Protein Structure
1/14
Earn XP
Description and Tags
There are two common types of covalent bonds between amino acids in proteins: the peptide bonds that link amino acids together into polypeptide chains and disulfide bridges between cysteine R-groups
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
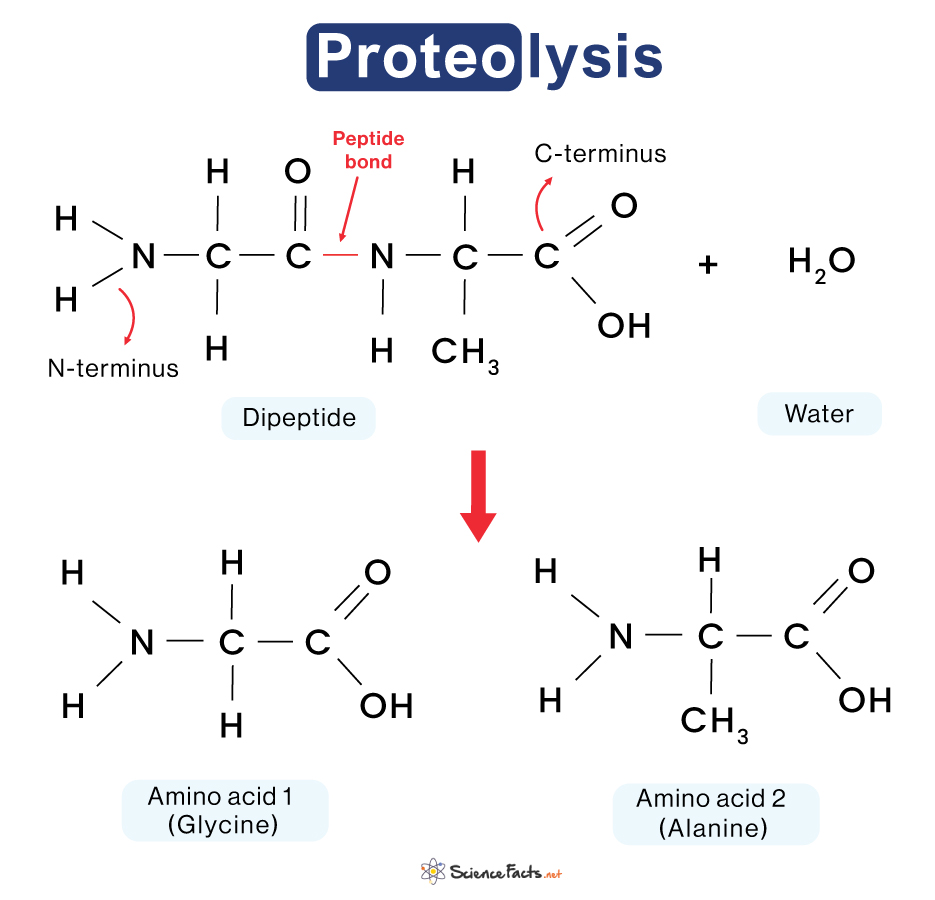
What are peptide bonds?
Covalent bonds that link amino acids together in polypeptide chains, formed between the carboxyl group of one amino acid and the α-amino group of another, with the loss of a water molecule.

What is the difference between a peptide bond vs a dipeptide bond?
Peptide: A peptide is a short chain of amino acids linked together by peptide bonds. It typically refers to a sequence of three or more amino acids.
Dipeptide: A dipeptide is the simplest form of peptide, consisting of just two amino acids linked together by a single peptide bond.
What is the structure formed by amino acids in a polypeptide chain?
The repeating N–C–C pattern along the polypeptide chain is known as the polypeptide backbone, with each amino acid residue contributing to its structure.

What are disulfide bridges in proteins?
Covalent bonds formed between the sulfur atoms of two cysteine residues within a protein.
They provide stability to protein structures, especially in extracellular environments.

What is a residue?
When two or more amino acids combine to form a peptide, the elements of water are removed, and what remains of each amino acid is called an amino-acid residue
What is proteolysis?
Proteolysis, or proteolytic cleavage, is the process of breaking peptide bonds within proteins. It is often catalyzed by proteolytic enzymes or proteases.
What are proteolytic enzymes?
Proteolytic enzymes, or proteases, are enzymes that catalyze the hydrolysis (water used to break down chemical bond) of peptide bonds in proteins. They have specificity for certain amino acids adjacent to the peptide bond they cleave.
What are the most common proteolytic enzymes?
The three main proteolytic enzymes produced naturally in your digestive system are pepsin, trypsin and chymotrypsin. Your body produces them to help break down dietary proteins like meat, eggs and fish into
What is denaturation in the context of proteins?
Denaturation refers to the disruption of a protein's shape without breaking peptide bonds, caused by factors like urea, extremes of pH, temperature, or salt concentration.
What is the primary structure of a protein?
Refers to the linear sequence of amino acids in a polypeptide chain, connected by peptide bonds.

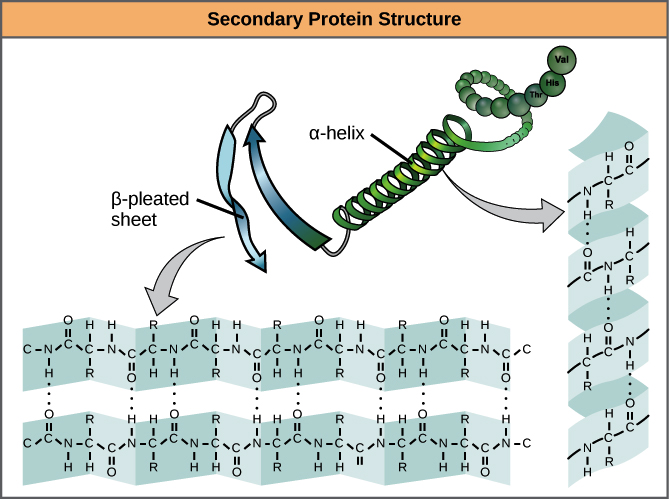
How are secondary structures of proteins stabilized?
Secondary structures, like α-helices and β-pleated sheets, are stabilized by hydrogen bonds between backbone NH and CO groups.
What is the tertiary structure of a protein?
Overall three-dimensional shape of a single polypeptide chain, stabilized by interactions between R-groups (e.g., hydrophobic interactions, hydrogen bonds).

Describe quaternary structure in proteins.
Arrangement and interactions between multiple polypeptide chains (subunits) in a multi-subunit protein complex, stabilized by non-covalent interactions and sometimes by covalent bonds.

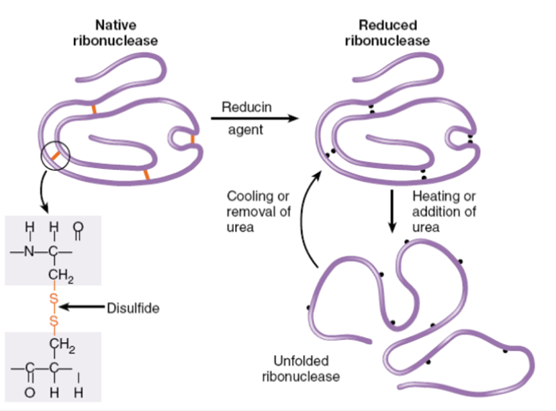
What did Anfinsen's experiment demonstrate about protein structure?
Anfinsen's experiment demonstrated that the specific shape (structure) of a protein, which determines its function, depends on its sequence of amino acids (primary structure).
Even after being unfolded (denatured), a protein like ribonuclease can refold into its original functional shape under the right conditions.
This shows that the sequence of amino acids directs how a protein folds and works, highlighting the importance of the primary structure in determining a protein's function.
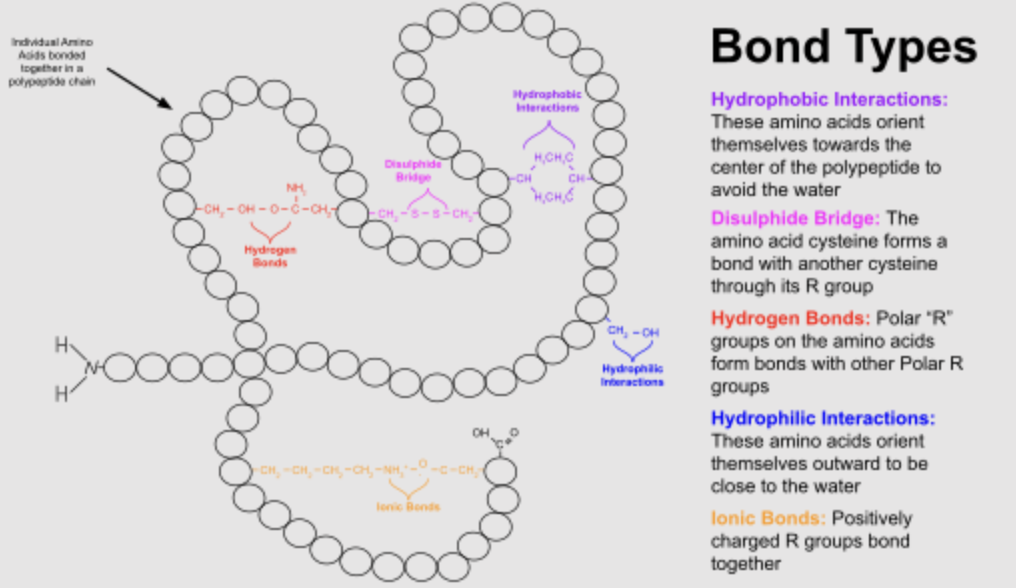
What 5 bonds are involved in protein folding?
Hydrophobic interactions: tend to cluster together in the interior of the protein structure away from H2O — helps minimize contact with water to help protein stability
Disulfide bridges: forms between cysteine residues where sulfur atoms in their R groups bond covalently — strong structural support for proteins that need to be in harsh environments
Hydrogen bonds: weak attraction between H atom and electronegative atom (O/N) in polar chains — stabilizes secondary structure like alpha helix and beta sheet
Hydrophilic interaction: oriented towards protein surface and interact with H2O — crucial for solubility and facilitate interactions with other molecules
Ionic Bonds: strong interaction between + and - amino side chains — stability of protein in aqueous environments where ions are prevalent