site of the enzyme
\
2. Enzyme and substrate change shape slightly to better catalyze the reaction ==**(induced fit model)**==
1. When the substrate binds the enzyme → ==**Enzyme-Substrate Complex**==
\
3. The enzyme facilitates the rxn by lowering the activation energy
\
4. Products are released and the cycle repeats
69
New cards
Exergonic rxn
* Free energy is released
* Spontaneous with -𝛥G
\
* Can be used to drive otherwise nonspontaneous rxns (ATP hydrolysis - exergonic - to facilitate endergonic rxns)
* Spontaneous with -𝛥G
\
* Can be used to drive otherwise nonspontaneous rxns (ATP hydrolysis - exergonic - to facilitate endergonic rxns)
70
New cards
Endergonic rxn
* Free energy is absorbed
* Nonspontaneous with +𝛥G
* Nonspontaneous with +𝛥G
71
New cards
PCR
* Creates a large amount of DNA by amplifying a DNA sample:
1\. Denaturation: High heat separates ds DNA
2\. Annealing: Sample is cooled so primers attach to separated strands
3\. Elongation: Polymerase synthesize new strands
* Cycle repeats to increase amount of DNA exponentially
1\. Denaturation: High heat separates ds DNA
2\. Annealing: Sample is cooled so primers attach to separated strands
3\. Elongation: Polymerase synthesize new strands
* Cycle repeats to increase amount of DNA exponentially
72
New cards
Reverse Transcriptase
* Used to synthesize DNA from an RNA template
* Sometimes used to create complementary DNA (cDNA) off an mRNA template
* cDNA lacks introns
* Naturally used by viruses
* Sometimes used to create complementary DNA (cDNA) off an mRNA template
* cDNA lacks introns
* Naturally used by viruses
73
New cards
DNA sequencing
* Used to determine the sequence of base pairs in a DNA or RNA molecule
\
* Example: ==**Dideoxy Chain Termination**== is based on the principle that during DNA synthesis, addition of a nucleotide requires a free OH group on the 3ʹ carbon of the sugar of the last nucleotide of the growing DNA strand
\
* Example: ==**Dideoxy Chain Termination**== is based on the principle that during DNA synthesis, addition of a nucleotide requires a free OH group on the 3ʹ carbon of the sugar of the last nucleotide of the growing DNA strand
74
New cards
Blotting techniques
* Used for identifying specific fragments of DNA, RNA or protein
\
### Types:
* Southern: DNA
* Northern: RNA
* Western: Protein
\
### Method:
1\. Electrophoresis: separates sample
2\. Sample is transferred to nitrocellulose gel
3\. Probe is added to hybridize and mark target fragment
\
### Types:
* Southern: DNA
* Northern: RNA
* Western: Protein
\
### Method:
1\. Electrophoresis: separates sample
2\. Sample is transferred to nitrocellulose gel
3\. Probe is added to hybridize and mark target fragment
75
New cards
Hybridization
Nucleic acids form complementary base pairs with nucleic acids of different strand during blotting
76
New cards
Gel electrophoresis
* Used to separate DNA molecules by size and charge: the smaller the molecule, the farther it travels down the gel
* After separating the DNA sample, it can be sequenced or probed to find location of specific sequence
* After separating the DNA sample, it can be sequenced or probed to find location of specific sequence
77
New cards
Microarray Assays
Used to monitor the expression of large groups of genes across a genome
78
New cards
Recombinant DNA & Gene Libraries
* Recombinant DNA contains segments from multiple sources
\
* Recombinant DNA is vital for creating ==**gene libraries**== (collections of DNA pieces from a genome)
\
* **Process to make and use recombinant DNA:**
1. Using ==**restriction endonucleases**== (restriction enzymes) to cut specific segments of DNA called ==**restriction sites**==
* These enzymes create sticky ends, which allow new DNA pieces to bind
2. ==**DNA ligase**== connects the different fragments together
3. A vector can then be used to transfer foreign DNA into another cell
* ==**Vectors:**== plasmids and bacteriophages
\
* Recombinant DNA is vital for creating ==**gene libraries**== (collections of DNA pieces from a genome)
\
* **Process to make and use recombinant DNA:**
1. Using ==**restriction endonucleases**== (restriction enzymes) to cut specific segments of DNA called ==**restriction sites**==
* These enzymes create sticky ends, which allow new DNA pieces to bind
2. ==**DNA ligase**== connects the different fragments together
3. A vector can then be used to transfer foreign DNA into another cell
* ==**Vectors:**== plasmids and bacteriophages
79
New cards
Recombinant DNA in bacterial cloning
1. ==**Restriction enzyme**== is applied to __**both**__ the bacterial plasmid and the foreign DNA to create the same sticky ends
2. ==**DNA ligase**== attaches the fragments to create plasmid with new DNA
3. Plasmid is introduced into bacteria using ==**transformation**==
4. Bacteria can be grown to produce a product or form a colony
1. To ensure that the bacteria has included the plasmid, a gene for antibiotic resistance is added to the plasmid. Bacteria without the plasmid will perish under antibiotic conditions
80
New cards
Competitive inhibition
* Substance that mimics the substrate and inhibits the enzyme by binding at the active site
* Effect can be overcome by ↑ \[substrate\]
* Km ↑
* Vmax stays same
* Effect can be overcome by ↑ \[substrate\]
* Km ↑
* Vmax stays same
![* Substance that mimics the substrate and inhibits the enzyme by binding at the active site
* Effect can be overcome by ↑ \[substrate\]
* $$Km ↑$$
* $$Vmax stays same$$](https://knowt-user-attachments.s3.amazonaws.com/26b7bc1dcfd44854a8a3a652356c326c.jpeg)
81
New cards
Vmax
Max velocity of a rxn at peak substrate saturation
82
New cards
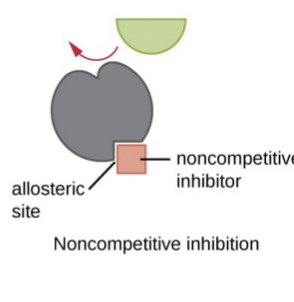
Noncompetitive inhibition
* Substance inhibits enzyme by binding elsewhere than the active site → substrate still binds but the rxn is prevented from completing
* Vmax ↓
* Km stays same
* Vmax ↓
* Km stays same
83
New cards
Uncompetitive / anti-competitive inhibition
Occurs when an enzyme inhibitor binds only to the formed enzyme-substrate (ES) complex → prevents formation of product
84
New cards
Cooperativity
When an enzyme become more receptive to additional substrate molecules after one substrate molecule binds to the active site
85
New cards
Peripheral membrane proteins
* Loosely attached to the surface of one side of the membrane
\
* Hydrophilic
\
* Held in place by H-bonds and electrostatic interactions
\
* Can disrupt/detach them by changing \[salt\] or pH
\
* Hydrophilic
\
* Held in place by H-bonds and electrostatic interactions
\
* Can disrupt/detach them by changing \[salt\] or pH
86
New cards
Integral membrane proteins
* Embedded in the cell membrane
* Hydrophobic
* Can be destroyed using detergent
* Hydrophobic
* Can be destroyed using detergent
87
New cards
Transmembrane proteins
Type of integral membrane that travels all the way through the cell membrane
88
New cards
Membrane proteins
* Channel proteins
* Ion channels
* Porins
* Recognition proteins
* Carrier proteins
* Transport proteins
* Adhesion proteins
* Receptor proteins
* Ion channels
* Porins
* Recognition proteins
* Carrier proteins
* Transport proteins
* Adhesion proteins
* Receptor proteins
89
New cards
Channel proteins
Provide a passageway through the membrane for hydrophilic, polar and charged substances
90
New cards
Recognition proteins
* Glycoprotein (have an attached oligosaccharide) used to distinguish b/w self and foreign
* Example: MHC on macrophages
* Example: MHC on macrophages
91
New cards
Ion channels
* Used to pass ions across the membrane
* Gated channels in nerve and muscle cells
* Voltage gated: respond to difference in membrane potential
* Ligand-gated: chemical binds to open channel
* Mechanically gated: respond to pressure or vibration
* Gated channels in nerve and muscle cells
* Voltage gated: respond to difference in membrane potential
* Ligand-gated: chemical binds to open channel
* Mechanically gated: respond to pressure or vibration
92
New cards
Porins
* Allow passage of certain ions and small polar molecules
* Increase rate of H2O passing in kidney and plant root cells
* Less specific
* Increase rate of H2O passing in kidney and plant root cells
* Less specific
93
New cards
Carrier proteins
* Specific to mvt across membrane via integral membrane protein
* Changes shape after binding to specific mol. (glucose into cell)
* Changes shape after binding to specific mol. (glucose into cell)
94
New cards
Transport proteins
* Use ATP to transport materials across the membrane
* ==**Active transport (Na+/K+ pump):**== reqiures ATP
* ==**Facilitated diffusion:**== doesn’t require ATP
* ==**Active transport (Na+/K+ pump):**== reqiures ATP
* ==**Facilitated diffusion:**== doesn’t require ATP
95
New cards
Adhesion proteins
Attach cells to neighbouring cells and provide anchors for stability via internal filaments and tubules
96
New cards
Receptor proteins
Serve as binding sites for hormones and other trigger molecules
97
New cards
Membrane properties
* ==**Phospholipid membrane permeability:**== allows small, uncharged, hydrophobic molecules to freely pass the membrane. Other molecules that are large, polar, or charged require a transporter
* Polar molecules can cross if they are small and uncharged
\
* ==**Cholesterol:**== regulates fluidity of cell membrane (↑ temp = ↓ fluidity)
* Prokaryotes don’t have cholesterol in their membranes
\
* ==**Glycocalyx**==: carbohydrate coat covering the outer face of the cell wall of bacteria and of the plasma membrane in animal cells
* **Fxns:** adhesive capabilities, barrier to infection, markers for cell-cell recognition
* Polar molecules can cross if they are small and uncharged
\
* ==**Cholesterol:**== regulates fluidity of cell membrane (↑ temp = ↓ fluidity)
* Prokaryotes don’t have cholesterol in their membranes
\
* ==**Glycocalyx**==: carbohydrate coat covering the outer face of the cell wall of bacteria and of the plasma membrane in animal cells
* **Fxns:** adhesive capabilities, barrier to infection, markers for cell-cell recognition
98
New cards
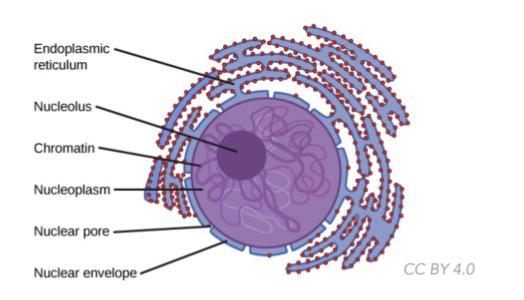
Nucleus
* Contains the cell’s DNA, and coordinates cell activities such as protein synthesis & reproduction
\
* ==**Chromatin**==: general packaging structure of DNA around proteins in eukaryotes
\
* ==**Chromosomes:**== tightly condensed chromatin when the cell is ready to divide
\
* ==**Histones:**== organize DNA which coil around it into bundles called nucleosomes
\
* ==**Nucleolus:**== inside the nucleus where ribosome is synthesized ==(using rRNA)==
\
* Surrounded by double layer nuclear envelope w/ nuclear pores for transport
\
* Contains nucleoplasm (no cytoplasm)
\
* ==**Chromatin**==: general packaging structure of DNA around proteins in eukaryotes
\
* ==**Chromosomes:**== tightly condensed chromatin when the cell is ready to divide
\
* ==**Histones:**== organize DNA which coil around it into bundles called nucleosomes
\
* ==**Nucleolus:**== inside the nucleus where ribosome is synthesized ==(using rRNA)==
\
* Surrounded by double layer nuclear envelope w/ nuclear pores for transport
\
* Contains nucleoplasm (no cytoplasm)
99
New cards
Nucleiod
Region w/n ==**prokaryotic cells**== containing genetic material
100
New cards
Nuclear lamina
* Dense fibrillar network inside of the nucleus of ==**eukaryotic cells**== that provides mechanical support
\
* Helps regulate DNA replication, cell division and chromatin organization
\
* Helps regulate DNA replication, cell division and chromatin organization